Abstract
The studies reported here identify propionyl coenzyme A (propionyl-CoA) as the common intermediate in the 1,2-propanediol and propionate catabolic pathways of Salmonella enterica serovar Typhimurium LT2. Growth on 1,2-propanediol as a carbon and energy source led to the formation and excretion of propionate, whose activation to propionyl-CoA relied on the activities of the propionate kinase (PduW)/phosphotransacetylase (Pta) enzyme system and the CobB sirtuin-controlled acetyl-CoA and propionyl-CoA (Acs, PrpE) synthetases. The different affinities of these systems for propionate ensure sufficient synthesis of propionyl-CoA to support wild-type growth of S. enterica under low or high concentrations of propionate in the environment. These redundant systems of propionyl-CoA synthesis are needed because the prpE gene encoding the propionyl-CoA synthetase enzyme is part of the prpBCDE operon under the control of the PrpR regulatory protein, which needs 2-methylcitrate as a coactivator. Because the synthesis of 2-methylcitrate by PrpC (i.e., the 2-methylcitrate synthase enzyme) requires propionyl-CoA as a substrate, the level of propionyl-CoA needs to be raised by the Acs or PduW-Pta system before 2-methylcitrate can be synthesized and prpBCDE transcription can be activated.
A question of great interest to cell physiologists in the postgenomic era is how the cell integrates the large number of metabolic pathways that make up its intermediary metabolism. The wealth of genomic information and new technologies developed in recent years has attracted attention to this area of metabolism research.
In the recent past, work in several laboratories has shown that the 2-methylcitric acid pathway for propionate catabolism is widely distributed among prokaryotes (5, 6, 11, 15, 22-25, 39). Studies of the role of sirtuin function (encoded by the cobB gene) in propionate catabolism in Salmonella enterica serovar Typhimurium uncovered an interaction between the pathways required for the degradation of propionate and 1,2-propanediol (1,2-PDL) (41). Briefly, growth conditions were reported that restored the growth of a cobB mutant strain of S. enterica serovar Typhimurium on propionate. Key to this phenotypic correction was the induction of the genes encoding functions required for the catabolism of 1,2-PDL, i.e., the 1,2-PDL utilization (pdu) operon, which contains 21 genes (pduABCDEGHJKLMNOPQSTUVWX) (3); catabolism of 1,2-PDL was not required to correct the inability of the cobB mutant strain to grow on propionate.
Studies reported here identify pduW as the gene encoding a propionate kinase enzyme that can compensate for the lack of CobB during the growth of a cobB pduW mutant strain on propionate. PduW activity is important for the synthesis of propionyl coenzyme A (Pro-CoA) during growth on 1,2-PDL. Pro-CoA is the precursor of 2-methylcitrate, the coactivator needed for transcriptional activation of the prpBCDE operon by the PrpR activator protein (23, 31, 42). The PrpB, -C, -D, and -E and aconitase enzymes (AcnA, AcnB) convert Pro-CoA to pyruvate via the 2-methylcitric acid cycle (see Fig. 4) (22, 25).
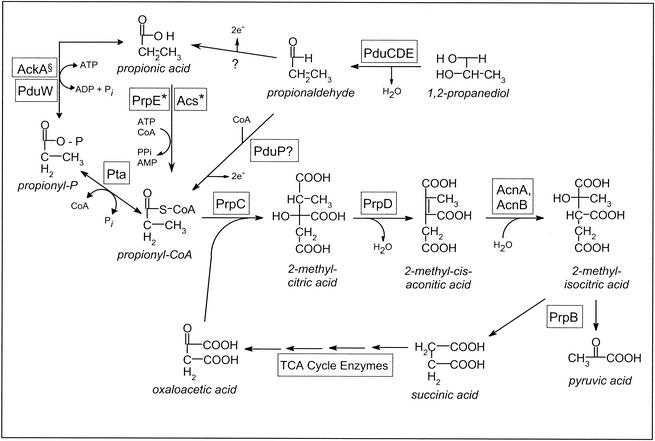
FIG. 4.
Pro-CoA synthesis during growth on 1,2-PDL. PduCDE, coenzyme B12-dependent propanediol dehydratase; PduP, putative propionaldehyde dehydrogenase; PduW, propionate kinase; Pta, phosphotransacetylase; PrpE, Pro-CoA synthetase; Acs, Ac-CoA synthetase; PrpC, 2-methylcitrate synthase; PrpD, 2-methylcitrate dehydratase; AcnA and AcnB, aconitases A and B, respectively; PrpB, 2-methylisocitrate lyase; TCA, tricarboxylic acid; *, under the control of the CobB sirtuin system; §, the chromosomal level does not compensate for the lack of PduW.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All of the strains used in this work were derivatives of S. enterica serovar Typhimurium LT2. The genotypes of the strains and plasmids used are listed in Table 1. The minimal medium (no-carbon medium E [NCE]) (2) was supplemented with succinate at 50 mM, propionate at 30 mM, and 1,2-PDL at 82 mM for growth as a carbon and energy source and 12 mM for induction of the pdu operon, glycerol at 1 mM, l-methionine at 0.5 mM, MgSO4 at 1 mM, vitamin B12 at 150 nM, Na tetrathionate at 10 mM (33), and trace minerals (1). Growth was monitored with a Spectronic 20D spectrometer (Spectronic Instruments). Antibiotics were present at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 20 μg/ml; tetracycline, 10 μg/ml; kanamycin, 40 μg/ml. All chemicals were purchased from Sigma Chemical Co. unless otherwise stated.
TABLE 1.
Genotypes of the strains and plasmids used in this work
| Strain or plasmid | Genotype | Reference or sourcea |
|---|---|---|
| Strains | ||
| S. enterica | ||
| JR501 | hsdSA29 hsdSB121 hsdL6 metE551 trpC2 ilv-452 rpsL120 xyl-404 galE719 H1-b H2-en,n,x [cured of Fels2(−)] fla-66 nml | 40 |
| TR6583 | metE205 ara-9 | K. Sanderson via J. Roth |
| TN4502 | put-521 oxd-2(tdcD)::Mud(cat+) | 45 |
| DM6903 | pduW505::MudJ | D. Downs |
| Derivatives of TR6583 | ||
| JE2445 | cobB1176::Tn10d(tet+) | Laboratory collection |
| JE2506 | pduN8::MudJ(kan+) | Laboratory collection |
| JE2845 | cobB1206::MudJ(kan+)b | Laboratory collection |
| JE4175 | pBAD30 bla+ | |
| JE4313 | Δ1231acs prpE213::kan+ | |
| JE4597 | pta-209::Tn10 | Laboratory collection |
| JE4718 | cobB1176::Tn10d(tet+) pta-102::MudJ(kan+) | |
| JE5330 | pduN8::MudJ cobB1176::Tn10d(tet+)c | |
| JE5913 | cobB1176::Tn10d(tet+) pduN8::MudJ(kan+)/pPDU1 pduW+ | |
| JE5945 | cobB1176::Tn10d(tet+) pduN8::MudJ(kan+)/pBAD30 bla+ | |
| JE6176 | cobB1176::Tn10d(tet+) pduN8::MudJ(kan+)/pTDC1 tdcD+ | |
| JE6284 | pta::cat+ | R. Maurer |
| JE6290 | Δ1231acs prpE213::kan+pta-209::Tn10 | |
| JE6291 | pduW504::kan+ | |
| JE6622 | pduW504::kan+cobB1176::Tn10d(tet+) | |
| JE6923 | pta-102::MudJ/pBAD30 bla+ | |
| JE6924 | pta-102::MudJ/pTDC1 tdcD+ | |
| JE6925 | pta-102::MudJ/pPDU1 pduW+ | |
| JE6952 | cobB1176::Tn10d(tet+) pduN8::MudJ(kan+) pta::cat+ | |
| JE6953 | cobB1176::Tn10d(tet+) pduN8::MudJ(kan+) pta::cat+/pBAD30 bla+ | |
| JE6954 | cobB1176::Tn10d(tet+) pduN8::MudJ(kan+) pta::cat+/pPDU1 pduW+ | |
| JE6955 | cobB1176::Tn10d(tet+) pduN8::MudJ(kan+) pta::cat+/pTDC1 tdcD+ | |
| JE7045 | pduW505::MudJ | |
| JE7053 | pduW505::MudJ/pBAD30 bla+ | |
| JE7054 | pduW505::MudJ/pPDU1 pduW+ | |
| JE7058 | cobB1176::Tn10d(tet+) pduW505::MudJ/pPDU1 pduW+ | |
| JE7259 | pduW505::MudJ/pACK3 ackA+ | |
| JE7260 | pduW505::MudJ/pTDC1 tdcD+ | |
| E. coli | ||
| DB3.1 | RR1 gyrA462 endA (recA) | Invitrogen |
| DH5α | F′/endA1 hsdR17(rK− mK+) supE44 thi-1 recA1 gyrA (Nalr) relA1 Δ(lacZYA-argF)U169 deoR [φ80d/acΔ(lacZ)M15] | |
| Plasmids | ||
| pPDU1 | pduW+ in pBAD30 bla+ | |
| pPDU2 | pduO+ in pBA18s bla+ | |
| pPDU3 | pduP+ in pBAD18s bla+ | |
| pPDU4 | pduQ+ in pBAD18s bla+ | |
| pPDU15 | pduX+ in pBAD30 bla+ | |
| pPDU16 | pduW+pduX+ in pBAD30 bla+ | |
| pTDC1 | tdcD+ in pBAD18s bla+ | |
| pENTR1A | Entry vector; kan+ccdB+ | Invitrogen |
| pBAD30-dest | Converted destination vector; bla+cat+ccdB+ | |
| pBAD18s-dest | Converted destination vector; bla+cat+ccdB+ | |
| pBAD18s | Cloning vector | 18 |
| pBAD30 | Cloning vector | 18 |
High-pressure liquid chromatography (HPLC) analysis of propionate excreted into the medium.
To determine the amount of propionate excreted by specific strains, cultures (20 ml of medium in a 125-ml Klett flask) were grown in NCE containing 1,2-PDL as the sole source of carbon and energy. Cultures were grown for 40 h at 37°C with shaking, and cell density was monitored with a Klett colorimeter (Manostat Corp.). Samples (1 ml each) were removed from the culture as a function of time; cells were removed by centrifugation, and the supernatant was prepared for HPLC analysis. For this purpose, a 500-μl sample was filtered through a Costar Spin-X 0.45-mm cellulose acetate centrifuge tube filter (Corning Inc., Corning, N.Y.), acidified with 2.5 μl of 5N H2SO4, and stored at −20°C until used. The amount of propionate in each sample was determined with a computer-controlled Waters HPLC system equipped with a model 600 quaternary solvent delivery system and a model 996 photodiode array detector. Components of the sample (200 μl) were resolved with an Aminex HPX-87H (300 by 7.8 mm) organic acid analysis column (Bio-Rad Laboratories) heated at 45°C and developed isocratically with a 5 mM H2SO4 at a flow rate of 0.6 ml/min; elution from the column was monitored at 210 nm. Propionate eluted 17 min after injection. The amount of propionate was determined by measuring the area under the curve with the Waters Millennium32 software package. NCE containing 0.5, 1, 2, 4.1, 8.2, or 16 μmol of propionate/ml of medium was used to generate a standard curve. Regression analysis of standard-curve data showed a correlation coefficient of 0.99988.
Complementation studies.
Plasmids were introduced into strains by transformation (35). For complementation analysis on solid medium, transformants were patched on Luria-Bertani (LB) solid-agar plates supplemented with ampicillin, incubated for 2 to 3 h at 37°C, and replica printed onto minimal-medium plates supplemented with the appropriate nutrients. Plates were incubated at 37°C for 24 to 36 h, until growth was observed. For complementation analysis in liquid medium, overnight cultures (37°C, 16 h) of strains grown in LB broth supplemented with antibiotics were used to inoculate minimal medium containing appropriate nutrients. The volume of inoculum was 1/100 of the total volume of the culture. Growth was monitored in 96-well microtiter dishes (Falcon; Becton Dickinson & Co., Franklin Lakes, N.J.) as a function of time with a computer-controlled SpectraMAX Plus high-throughput spectrophotometer (Molecular Devices). Optical density readings at 650 nm were collected every 10 min for 50 h with 5-min shaking periods between measurements. The incubation chamber inside the spectrophotometer was maintained at 37°C. Each culture was analyzed in octuplicate.
Transductions.
Transductions involving the generalized transducing phage P22 HT105 int-201 were performed as previously described (10, 13, 36, 43).
DNA isolations and manipulations.
Plasmids transferred into S. enterica serovar Typhimurium were first introduced into the recombination-deficient JR501 strain by transformation (35). Plasmid DNA was isolated with the Wizard Plus SV Plasmid Miniprep kit from Promega (Madison, Wis.) in accordance with the manufacturer's instructions. DNA fragments were isolated from 1% (wt/vol) agarose gels and purified with the QIAquick Gel Extraction kit; PCRs were purified with the QIAquick PCR purification kit from Qiagen (Chatsworth, Calif.) as suggested by the manufacturer.
PCR amplifications.
Enzyplus polymerase from Stratagene (La Jolla, Calif.) was used in all PCR amplification protocols. A Perkin-Elmer 2400 GeneAmp PCR System was used to perform the reactions (Perkin-Elmer, Branchburg, N.J.). All primers were obtained from Integrated DNA Technologies, (Coralville, Iowa).
Mapping of the pdu-8::MudJ insertion.
Arbitrary-primer PCR was used to determine the precise location of the pdu-8::MudJ insertion in the chromosome of strain JE2504 (pdu-8::MudJ) (8, 30, 32). The insertion was mapped with a PCR product amplified from strain JE2504 genomic DNA obtained with the Epicentre Genomic DNA extraction kit (Epicentre, Madison, Wis.). For the first round of PCR, a standard 50-μl reaction mixture was prepared with 1 μM primer ARB1 [GGCCACGCGTCGACTAGTACN (10)GATAT] and internal transposon primer MuL (ATCCCGAATAATCCAATGTCC). The amplification profile for the reaction was 95°C for 5 min; 5 cycles of 94°C for 30 s, 30°C for 30 s, and 72°C for 1 min; 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min; and then 5 min at 72°C. Amplified DNA was purified and used as the template for the second round of PCR amplification. A similar reaction mixture was prepared with primers MuL and ARB2 (GGCCACGCGTCGACTAGTAC). The amplification profile was the following: 29 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min and then 5 min at 72°C at the end. Amplified DNA was visualized by gel electrophoresis (2% [wt/vol] agarose-1× Tris-borate-EDTA gel) and purified from the gel in accordance with the manufacturer's instructions. Sequencing PCRs were prepared with the ABI PRISM dye terminator cycle sequencing kit (Perkin-Elmer Life Sciences) in accordance with the manufacturer's instructions. DNA in the reaction mixtures was purified with AutoSeq G-50 columns (Amersham Pharmacia Biotech), dried under vacuum, and sequenced at the Biotechnology Center of the University of Wisconsin, Madison.
Construction of plasmids carrying pdu genes.
All pPDU and pTDC plasmids (Table 1) were constructed with the Gateway cloning system (Invitrogen, Carlsbad, Calif.) in accordance with the manufacturer's protocols. The pduO, pduP, pduQ, pduW, pduX, and tdcD genes were individually amplified with primers carrying the attB1 and attB2 sites for the 5′ and 3′ ends (Table 2), respectively, and genomic DNA as the template. PCR amplification products were cloned into entry vector pENTR1A and used to transform DH5α cells with selection for kanamycin resistance. Plasmid DNA isolated from a kanamycin-resistant transformant was used to transfer the genes into destination plasmids pBAD18s-dest (cat+ bla+) and pBAD30-dest (cat+ bla+). These plasmids were derivatives of pBAD30/18s converted into destination vectors with the Gateway vector conversion system. This conversion was accomplished by cloning a 1.7-kb insert containing attR, the ccdB lethal toxin gene, and a chloramphenicol resistance marker (provided by the manufacturer) into the SmaI and EcoRI restriction sites of pBAD30 and pBAD18s (18), respectively. The resulting plasmids were introduced into Escherichia coli DB3.1 cells for propagation. Transfer of the insert from the entry to the destination vector was performed in accordance with the manufacturer's instructions. Plasmid DNA from the insert transfer reaction was used to transform DH5α cells with selection for resistance to ampicillin. The presence of inserts in destination vectors was verified by PCR amplification of specific genes with the primers listed in Table 2 for each gene. Vector DNA was used as a negative control. Genes pduO, pduP, pduQ, and tdcD were cloned into plasmid pBAD18s, since the amplified fragment did not include the native ribosome-binding site, and pduW, pduX, and pduWX were cloned into pBAD30. For pduW, the coding sequence could not be cloned by itself, since a predicted 14-bp secondary structure of the mRNA downstream of the ATG start codon prevented efficient amplification of pduW. This problem was circumvented by extending the primer 13 bp 5′ to the pduW start codon, thus including the ribosome-binding site.
TABLE 2.
Primers used in this work
| Plasmid | Primer | Sequence |
|---|---|---|
| pPDU1 | pduW5′attB1 | 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCGAAGGAACCTTTATG-3′ |
| pduW3′attB2 | 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCAGGCTGGTACACAAA-3′ | |
| pPDU2 | pduO5′attB1 | 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTTATGGCGATTTATAC-3′ |
| pduO3′attB2 | 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCATTGATGAGTCCCC-3′ | |
| pPDU3 | pduP5′attB1 | 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCAATGAATACTTCTA-3′ |
| pduP3′attB2 | 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTAGCGAATAGAAAAGC-3′ | |
| pPDU4 | pduQ5′attB1 | 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCGATGAATACCTTCTC-3′ |
| pduQ3′attB2 | 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCATAGCAGTTCTTCCA-3′ | |
| pPDU15 | pduX5′RBSattB1 | 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTTTACCATGACACTAT-3′ |
| pduX3′attB2 | 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTATATCAGGCTCTCACTG-3′ | |
| pPDU16 | pduW5′attB1 | 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCGAAGGAACCTTTATG-3′ |
| pduX3′attB2 | 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTATATCAGGCTCTCACTG-3′ | |
| pTDC1 | tdcD5′attB1 | 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTACATGAATGAATTTCCG-3′ |
| tdcD3′attB2 | 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTGTTATGCAAATTCCACTGG-3′ |
Preparation of cell extracts.
Strains JE1208, JE6813, and JE6814 were grown for 16 h in 4 ml of LB medium containing ampicillin (100 μg/ml). These cultures were used to inoculate 400 ml of minimal medium supplemented with succinate (50 mM), MgSO4 (1 mM), l-methionine (0.5 mM), ampicillin (100 μg/ml), arabinose (500 μg/ml), and 1,2-PDL (12 mM), as indicated. Cultures were grown overnight at 37°C with shaking. Cells were harvested by centrifugation at 8,000 × g for 10 min. Cells were resuspended in 10 ml of 50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffer (pH 7.5) containing 0.1 mM dithiothreitol. Cells were disrupted with a French pressure cell (104 kPa), and cell extracts were transferred into Snakeskin dialysis tubing (10,000 MWCO; Pierce, Rockford, Ill.) and dialyzed against two 2-liter changes of 50 mM HEPES buffer (pH 7.5) containing 0.1 mM dithiothreitol at 4°C. Cell extracts were dialyzed initially for 4 h, placed on fresh buffer, and dialyzed overnight. Protein concentrations were determined with the Bio-Rad protein assay kit.
Propionate kinase activity assay.
Propionate kinase activity in cell extracts was assayed in a reaction mixture (final volume, 1 ml) containing: propionate (5 mM), HEPES buffer (pH 7.5; 50 mM), phosphoenolpyruvate (3 mM), ATP (2.5 mM), MgCl2 (12.5 mM), pyruvate kinase (20 U), lactate dehydrogenase (25 U), reduced nicotinamide, adenine dinucleotide (NADH; 250 μM). The reaction mixture was preincubated at 37°C for 2 min in a computer-controlled Perkin-Elmer Lambda Bio-40 scanning spectrophotometer (Perkin-Elmer, Norwalk, Conn.) equipped with a circulating-water jacket. Assays were started by the addition of cell extract (10 to 20 μg of protein), and changes in absorbance at 340 nm were monitored as a function of time. Purified TdcD propionate kinase (1 μg) (V. J. Starai and J. C. Escalante-Semerena, unpublished results) was used as a positive control. Background NADH oxidation in extracts was determined with equivalent amounts of protein of a cell extract of strain JE6923, which carried the cloning vector pBAD30. The specific activity of NADH oxidation with JE6923 cell extract was ≤0.3 μmol of NADH/min/mg of protein. Activities were calculated on the basis of the molar extinction coefficient of NADH at 37°C (6,110 M−1 cm−1) (14). One unit of pyruvate kinase activity was defined as the amount of enzyme needed to convert 1.0 μmol of phosphoenolpyruvate to pyruvate per min at pH 7.5 and 37°C. One unit of lactate dehydrogenase activity was defined as the amount of enzyme needed to convert 1.0 μmol of pyruvate to l-lactate per min at pH 7.5 and 37°C.
RESULTS
The 1,2-PDL-inducible function that allows a cobB mutant strain to grow on propionate is encoded by the pduW gene.
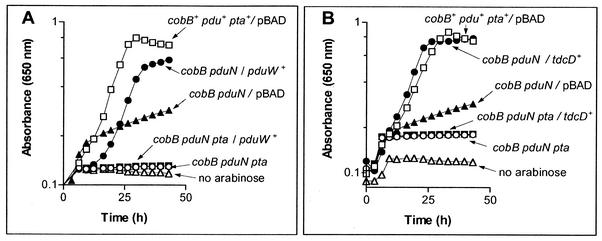
It was previously reported that expression of the pdu operon in a strain carrying the pdu-8::MudJ element failed to compensate for the absence of CobB protein in a cobB mutant strain during growth on propionate as the source of carbon and energy and low concentrations of 1,2-PDL in the absence of cobalamin (41). This result indicated that the pdu gene responsible for allowing cobB mutants to grow on propionate was either interrupted by or located downstream of the pdu-8::MudJ element. Analysis of the DNA sequence flanking the MudJ element revealed that it was located within the pduN gene (data not shown). Downstream of the pduN8::MudJ insertion, there are nine genes, namely, pduOPQSTUVWX. Except for pduO (26), the function of these genes remains speculative. The pduP, pduQ, and pduW genes were considered to be proteins that could have enzymatic activity. Each gene was individually cloned into arabinose-inducible pBAD plasmids, and the resulting plasmids (pPDU1 to -4; Table 1) were transformed into strain JE5330 [pduN8::MudJ cobB1176::Tn10d(tet+)]. The resulting strains were tested for the ability to grow on propionate as a source of carbon and energy with arabinose to induce expression of the plasmid-encoded pdu genes. Arabinose-dependent induction of the pduW+ allele from plasmid pPDU1 in strain JE5913 [pduN8::MudJ cobB1176::Tn10d(tet+)/pPDU1 ParaBAD-pduW+] allowed this strain to grow on propionate (Fig. 1A). The doubling time for strain JE5913 (16 h; Fig. 1B, closed circles) was twice as long as that of the positive control strain JE4175 (cobB+ pduW+/pBAD) (7 h; Fig. 1A, open squares) but was half of the doubling time of negative control strain JE5945 [cobB1176::Tn10d(tet+) pduN8::MudJ/pBAD30 bla+] (35 h; Fig. 1A, solid triangles). Further evidence that pduW function was sufficient to allow growth of the cobB mutant strain on propionate was obtained by introducing plasmid pPDU1 ParaBAD-pduW+ into strain JE7044 [cobB1176::Tn10d(tet+) pduW505::MudJ]. The resulting strain (JE7058) grew as well as strain TR6583 (pdu+ cobB+) on propionate and 1,2-PDL (data not shown). Plasmids carrying pduO+, pduP+, or pduQ+ failed to correct the Prp− phenotype of strain JE5330 to any extent (data not shown).
FIG. 1.
In vivo evidence that proteins with propionate kinase activity restore the growth of a cobB mutant strain on propionate. In the genotypes described below, pBAD refers to cloning vector pBAD30 (18), cobB represents allele cobB1176::Tn10d(tet+), and pduN represents allele pduN8::MudJ(kan+). In both panels, when added to the medium, arabinose was at 200 μM. (A) Complementation by pduW+. Strains (genotype; symbol): JE4175 (cobB+ pdu+ pta+/pBAD; open squares), JE5913 (cobB pduN/pduW+; solid circles), JE5945, (cobB pduN/pBAD; solid triangles), JE6954 (cobB pduN pta pduW+; solid squares), and JE6953 (cobB pduN pta/pBAD; open circles). The growth behavior of strains JE5913 and JE6954 in the absence of arabinose is represented by open triangles. (B) Complementation by tdcD+. Strains (genotype; symbol): JE4175 (open squares); JE6176 (cobB pduN tdcD+; solid squares), JE5945 (cobB pduN/pBAD; solid triangles), JE6955 (cobB pduN pta tdcD+; solid squares), JE6953 (open circles). The growth behavior of strains JE6176 and JE6955 in the absence of arabinose is represented by open triangles.
In vivo evidence that pduW encodes a protein with propionate kinase activity.
No in vitro data have been reported to support the annotation of pduW as encoding a propionate kinase enzyme. Evidence for the biochemical role of the PduW protein was obtained through in vivo analysis of the effect that expression of a bona fide propionate kinase had on the ability of a cobB mutant strain to grow on propionate. The tdcD gene of S. enterica serovar Typhimurium encodes a propionate kinase (19) (Starai and Escalante-Semerena, unpublished results). tdcD was cloned under the control of an arabinose-inducible promoter {plasmid pTDC1 (ParaBAD-tdcD+) was introduced into strain JE5330 [pduN8::MudJ cobB1176::Tn10d(tet+)]}, and growth of the resulting strain (JE6176) on propionate was assessed in the absence or presence of arabinose in the culture medium (Fig. 1B). Growth of strain JE6176 was compared to that of strain JE5913, which was constructed by introducing plasmid pPDU1 (ParaBAD-pduW+) into strain JE5330. Growth of strain JE5330 on propionate was restored when expression of either the pduW+ or tdcD+ gene was induced by arabinose (Fig. 1A and B, respectively, solid circles). In both cases, growth on propionate was dependent on a functional phosphotransacetylase (Pta) enzyme (Fig. 1A and B, open circles), a result consistent with the hypothesis that the poor but reproducible growth of the cobB pduN mutant strain on propionate (Fig. 1A, solid triangles) was due to the synthesis of Pro-CoA via the propionate kinase-phosphotransacetylase pathway. Providing a wild-type allele of pduW or tdcD in trans did not restore growth of the cobB pduN pta triple mutant (Fig. 1A and B, solid squares), consistent with a need for a functional propionate kinase-Pta system for the generation of Pro-CoA. Expression of the ackA gene (encodes acetate kinase) substituted for PduW during growth of cobB pduW mutant strain JE7259 on propionate. The growth of strain JE7259 was as good as that of the wild-type strain (both strains grew with a 7-h doubling time). These results led to the conclusion that the PduW protein had propionate kinase activity.
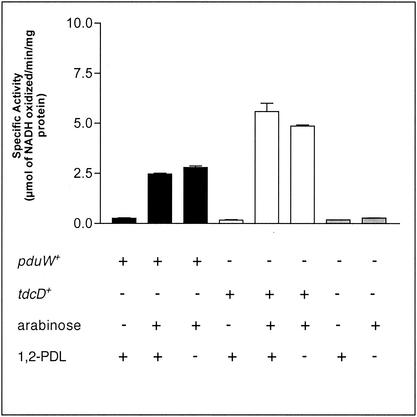
In vitro measurements of the propionate kinase activity of PduW.
Further support for the conclusion that PduW had propionate kinase enzymatic activity was obtained by measuring propionate kinase activity in cell extracts obtained from strains carrying a plasmid encoding the pduW or tdcD gene under the control of an arabinose-inducible promoter. Cell extracts enriched for the PduW or TdcD protein showed levels of propionate kinase activity that were ∼11- and 45-fold higher than the background, respectively (Fig. 2).
FIG. 2.
In vitro evidence that pduW encodes a protein with propionate kinase enzyme activity. Cells were grown in the presence or absence of arabinose (500 μM) and/or 1,2-PDL (12 mM), as indicated. Data shown as solid bars were obtained with extracts of strain JE6925, data shown as open bars were obtained with cell extracts of strain JE6924, and data shown as light gray bars were obtained with cell extracts of strain JE6923. Growth conditions are shown at the bottom. Error bars show the standard deviation of determinations performed in triplicate.
pduW and pduX functions are not needed for growth on 1,2-PDL.
A strain containing a chromosomal MudJ element inserted in the pduW gene (strain JE7045) grew as well as the strain carrying a wild-type pdu operon on 1,2-PDL as the source of carbon and energy under aerobic conditions or under anaerobic conditions with tetrathionate as the electron acceptor (data not shown). These results suggest that neither pduW nor pduX (downstream of pduW) is required for growth of S. enterica serovar Typhimurium on 1,2-PDL.
Pro-CoA synthetase and propionate kinase-phosphotransacetylase activities are required to activate propionate excreted during growth of S. enterica serovar Typhimurium on 1,2-PDL.
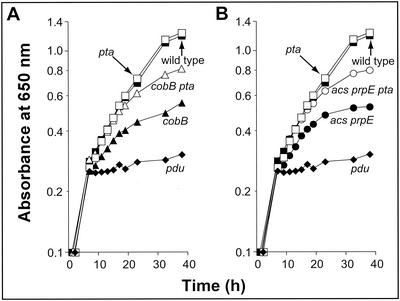
Wild-type growth of S. enterica serovar Typhimurium on 1,2-PDL required CobB protein deacetylase activity (Fig. 3A, solid triangles versus solid squares). This result suggested that, during growth on 1,2-PDL, the wild-type strain excreted a significant amount of propionate. Measurements of propionate levels in the culture medium during growth on 1,2-PDL showed that the wild-type strain excreted as much as 26% of the initial carbon load available as 1,2-PDL as propionate (Table 3, line 1). Since CobB deacetylase activity is needed to maintain acetyl CoA (Ac-CoA) and Pro-CoA synthetase (encoded by acs and prpE, respectively) in their deacetylated, active state (16, 37, 38), it follows that lack of CobB would block the conversion of propionate to Pro-CoA, thus reducing the amount of carbon and energy available to the cell for growth (Fig. 4).
FIG. 3.
In vivo evidence that the PduW-Pta and Acs systems are needed during growth on 1,2-PDL. (A) Strains (symbols): TR6383 (solid squares), JE4597 (open squares), JE4718 (open triangles), JE2445 (solid triangles), and JE2506 (solid diamonds). (B) Strains (symbols): TR6583 (solid squares), JE4597 (open squares), JE6290 (open circles), JE4313 (solid circles), and JE2506 (solid diamonds). Cultures (5 ml of each) were grown in tubes (16 by 150 mm) with shaking at 37°C. Growth was monitored at 650 nm. The culture medium contained: NCE buffer salts, glycerol (1 mM), 1,2-PDL (30 mM), vitamin B12 (150 nM), l-methionine (0.5 mM), and trace minerals.
TABLE 3.
Concentrations of propionate in medium
| Line no. | Strain | Relevant genotypea | Cell density (Klett units) after 40 h of incubationb | Concn of excreted propionate (mM) | % of carbon as propionatec |
|---|---|---|---|---|---|
| 1 | TR6583 | cobB+pta+pdu+acs+prp+ | 129 | 21 | 26 |
| 2 | JE7045 | pduW | 114 | 30 | 37 |
| 3 | JE4718 | cobB pta | 106 | 32 | 39 |
| 4 | JE4597 | pta | 114 | 30 | 37 |
| 5 | JE2445 | cobB | 85 | 21 | 26 |
| 6 | JE4313 | acs prpE | 85 | 20 | 24 |
In strain TR6583, pdu+ prp+ means that all of the genes in the pdu and prp operons are wild type. All strains are derivatives of strain TR6583 and carry metE205 and ara-9 mutations in their background. The natures of the mutations in pduW, cob, pta, acs, and prpE are listed in Table 1. Despite the polar effect of the mutation in pduW on pduX, the effect of the lack of pduW was corrected by a plasmid carrying only pduW+, indicating that pduW function was necessary and sufficient to correct the phenotype caused by the pduW insertion.
All cultures were in stationary phase after 40 h of incubation at 37°C with shaking.
The initial concentration of carbon in the medium was 82 mM 1,2-PDL. Strains JE2506 (pduN), JE5330 (cobB pduN), JE5913 (cobB pduN/pPDU1 pduW+), and JE6176 (cobB pduN/pTDC1 tdcD+) grew to cell densities of <30 Klett units, and the concentration of propionate in the medium was <2 mM.
If the effect of the lack of CobB activity on 1,2-PDL catabolism were due to acetylated, inactive Acs and PrpE enzymes, the growth behavior of the acs prpE pta+ cobB+ and acs prpE pta cobB+ strains on 1,2-PDL should mirror that of the cobB pta+ acs+ prpE+ and cobB pta acs+ prpE+ strains. In fact, the growth responses of the acs prpE and cobB mutant strains during growth on 1,2-PDL were very similar (Fig. 3A and B, closed triangles versus closed circles). Interestingly, in both cases, inactivation of pta function had a positive effect on the rate of growth and final cell density of the cultures (Fig. 3A and B, open triangles versus open circles).
Acyl-CoA synthetases and the PduW-Pta system work in concert to recapture exogenous propionate.
The amount of propionate excreted into the medium was also measured in strains JE2445 (cobB), JE4597 (pta), JE4718 (cobB pta), JE7045 (pduW), and JE4313 (acs prpE). Two patterns emerged from these measurements: (i) strains that reached cell densities slightly lower than that reached by the wild-type strain excreted 37 to 39% of the initial 1,2-PDL as propionate (i.e., strains JE4597, JE4718, and JE7045) and (ii) strains that grew to markedly reduced cell densities relative to the wild-type strain (JE2445 and JE4313) but excreted amounts of propionate equivalent to that of the wild-type strain (Table 3, lines 5 and 6). These data suggest that the positive effect that the inactivation of the pta gene has on the growth of the cobB and acs prpE mutants on 1,2-PDL (Fig. 3A and B) is inconsistent with the idea that pta mutants excrete less propionate, funnel more Pro-CoA into the 2-methylcitric acid cycle, and thus increase carbon flux through the tricarboxylic acid cycle. Instead, it appears that pta mutants are more efficient users of the Pro-CoA generated from 1,2-PDL catabolism. The efficient use of Pro-CoA in pta mutants did not require acyl-CoA synthetase (Acs and PrpE) activities, since loss of these activities did not result in a larger amount of propionate being excreted into the medium by the cobB pta double mutant relative to the pta mutant (Table 3, lines 3 and 4). The lack of PduW kinase activity also resulted in levels of propionate excreted equivalent to those measured in the pta and cobB pta mutant strains (Table 3, line 2), suggesting that chromosomal levels of acetate kinase (AckA), a homolog of PduW, did not compensate for the lack of PduW and that the PduW-Pta system is required to recapture exogenous propionate.
Taken together, the above data indicate that the PduW-Pta pathway contributes a substantial amount of the Pro-CoA generated from exogenous propionate during growth on 1,2-PDL. The data also support the existence of an as-yet-unidentified aldehyde dehydrogenase enzyme that oxidizes propionaldehyde derived from 1,2-PDL to propionate. Whether the putative aldehyde dehydrogenase is encoded by the pduP gene remains to be determined.
The low cell densities reached by strains that lack acyl-CoA synthetase functions but have a functional PduW-Pta system (Table 3, lines 5 and 6) are indicative of a role for the acyl-CoA synthetases in the activation of exogenous propionate during growth on 1,2-PDL. The low cell densities of cultures of strains JE2445 and JE4313 are likely a consequence of the low affinity of the PduW-Pta system for propionate.
DISCUSSION
Pro-CoA is central to the integration of the 1,2-PDL and propionate catabolic pathways in S. enterica serovar Typhimurium.
The data reported in this paper identify Pro-CoA as a common intermediate of the 1,2-PDL and propionate pathways in S. enterica serovar Typhimurium. During growth of this bacterium on 1,2-PDL, three systems synthesize Pro-CoA, all of which ensure that transcription of the prpBCDE operon encoding the propionate catabolic enzymes can occur.
The scheme shown in Fig. 4 is supported by the data reported here. In this scheme, carbon flux from 1,2-PDL to pyruvate via the 2-methylcitric acid cycle relies on Pro-CoA-synthesizing systems. Pro-CoA is derived from 1,2-PDL catabolism by the concerted action of 1,2-PDL dehydratase (PduCDE) and the putative propionaldehyde dehydrogenase (PduP) enzymes (4). Figure 4 shows that propionate is generated via two routes. One route is the direct oxidation of propionaldehyde to propionate by an as-yet-unidentified aldehyde dehydrogenase. A second route is the reactions catalyzed by the propionate kinase PduW-phosphotransacetylase (Pta) enzymes running in reverse. We speculate that these two routes are needed to maintain a concentration of Pro-CoA in the cell that would not be deleterious to cell function. Although acetate kinase (AckA) can synthesize propionyl-phosphate, chromosomal levels of AckA fail to compensate for PduW during growth of the cobB pduW mutant on propionate. This observation may reflect a higher affinity of PduW for propionate and/or a higher turnover value for PduW than for the AckA enzyme.
Since a substantial amount of propionate is excreted during growth on 1,2-PDL (Table 3), it is important to the cell to recapture propionate as the level of Pro-CoA in the cell decreases because of its catabolism via the 2-methylcitric acid cycle. To activate propionate, the cell relies on the PduW-Pta enzymes and the Ac-CoA (Acs) and Pro-CoA synthetase (PrpE) enzymes. Multiple Pro-CoA-synthesizing systems are needed to ensure the cell an adequate supply of Pro-CoA, a substrate of the 2-methylcitrate synthase (PrpC) enzyme. PrpC catalyzes the condensation of Pro-CoA and oxaloacetate to yield 2-methylcitrate, the coactivator molecule sensed by the PrpR regulatory protein that controls the expression of the prpBCDE operon encoding propionate catabolic enzymes (31, 42).
Role of the CobB protein deacetylase in propionate catabolism.
The negative effect that the lack of NAD+-dependent CobB protein deacetylase activity has on 1,2-PDL catabolism is indirect and is caused by the need for CobB to maintain both Acs and PrpE in their deacetylated, active state (37, 38). Although expression of the prpBCDE operon in a cobB mutant can be induced by exogenous 2-methylcitrate (S. Palacios and J. C. Escalante-Semerena, unpublished results), growth of the cobB mutant on 1,2-PDL is still impaired because Acs and PrpE remain acetylated and hence inactive.
Why is the propionate kinase-phosphotransacetylase or acyl-CoA synthetase pathway of Pro-CoA synthesis needed for the catabolism of propionate?
The question of why the propionate kinase-phosphotransacetylase or acyl-CoA synthetase pathway of Pro-CoA synthesis is needed for the catabolism of propionate is an important question in light of the fact that S. enterica serovar Typhimurium already has a specific, efficient Pro-CoA synthetase enzyme (encoded by the prpE gene [21, 24]). However, synthesis of PrpE can occur only when 2-methylcitrate is present to induce prpBCDE operon expression (31, 42). To circumvent the problem of not having PrpE available to activate propionate, S. enterica serovar Typhimurium uses Ac-CoA synthetase (Acs) to generate Pro-CoA so that the 2-methylcitrate synthase (PrpC) enzyme can condense it with oxaloacetate to make 2-methylcitrate (Fig. 4) (24).
We propose that the role of the propionate kinase-phosphotransacetylase- and CobB-dependent pathways is to ensure that a sufficient level of Pro-CoA is synthesized so that the 2-methylcitrate synthase (PrpC) enzyme can generate enough 2-methylcitrate to activate the expression of the prpBCDE operon (25). This argument assumes that, in the absence of 2-methylcitrate, there is residual transcription of the prpBCDE operon. If this were correct, it would mean that the level of PrpE enzyme synthesized in the absence of 2-methylcitrate is insufficient to raise the concentration of Pro-CoA enough so that residual PrpC levels can generate 2-methylcitrate to activate prpBCDE operon expression by PrpR (25). An alternative scenario is that there is no residual transcription of prpBCDE but the GltA (citrate synthase) enzyme, which is known to synthesize 2-methylcitrate from Pro-CoA and oxaloacetate (20), provides sufficient 2-methylcitrate for the activation of prpBCDE expression by PrpR. Work addressing these questions is in progress.
Why is propionate excreted during growth on 1,2-PDL?
While it is clear that a large percentage of the initial carbon available to the cell as 1,2-PDL is excreted into the medium as propionate, it is unclear why this happens. Possible explanations for why S. enterica serovar Typhimurium excretes propionate during growth on 1,2-PDL are (i) that a pool of free CoA needs to be maintained in the cell for the synthesis of other acyl-CoA metabolites; (ii) that a rate-limiting step beyond the dehydration reaction that converts 1,2-PDL to propionaldehyde leads to the accumulation of this toxic, reactive aldehyde (the cell solves the danger posed by propionaldehyde by oxidizing it to propionate, which can then be extruded into the medium for later use); and (iii) that accumulation of Pro-CoA may lead to the synthesis of toxic metabolites that can inhibit enzymes required for survival.
Previous work from our laboratory showed that glutathione (GSH) is needed for wild-type growth of S. enterica serovar Typhimurium on 1,2-PDL (34). GSH might be used by this bacterium to neutralize the toxic effects of propionaldehyde by forming hydroxypropyl-GSH, which could serve as a substrate for an aldehyde dehydrogenase. GSH-dependent aldehyde dehydrogenases are known to exist in S. enterica serovar Typhimurium's close relative E. coli (17), and GSH-dependent detoxification of reactive aldehydes is best illustrated by the metabolism of pyruvaldehyde (also known as methylglyoxal) (12, 27). It is unknown whether the putative propionaldehyde dehydrogenase (PduP) enzyme of S. enterica serovar Typhimurium is a GSH-dependent enzyme or whether additional GSH-dependent aldehyde dehydrogenases that can use propionaldehyde as a substrate exist in this bacterium.
Accumulation of Pro-CoA may also be deleterious to S. enterica serovar Typhimurium. Pro-CoA has been shown to inhibit pyruvate dehydrogenase in Rhodobacter sphaeroides (29) and to be a competitive inhibitor of citrate synthase in E. coli (28). The negative effect of Pro-CoA, however, appears to be indirect. Studies in S. enterica serovar Typhimurium suggest that 2-methylcitrate formed by citrate synthase is toxic to the cell, although it is unclear whether the negative effects of this compound are due to it or to a derivative of it (20). 2-Methylcitrate is also thought to have toxic effects in Aspergillus niger (7).
In summary, regardless of why S. enterica serovar Typhimurium excretes propionate, this bacterium needs to reactivate propionate to Pro-CoA before it can be used as a carbon and energy source. To do this, the cell relies on the Ac-CoA (Acs) and PduW (kinase)-Pta (phosphotransacetylase) enzymes. Without Pro-CoA, the cell cannot synthesize 2-methylcitrate, the metabolite that signals the presence of propionate and 1,2-PDL in the environment. Since the lack of PduW kinase activity does not arrest growth on 1,2-PDL, it appears that this function is dedicated to the integration of the 1,2-PDL and propionate catabolic pathways. The role of the PduX protein remains unclear.
Acknowledgments
This work was supported by grant GM62203 to J.C.E.-S. S.P. was supported in part by a predoctoral fellowship from CONACyT (Mexico), and V.J.S. was supported by the Jerome Stefaniak and Pfizer predoctoral fellowships.
We thank R. Maurer, C. Miller, and D. Downs for strains.
REFERENCES
- 1.Balch, W. E., and R. S. Wolfe. 1976. New approach to the cultivation of methanogenic bacteria: 2-mercaptoethanesulfonic acid (HS-CoM)-dependent growth of Methanobacterium ruminantium in a pressurized atmosphere. Appl. Environ. Microbiol. 32:781-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berkowitz, D., J. M. Hushon, H. J. Whitfield, Jr., J. Roth, and B. N. Ames. 1968. Procedure for identifying nonsense mutations. J. Bacteriol. 96:215-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bobik, T. A., G. D. Havemann, R. J. Busch, D. S. Williams, and H. C. Aldrich. 1999. The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B12-dependent 1,2-propanediol degradation. J. Bacteriol. 181:5967-5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bobik, T. A., Y. Xu, R. M. Jeter, K. E. Otto, and J. R. Roth. 1997. Propanediol utilization genes (pdu) of Salmonella typhimurium: three genes for the propanediol dehydratase. J. Bacteriol. 179:6633-6639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brämer, C. O., L. F. Silva, J. G. C. Gomez, H. Priefert, and A. Steinbüchel. 2002. Identification of the 2-methylcitrate pathway involved in the catabolism of propionate in the polyhydroxyalkanoate-producing strain Burkholderia sacchari IPT101T and analysis of a mutant accumulating a copolyester with higher 3-hydroxyvalerate content. Appl. Environ. Microbiol. 68:271-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brämer, C. O., and A. Steinbüchel. 2001. The methylcitric acid pathway in Ralstonia eutropha: new genes identified involved in propionate metabolism. Microbiology 147:2203-2214. [DOI] [PubMed] [Google Scholar]
- 7.Brock, M., R. Fischer, D. Linder, and W. Buckel. 2000. Methylcitrate synthase from Aspergillus nidulans: implications for propionate as an antifungal agent. Mol. Microbiol. 35:961-973. [DOI] [PubMed] [Google Scholar]
- 8.Caetano-Annoles, G. 1993. Amplifying DNA with arbitrary oligonucleotide primers. PCR Methods Appl. 3:85-92. [DOI] [PubMed] [Google Scholar]
- 9.Castilho, B. A., P. Olfson, and M. J. Casadaban. 1984. Plasmid insertion mutagenesis and lac gene fusions with mini-Mu bacteriophage transposons. J. Bacteriol. 158:488-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan, R. K., D. Botstein, T. Watanabe, and Y. Ogata. 1972. Specialized transduction of tetracycline resistance by phage P22 in Salmonella typhimurium. II. Properties of a high transducing lysate. Virology 50:883-898. [DOI] [PubMed] [Google Scholar]
- 11.Claes, W. A., A. Pühler, and J. Kalinowski. 2002. Identification of two prpDBC gene clusters in Corynebacterium glutamicum and their involvement in propionate degradation via the 2-methylcitrate cycle. J. Bacteriol. 184:2728-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooper, R. A. 1984. Metabolism of methylglyoxal in microorganisms. Annu. Rev. Microbiol. 38:49-68. [DOI] [PubMed] [Google Scholar]
- 13.Davis, R. W., D. Botstein, and J. R. Roth. 1980. A manual for genetic engineering: advanced bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 14.Dawson, R. M. C., D. C. Elliott, W. H. Elliott, and K. M. Jones. 1986. Data for biochemical research, third edition. Oxford University Press, Oxford, United Kingdom.
- 15.Gerike, U., D. W. Hough, N. J. Russell, M. L. Dyall-Smith, and M. J. Danson. 1998. Citrate synthase and 2-methylcitrate synthase: structural, functional and evolutionary relationships. Microbiology 144:929-935. [DOI] [PubMed] [Google Scholar]
- 16.Gulick, V. J. Starai, A. R. Horswill, K. M. Homick, and J. C. Escalante-Semerena. 2003. Structure of acetylation-controlled acetyl-CoA synthetase bound to coenzyme A. Biochemistry 42:2866-2873. [DOI] [PubMed]
- 17.Gutheil, W. G., B. Holmquist, and B. L. Vallee. 1992. Purification, characterization, and partial sequence of the glutathione-dependent formaldehyde dehydrogenase from Escherichia coli: a class III alcohol dehydrogenase. Biochemistry 31:475-481. [DOI] [PubMed] [Google Scholar]
- 18.Guzman, L.-M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hesslinger, C., S. A. Fairhurst, and G. Sawers. 1998. Novel keto acid formate-lyase and propionate kinase enzymes are components of an anaerobic pathway in Escherichia coli that degrades l-threonine to propionate. Mol. Microbiol. 27:477-492. [DOI] [PubMed] [Google Scholar]
- 20.Horswill, A. R., A. R. Dudding, and J. C. Escalante-Semerena. 2001. Studies of propionate toxicity in Salmonella enterica identify 2-methylcitrate as a potent inhibitor of cell growth. J. Biol. Chem. 276:19094-19101. [DOI] [PubMed] [Google Scholar]
- 21.Horswill, A. R., and J. C. Escalante-Semerena. 2002. Characterization of the propionyl-CoA synthetase (PrpE) enzyme of Salmonella enterica: residue Lys592 is required for propionyl-AMP synthesis. Biochemistry 41:2379-2387. [DOI] [PubMed] [Google Scholar]
- 22.Horswill, A. R., and J. C. Escalante-Semerena. 2001. In vitro conversion of propionate to pyruvate by Salmonella enterica enzymes: 2-methylcitrate dehydratase (PrpD) and aconitase enzymes catalyze the conversion of 2-methylcitrate to 2-methylisocitrate. Biochemistry 40:4703-4713. [DOI] [PubMed] [Google Scholar]
- 23.Horswill, A. R., and J. C. Escalante-Semerena. 1997. Propionate catabolism in Salmonella typhimurium LT2: two divergently transcribed units comprise the prp locus at 8.5 centisomes, prpR encodes a member of the sigma-54 family of activators, and the prpBCDE genes constitute an operon. J. Bacteriol. 179:928-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horswill, A. R., and J. C. Escalante-Semerena. 1999. The prpE gene of Salmonella typhimurium LT2 encodes propionyl-CoA synthetase. Microbiology 145:1381-1388. [DOI] [PubMed] [Google Scholar]
- 25.Horswill, A. R., and J. C. Escalante-Semerena. 1999. Salmonella typhimurium LT2 catabolizes propionate via the 2-methylcitric acid cycle. J. Bacteriol. 181:5615-5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson, C. L. V., E. Pechonick, S. D. Park, G. D. Havemann, N. A. Leal, and T. A. Bobik. 2001. Functional genomic, biochemical, and genetic characterization of the Salmonella pduO gene, an ATP:cob(I)alamin adenosyltransferase gene. J. Bacteriol. 183:1577-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lo, T. W. C., M. E. Westwood, A. C. McLellan, T. Selwood, and P. J. Thornalley. 1994. Binding and modification of proteins by methylglyoxal under physiological conditions. J. Biol. Chem. 269:32299-32305. [PubMed] [Google Scholar]
- 28.Man, W.-J., Y. Li, C. D. Connor, and D. C. Wilton. 1995. The binding of propionyl-CoA and carboxymethyl-CoA to Escherichia coli citrate synthase. Biochim. Biophys. Acta 1250:69-75. [DOI] [PubMed] [Google Scholar]
- 29.Maruyama, K., and H. Kitamura. 1985. Mechanisms of growth inhibition by propionate and restoration of the growth by sodium bicarbonate or acetate in Rhodopseudomonas sphaeroides S. J. Biochem. (Tokyo) 98:819-824. [DOI] [PubMed] [Google Scholar]
- 30.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signaling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 31.Palacios, S., and J. C. Escalante-Semerena. 2000. prpR, ntrA, and ihf functions are required for expression of the prpBCDE operon, encoding enzymes that catabolize propionate in Salmonella enterica serovar Typhimurium LT2. J. Bacteriol. 182:905-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parker, J. D., P. S. Rabinovitch, and G. C. Burmer. 1991. Targeted gene walking polymerase chain reaction. Nucleic Acids Res. 19:3055-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Price-Carter, M., J. Tingey, T. A. Bobik, and J. R. Roth. 2001. The alternative electron acceptor tetrathionate supports B12-dependent anaerobic growth of Salmonella enterica serovar Typhimurium on ethanolamine or 1,2-propanediol. J. Bacteriol. 183:2463-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rondon, M. R., R. Kazmierczak, and J. C. Escalante-Semerena. 1995. Glutathione is required for maximal transcription of the cobalamin biosynthetic and 1,2-propanediol utilization (cob/pdu) regulon and for the catabolism of ethanolamine, 1,2-propanediol, and propionate in Salmonella typhimurium LT2. J. Bacteriol. 177:5434-5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, second ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Schmieger, H., and H. Bakhaus. 1973. The origin of DNA in transducing particles of P22 mutants with increased transduction frequencies (HT-mutants). Mol. Gen. Genet. 120:181-190. [DOI] [PubMed] [Google Scholar]
- 37.Starai, V. J., I. Celic, R. N. Cole, J. D. Boeke, and J. C. Escalante-Semerena. 2002. Sir2-dependent activation of acetyl-coenzyme A synthetase by deacetylation of an active lysine. Science 298:2390-2392. [DOI] [PubMed] [Google Scholar]
- 38.Starai, V. J., H. Takahashi, J. D. Boeke, and J. C. Escalante-Semerena. 2003. Short-chain fatty acid activation by acyl-coenzyme A synthetases requires SIR2 protein function in Salmonella enterica and Saccharomyces cerevisiae. Genetics 163:545-555. [DOI] [PMC free article] [PubMed]
- 39.Textor, S., V. F. Wendisch, A. A. De Graaf, U. Müller, M. I. Linder, D. Linder, and W. Buckel. 1997. Propionate oxidation in Escherichia coli: evidence for operation of a methylcitrate cycle in bacteria. Arch. Microbiol. 168:428-436. [DOI] [PubMed] [Google Scholar]
- 40.Tsai, S. P., R. J. Hartin, and J. Ryu. 1989. Transformation in restriction-deficient Salmonella typhimurium LT2. J. Gen. Microbiol. 135:2561-2567. [DOI] [PubMed] [Google Scholar]
- 41.Tsang, A. W., and J. C. Escalante-Semerena. 1996. cobB function is required for the catabolism of propionate in Salmonella typhimurium LT2: evidence for existence of a substitute function for CobB within the 1,2-propanediol utilization (pdu) operon. J. Bacteriol. 178:7016-7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsang, A. W., A. R. Horswill, and J. C. Escalante-Semerena. 1998. Studies of regulation of expression of the propionate (prpBCDE) operon provide insights into how Salmonella typhimurium LT2 integrates its 1,2-propanediol and propionate catabolic pathways. J. Bacteriol. 180:6511-6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watanabe, T., Y. Ogata, R. K. Chan, and D. Botstein. 1972. Specialized transduction of tetracycline resistance by phage P22 in Salmonella typhimurium. I. Transduction of R factor 222 by phage P22. Virology 50:874-882. [DOI] [PubMed] [Google Scholar]
- 44.Way, J. C., M. A. Davis, D. Morisato, D. E. Roberts, and N. Kleckner. 1984. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene 32:369-379. [DOI] [PubMed] [Google Scholar]
- 45.Wei, Y., and C. G. Miller. 1999. Characterization of a group of anaerobically induced fnr-dependent genes of Salmonella typhimurium. J. Bacteriol. 181:6092-6097. [DOI] [PMC free article] [PubMed] [Google Scholar]