Abstract
Escherichia coli is a clonal species. The best-understood components of its clonal variation are the flagellar (H) and polysaccharide (O) antigens, both well documented since the mid-1930s because of their use in serotyping. Flagellin is the protein subunit of the flagellum that carries H-antigen specificity. We show that 43 of the 54 H-antigen specificities of E. coli map to the flagellin gene at fliC and sequenced all 43 forms and confirmed specificity of each by cloning and expression. This is, to our knowledge, the first time that all known forms of such a highly polymorphic gene have been fully sequenced and characterized for any species. The established distinction between a highly variable central region and more conserved flanking regions is upheld. The sequences fall into two groups, one of which may be derived from the fliC gene of the E. coli/Salmonella enterica common ancestor, the other perhaps obtained by lateral transfer since species divergence. Comparison of sequences revealed that both horizontal DNA transfer and fixation of mutations under diversifying selection pressure contributed to polymorphism in this locus.
The O polysaccharide and flagellin are the two major antigens of gram-negative bacteria, also known respectivly as the O and H antigens. Both are highly polymorphic, and Escherichia coli, if one includes the Shigella strains, has 187 O and 53 H forms defined by serology (4, 6, 15, 21). In this study, we show that 43 of the 53 H forms map to the fliC locus and have sequenced all 43 alleles. In some strains the H-antigen phenotype maps to alternative loci, so we cloned, sequenced, and expressed the fliC gene from type strains to relate definitively H-antigen specificity and sequence. These data supplement the genome sequence data of E. coli K-12 and O157:H7 to give more comprehensive genetic information on the species and, in conjunction with the recently published structure (31) of one flagellin form, will allow analysis of the structural basis of the antigenic variation and development of a molecular typing scheme for the H antigen.
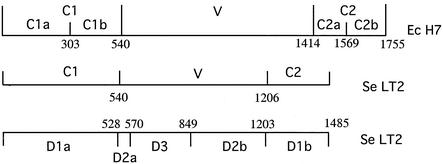
The bacterial flagellum projects well beyond the surface of the cell and is rotated to provide motive power. The flagellar filament is composed of a single protein, flagellin. The flagellin proteins of E. coli and several other species are conserved in their terminal regions, while the central region is variable and carries H-serotype-specific epitopes (9, 17, 22, 39, 40). The structure of the Salmonella enterica LT2 flagellum is known from electron microscopy, X-ray fiber diffraction, and X-ray crystallography. Three domains are recognized (Fig. 1). The conserved terminal segments form the D1 domain located in the center of the flagellum, while the central region of the protein forms two domains (D2 and D3) exposed on the surface (31). The boundaries between D1 and D2 correspond quite well to the boundaries between the central and terminal regions of the protein as determined by alignment of sequences of different forms. However, because we are dealing mostly with sequence data, we refer to conserved (C) and variable (V) regions (also shown in Fig. 1), except where discussing the domains based on structural data.
FIG. 1.
Diagram showing protein domains and sequence-based regions of fliC. Top: the C1, V, and C2 regions and C1a, C1b, C2a, and C2b subregions of E. coli H7, recognized in comparisons of E. coli DNA sequences described in this paper. Center: the C1, V, and C2 regions of S. enterica LT2 located by homology with E. coli H7. Bottom: the domains (D1, D2, and D3) identified in the flagellin protein of S. enterica serovar Typhimurium LT2 (31).
More than 40 genes are needed for flagellar assembly, structure, and function and the associated sensory reception and transduction of the information used to influence the operation of flagella (17). In both E. coli and S. enterica these genes are mostly in four gene clusters, with fliC in one of them (17). Most S. enterica have two flagellin genes, fliC and fljB, expressed alternatively to give the phase 1 and phase 2 H antigens, respectively. The fljB gene is in a fifth gene cluster, not known to occur in E. coli, that contains only fljB and fljA (32). fljB and fljA are coexpressed, with fljA encoding a repressor of fliC, thus ensuring that only one flagellin gene is expressed at any time (35, 36).
The single H antigen of E. coli K-12 is encoded at the fliC locus, and until recently this was thought to apply to all 53 forms (16, 17, 27). However, Ratiner (27) showed that some H-antigen genes are at loci other than fliC, although none have yet been found at fljB. The new loci, flkA, fllA, and flmA, have not been mapped. The 53 H types in E. coli are numbered from 1 to 56, with numbers 13, 22, and 50 not in use (6, 21). In this paper we show by cloning and expression that 43 of the 53 H-antigen types of E. coli are encoded by genes at the fliC locus and that 10 map to other loci.
MATERIALS AND METHODS
Bacterial strains.
The E. coli H-antigen type strains (6, 21) were obtained from the Institute of Medical and Veterinary Science, Adelaide, Australia, or from Karl Bettelheim of the Victorian Infectious Diseases Reference Laboratory, Victoria, Australia. E. coli K-12 fliC mutant strain KS01 (fliC::Tn10) was kindly provided by B. Westerlund-Wikstrom, University of Helsinki, Helsinki, Finland. Antisera against E. coli H antigens were obtained from Denka Seiken Co. Ltd., Tokyo, Japan, and the Institute of Medical and Veterinary Science.
PCR and sequencing, cloning, and expression of fliC genes.
The fliC gene from each H-antigen type strain was PCR amplified, and the product was sequenced. Primers were based on sequences flanking or within sequenced E. coli fliC genes (see Supplementary Tables 1 and 2 at http://www.mmb.usyd.edu.au/archives/). Each fliC gene was cloned into plasmid vector pTRC99A (Pharmacia), and expression was checked by agglutination and microscopic examination of a fresh culture for motility.
TABLE 1.
KS and KA values for comparisons between Ec2 and Se1 fliC sequencesa
| Sequence | KS | KA | KS/KA |
|---|---|---|---|
| C1a | 0.647 | 0.0259 | 24.97 |
| C1b | 1.007 | 0.140 | 7.18 |
| C2a | 3.857 | 0.138 | 27.96 |
| C2b | 0.895 | 0.053 | 16.90 |
Data are averages for all Ec2 sequences compared with all Se1 sequences (one sequence per H specificity).
TABLE 2.
KS and KA values of conserved (C1 and C2) and variable (V) regions of selected pairs of E. coli fliC genes
| Gene pair or avg | Region | KS | KA | KS/KA |
|---|---|---|---|---|
| H1/H12 | C1 | 0.162 | 0 | |
| V | 0.051 | 0.012 | 4.15 | |
| C2 | 0.101 | 0 | ||
| H30/H32 | C1 | 0.202 | 0.005 | 39.44 |
| V | 0.181 | 0.034 | 5.31 | |
| C2 | 0.164 | 0.005 | 35.07 | |
| H5/H56 | C1 | 0.270 | 0.002 | 120.37 |
| V | 0.223 | 0.063 | 3.52 | |
| C2 | 0.219 | 0 | ||
| Avg for Ec1 | C1a | 0.297 | 0.013 | 23.78 |
| C1b | 1.570 | 0.066 | 23.83 | |
| C2a | 0.466 | 0.083 | 5.64 | |
| C2b | 0.250 | 0.009 | 26.93 |
Bioinformatics.
DNA sequences were assembled and edited by using the programs Phred, Phrap, and Consed (7). The Stephens algorithm (37) was used to detect intragenic recombination, and nucleotide diversity (π) was calculated by the method described by Nei and Miller (19). Calculation of synonymous and nonsynonymous substitution rates used the program kindly provided by W. H. Li (14). Evolutionary trees were generated by the neighbor-joining method (30), based on distance estimated by using the two-parameter method of Kimura (12). Phylogenetic trees and bootstrap analysis to determine the statistical significance of each node were done by using PHYLIP (version 3.4; written by Joseph Felsenstein, Department of Genetics, University of Washington, Seattle).
Protein partial specific volume.
A calculated partial specific volume was obtained from high-resolution crystal structures by the following method with the CCP4 suite of programs (2). The nonhydrogen protein atoms from protein database entries were selected, and the volume of the protein was estimated by constructing a surface that is accessible to water by use of a 0.14-nm probe and by use of the surface to calculate a cavity volume. The amino acid sequence was used to calculate the molecular weight, and a calculated partial specific volume was derived from the values for cavity volume and molecular weight. The calculated specific volume of domains D2 plus D3 of flagellin (residues 176 to 403 of 1IO1) was 0.79. The calculated values for four selected proteins were as follows: bovine alpha chymotrypsin (4CHA), 0.83; hen egg white lysozyme (193L), 0.75; human serum albumin (1A06), 0.79; and porcine pepsinogen (3PSG), 0.80. Designations in parentheses are from the Protein Data Bank (www.rcsb.org/pdb/) for the protein structures used.
Nucleotide sequence accession number.
Sequence data from this article have been deposited with GenBank under accession no. AY249138 and AY249989 to AY250029.
RESULTS
Sequence of fliC genes.
We were able to PCR amplify flagellin genes from type strains for H antigens 1, 6, 7, 9, 10, 12, 14, 15, 16, 18, 19, 20, 23, 26, 28, 30, 31, 32, 34, 41, 43, 45, 46, 48, 49, 51, and 52 by using primers 1575 and 1576 (see Supplementary Table 1 at http://www.mmb.usyd.edu.au/archives/) based on sequences 51 to 34 bp upstream and 37 to 54 bp downstream, respectively, of the E. coli K-12 fliC gene. The full gene sequence was obtained for these 27 strains, and use of flanking sequence for primers established that they are at the fliC locus.
For the other type strains, we used various combinations of primers based on the sequence within the fliC gene and obtained sequence from the type strains for H antigens 2, 3, 4, 5, 8, 11, 17, 21, 24, 25, 27, 29, 33, 35, 37, 38, 39, 40, 42, 44, 47, 54, 55, and 56. The central variable region was obtained for all, and the details are shown in Supplementary Table 2 at http://www.mmb.usyd.edu.au/archives/. These data do not distinguish flagellin genes at the fliC locus from those at one of the alternative loci, as no flanking DNA sequence was obtained at this stage. To determine which of these flagellin genes are at the fliC locus, we carried out PCR by using primers specific to each gene in conjunction with primers based on fliD (primer no. 2650; see Supplementary Table 1 at http://www.mmb.usyd.edu.au/archives/) or on fliA (primer no. 648) that flank fliC in E. coli K-12 (3, 11, 18). Two primers specific to each gene were chosen, none having more than 85% identity with any other flagellin sequence (see Supplementary Table 3 at http://www.mmb.usyd.edu.au/archives/). A PCR product of the expected size was found for PCR across the junction of fliC with one of or both fliD and fliA for each gene, establishing that all are at the fliC locus. Some of the PCR products were also sequenced to give full fliC gene sequences.
Only for the H36 and H53 type strains did we not obtain flagellin gene sequence by the above methods. Both are known to be encoded by genes at loci other than fliC (27), and as we were unable to get amplification by using any of the fliC primer pairs, we presume that the fliC gene is at least partially deleted.
Before the start of our work, the fliC genes from the type strains for H antigens 1, 7, and 12 had been sequenced (33). Our sequences for these genes differ from the respective published sequence at from one to four sites, and we carefully rechecked our data.
Specificity of E. coli fliC genes.
The H3, H36, H47, and H53 flagellin genes have been shown to be at a locus called flkA, the H44 and H55 genes at fllA, and the H54 gene at flmA (27). However, the strains expressing some of these H antigens are known to carry a fliC gene in addition to a flkA, fllA, or flmA gene (27). For most other H antigens, it is not established that the expressed flagellin gene is at fliC as is generally assumed, and it was necessary to determine for each of the fliC genes sequenced if it carried the H specificity of the strain from which it was obtained.
The full fliC genes from the H type strains, except those for antigens 4, 17, 35, 36, 44, 53, and 54 (see below), were successfully PCR amplified and cloned into expression vector pTRC99A (for details see Supplementary Table 4 at http://www.mmb.usyd.edu.au/archives/). For H4, H17, and H44, we lacked sufficient sequence information to clone the full gene, so we made an intermediate plasmid (pPR1951) with the terminal sequences of the H7 fliC gene (76 and 93 bp) separated by BamHI and XbaI sites and cloned into it the appropriate part of fliC (see Supplementary Table 4 at http://www.mmb.usyd.edu.au/archives/). These plasmids were transformed into strain KS01, and motility and H-antigen expression were tested for each. In total, 43 of the 49 cloned fliC genes confer motility and encode the H antigen expressed in that strain, i.e., those for H antigens 1, 2, 4, 5, 6, 7, 8, 9, 10, 11, 12, 14, 15, 16, 18, 19, 20, 21, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 37, 38, 39, 41, 42, 43, 45, 46, 48, 49, 51, 52, and 56.
The fliC genes from the H3, H40, H47, and H55 type strains express H antigens 16, 8, 21, and 38, respectively, while those from strains H17 and H44 both express H antigen 4. The deduced amino acid sequences of these fliC genes differ in up to 8 amino acids from those of the corresponding type strains (for details see Supplementary Table 5 at http://www.mmb.usyd.edu.au/archives/). There were four H type strains for which we could not express the fliC gene. Those of the H35 and H54 type strains carried an insertion sequence in a gene otherwise nearly identical to the H11 and H21 fliC genes, respectively. We were unable to PCR amplify and clone the fliC gene from the H36- and H53 type strains.
Thus, of the 53 type strains, 43 have a fliC gene for their H antigen, while six have a fliC gene that expresses an alternative H antigen when cloned but not when situated on the chromosome, as cross-reaction is not reported. The four strains for which we could not express the fliC gene are all known to have the H antigen expressed from a locus other than fliC. Two carry insertion sequences, and two could not be amplified by PCR probably because they have undergone deletion. It seems clear that the flagellin genes for exactly 43 of the 53 H antigens are located at the fliC locus.
Comparison of fliC sequences.
We first used the S. enterica LT2 sequence as outgroup for analysis of the relationships of E. coli fliC genes. It became clear that there was a complex relationship between the LT2 and E. coli sequences, and for this reason, in the trees to be presented, we included Proteus, Yersinia, Bordetella, and Serratia sequences, with the Bordetella sequence as the outgroup (Fig. 1). There are 35 and 9 S. enterica fliC and fljB full gene sequences, respectively, available in GenBank, but there is considerable duplication, and they encode, respectively, only nine and two different H antigens. A representative of each was included in our analysis (for GenBank entries, see Supplementary Table 6 at http://www.mmb.usyd.edu.au/archives/).
As expected, the N- and C-terminal regions of the predicted E. coli flagellin sequences are strongly conserved. The junctions between them and the central variable regions were readily apparent in whole-protein alignments (see Supplementary Table 7 at http://www.mmb.usyd.edu.au/archives/). DNA alignment was based on the amino acid alignment (only whole-codon gaps were allowed). We divided each gene into three regions for separate phylogenetic analysis. C1 (conserved region 1) includes codons 1 to 176 (numbering based on that of the H7 sequence), V (variable region, codons 177 to 491), and C2 (conserved region 2, codons 492 to 585). The junctions (Fig. 1) agreed well with the structural data for S. enterica LT2 flagellin, which show that the two terminal conserved segments form a single domain located in the center of the flagellum (31).
The C1 and C2 sequences were readily aligned, and there were few indels (see Supplementary Table 7 at http://www.mmb.usyd.edu.au/archives/). However, the V region sequences were much more difficult to align even within E. coli, as there were areas of very low-level identity. Many indels were seen, some of which were very long, but it should be noted that, although they appear as indels in alignments, many are most likely due to substitutions by sequences so divergent that the program finds insufficient similarity for alignment.
DISCUSSION
A complete set of E. coli fliC sequences.
There are 53 H antigens in E. coli, and we have established that 43 are encoded by genes in the fliC locus. We present here the sequences of all 43 E. coli fliC H-antigen alleles.
After the sequences reported here were obtained, fliC sequences from the type strains for H antigens 4, 5, and 12 (GenBank accession numbers AB028472, AB028473, and AB028475, respectively) and 12 nontype strains (H antigens 6, 9, 10, 11, 14, 23, 42, 43, 45, 46, 48, and 49) were reported to GenBank (AF128945, AF079163, AF169320, EC0243796, AF169321, AB028476, AF169322, AF169323, AB028477, AB028478, U00096, and AB028480, respectively). Our sequences for the type strains are 100% identical to those published and are more than 99% identical to those of nontype strains.
E. coli H antigens not encoded by fliC genes.
It is known that genes encoding H antigens 3, 36, 44, 47, 53, 54, and 55 are located outside fliC (27), with three loci, flk, fll, and flm, being named. We show here that genes for H17, H35, and H40 are also located in loci other than fliC but were unable to determine if they mapped to flk, fll, or flm, because these loci have not been mapped. The type strains for H antigens 3, 40, 47, 55, 17, and 44 all have fliC genes that are very similar to fliC alleles for other E. coli H antigens and in K-12 express those antigens. The fact that each expresses one of the 43 known fliC H antigens suggests that the E. coli H-antigen typing scheme is virtually complete.
It is not clear what regulates the expression of flagellin genes when there is more than one such gene in E. coli, although it has been shown in some but not all cases studied that a repressor acts on the fliC genes to give phase variation as in S. enterica (26, 27). The strains that we studied are the H-antigen type strains and are not reported to express an additional H antigen, but we did not subject those with the expressed gene not at fliC to selection for expression of an alternate phase. As the promoter region of fliC was not cloned in the expression experiment, it is also possible that this region is mutated in some strains.
The fliC genes of Shigella strains.
All Shigella serotypes except Boydii 13 are fully within E. coli (24, 25), and for this reason we refer to them as Shigella, Dysenteriae, or Sonnei, etc., strains of E. coli. Shigella strains are nonmotile, and the studies by Al Mamun et al. (1) show that the basis for loss of motility varies and for example can be due to deletion in the fliF operon or an IS1 insertion mutation in the flhD operon. Sequences available for fliC genes of Dysenteriae 1 (GenBank SHDFKICD), Sonnei (SHFFLIC SB3), Boydii 5 (SHFFLIC B), and Flexneri 2a (SHFFLIC SF1) are 96.6, 99.8, 97.9, and 96.7% identical to our sequences for H18, H16, H45, and H14, respectively. This confirms the well-recognized view that Shigella strains are forms of E. coli. The fact that the four Shigella strains have fliC genes for one of the 43 E. coli H antigens already mapped to fliC further supports our belief that the 43 fliC genes represent close to a complete set for E. coli.
Major subdivisions of the fliC genes of E. coli.
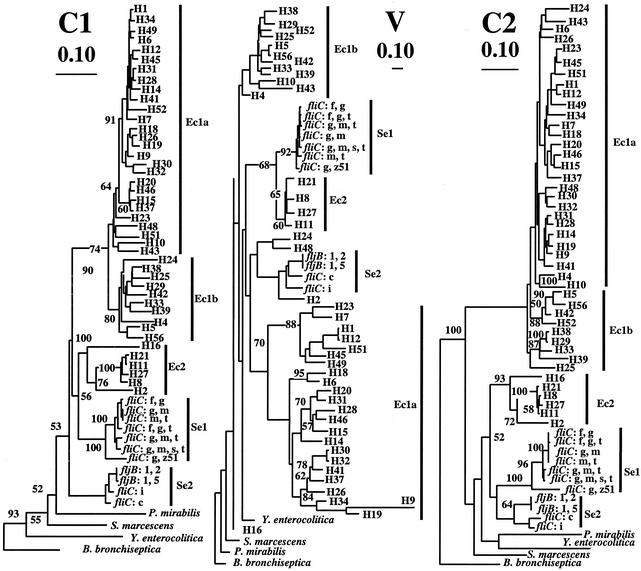
We compared the E. coli fliC genes with those of S. enterica and used other Enterobacteriaceae as outgroups. We first discuss the variation in E. coli but present only trees that include the other sequences. The E. coli fliC sequences for both C1 and C2 fell into two well-defined groups, which we have named Ec1 and Ec2 (Fig. 2). The Ec1 strains can be further divided into subgroups that we have named Ec1a and Ec1b, with the division consistent in the C1 and C2 regions, except for H4, H24, and H52. The Ec1a, Ec1b, and Ec2 groups and subgroups are also seen in the V region, but whereas the distribution of strains into Ec1 and Ec2 is the same for C1 and C2, there are exceptions for the V region that are discussed below with the S. enterica sequences. The Ec1a and Ec1b subgroups are also separated in the V region tree, and some forms lie outside the subgroups. For the C1 region pairwise identity levels within Ec1 or Ec2 range from 100 to 79.8% and 100 to 78.2%, respectively, and for the C2 region, from 100 to 75.5 and 100 to 79.1%, respectively. The average pairwise identity among all E. coli fliC genes is 66.1%. The division into Ec1 and Ec2 is strongly supported by high bootstrap values for the C1 and C2 sequences, although the bootstrap values for separation of Ec2 from the S. enterica groups are not so good (see below).
FIG. 2.
Phylogenetic trees for the three regions of the E. coli flagellin genes generated by the neighbor-joining method. The trees for the C1 and C2 regions were based on DNA alignment, and that for the V region was based on amino acid alignment. Eleven flagellin genes of S. enterica strains are included, and they are labeled with the locus (fliC or fljB) followed by the H-antigen form encoded, using antigen specificity designations traditional for S. enterica fliC and fliB genes (23). See text for details of groups of E. coli and S. enterica genes indicated in the figure. All E. coli fliC genes are from the type strain for that antigen. Flagellin genes of Proteus mirabilis (GenBank accession no. L07270), Yersinia enterocolitica (L33467), Bordetella bronchiseptica (L13043), and S. marcescens (M27219) were also included, and that of B. bronchiseptica was used as the outgroup. The value adjacent to a node indicates the percentage of 1,000 bootstrap trees that contain the node. Only those greater than 50% are shown.
Flagellin sequences and flagellar morphotypes.
Lawn et al. described the appearance under electron microscopy of flagella of E. coli type strains for all but H56 of the presently recognized H antigens and distinguished six morphotypes. The division into morphotypes correlates well with our division based on DNA sequence, with the V regions giving the best fit. Morphotype D comprises H21, H8, H27, and H11, which form the core strains of group Ec2. Seven of the eight morphotype B strains are in Ec1b for the V region, while the 8 morphotype E and 14 morphotype F strains comprise 22 of the 23 Ec1a forms for the V region. The six morphotype C strains are not grouped in the sequence trees, and the one morphotype A strain (H4) was not placed consistently in the C1, C2, and V region trees, but overall there is a very good correlation indicating that part of the sequence variation that we observe is responsible for the different structural forms. The eight H antigens that did not map to fliC had one each of five of the defined morphotypes. There were five H antigens not allocated to a morphotype, of which H9 and H16 map to fliC.
Relationships of E. coli and S. enterica flagellin genes.
The S. enterica flagellin sequences, like those of E. coli, fell into two discrete groups, with most fliC genes in Se1, but the fliC genes of serovars Typhimurium (fliC:i) and Choleraesuis (fliC:c) were grouped with the two fljB genes as Se2 (Fig. 2). However, as only two fljB sequences and a group of mostly closely related fliC sequences are available, it is premature to generalize about S. enterica flagellin genes.
The E. coli and S. enterica sequences do not form separate clades for each species, and neither does it appear that the two groups of flagellin genes in each species are derived from a gene duplication in a common ancestor. In general Se1, Se2, and Ec2 are more closely related to each other than any are to Ec1. The Proteus, Yersinia, Bordetella, and Serratia sequences are outside the E. coli/S. enterica sequences for C1, but for C2 the Proteus and Yersinia sequences are on long branches among the Ec2/S. enterica sequences, although bootstrap support for this arrangement is below 50%. The Se1, Se2, and Ec2 groups are supported by high bootstrap values for each region, but the relationship between them is less clear.
Origins of the two groups of E. coli flagellin genes.
The four flagellin gene clusters of E. coli, including the fliC cluster, are conserved in the Yersinia pestis genome (http://www.sanger.ac.uk/Projects/Y_pestis), and they map in similar locations in E. coli and S. enterica, indicating a long history in the Enterobacteriaceae and in particular a fliC locus in the E. coli/S. enterica common ancestor. The very high levels of divergence for the V region, both within E. coli and between E. coli and S. enterica, indicate that it has a very different history from that of housekeeping genes. However, for the C1 and C2 regions of fliC, the differences between E. coli Ec2 and S. enterica are comparable to those of housekeeping genes. E. coli and S. enterica housekeeping genes are, on average, 84% identical in the two species, ranging from 72.5 to 99% (34). Pairwise comparisons of the C1 regions of flagellin genes of Ec2 and S. enterica gave identity levels ranging from 63.7 to 80.3% and for the C2 regions identity levels ranging from 65.4 to 73.9%, both close to the divergence for homologous genes in general. We suggest that the conserved regions of the fliC genes in the E. coli Ec2 and Se1 groups were derived from the fliC genes of the common ancestor. This conclusion is supported by analysis of synonymous and nonsynonymous substitutions. Sharp (34) compared 67 housekeeping genes present in both E. coli K-12 and S. enterica Typhimurium and found average values for the number of synonymous substitutions per synonymous site (KS) and the number of nonsynonymous substitutions per nonsynonymous site (KA) of 0.94 and 0.039, respectively, and an average KS/KA ratio of 24. The average KS/KA ratio of 24 reflects the strong selection against nonsynonymous substitutions in the absence of selection for change, while synonymous substitutions are fixed by random genetic drift. We can compare our data with those for the 67 housekeeping genes. For this analysis of the relationship of Ec2 and S. enterica, the C1 and C2 regions were divided into C1a (bp 1 to 303, but note that 1 to 69 is not sequenced for most strains), C1b (bp 304 to 540), C2a (bp 1414 to 1569), and C2b (bp 1570 to 1755), as it became obvious that both conserved regions had segments with different KA values (Table 1; see also Fig. 1). The values of KS and KA (Table 1) for C1a and C2b are typical of the values for comparison of genes of E. coli and S. enterica, as is expected if the divergence in this part of the fliC gene was a result of random genetic drift. We conclude that the C1a and C2b segments have not been subject to selection for change but diverged only under random genetic drift over a period equivalent to that for divergence of E. coli and S. enterica. However, both the C1b and C2a segments gave higher KA values, and the C2a region also has a higher KS (see below).
The situation for Ec1 is quite different. Ec1 fliC sequences are much more divergent from S. enterica sequences and have KS and KA values that indicate saturation, such that they are not useful for indication of divergence time (data not shown). The Ec1 cluster is equidistant from the Ec2 and S. enterica clusters (Fig. 2) and was probably acquired by E. coli after the divergence of E. coli and S. enterica or alternatively was lost in S. enterica. However, only a fraction of the S. enterica flagellin genes have been sequenced, and it cannot be precluded that S. enterica flagellin genes corresponding to the Ec1 group will be found.
The difference between Se1 and Se2 is similar to that between either and Ec2, suggesting that Se1 and Se2 diverged soon after the species S. enterica and E. coli diverged. The situation is complicated by the fact that Se2 includes alleles of fliC and fljB, but it should be noted that gene conversion has been observed for fliC and fljB (20). However, we must again observe that the small number of S. enterica flagellin gene sequences presently available makes it premature to speculate on the origins of the variation in this species.
Recombination within E. coli.
While the Ec1 and Ec2 groups are distinct in the C1, V, and C2 regions, there are a few strains that fall into different groups in one of the regions, suggesting recombination within E. coli. H43 and H52 have Ec1a C1 regions but Ec1b V and C2 regions, while H10 has Ec1a C1 and C2 regions, but the V region is from Ec1b. H24 and H48 have V regions that group together at the base of Se2: H48 has Ec1a C1 and C2 regions, while H24 has one of each. The H2 V region is even closer to the Se2 cluster, while its C1 and C2 regions are at the base of the Se2 cluster. The depth of the Se2 V region cluster, if one includes H24, H48, and H2, is no greater than those for Ec1a and Ec1b. It is clearly possible that these V regions were derived from S. enterica, but the picture will become clearer as more S. enterica flagellin sequences become available. The V region of H16 is unique in being grouped with the outgroup strains of Yersinia pseudotuberculosis and Serratia marcescens, while the C1 and C2 regions are within the Ec1b group; it appears that the V region may have been acquired from a distantly related species. There are no cases of C2 or C2 sequences of E. coli or S. enterica not grouping with sequences from the same species. However, as relatively few of the S. enterica sequences have been published, it is not possible to preclude lateral transfer of whole fliC genes between these species.
There are several cases of sequence relationships of Ec1a or Ec1b being different in the V region from those found in the C1 and C2 regions, presumably due to recombination. The core of the Ec2 group includes the H8, H11, H21, and H27 alleles, which are closely related in their C1, V, and C2 regions (Fig. 2). However, for H16 and H2 the C1 and C2 regions are more distantly related but clearly within Ec2, whereas the H2 V regions cluster with the Se2 sequences and the H16 V region is in a poorly resolved group near the base of the tree with the outgroups. It seems clear that there has been recombination involving the V region only.
There are other cases where the C1 and C2 regions are in the same subgroup, but there are significant differences between the trees (Fig. 2). For example H26, H9, and H19 are clearly within Ec1a, but whereas their C1 regions are closely related, the C2 regions of H9 and H19 are closer to that of H14 than to that of H26 (Fig. 2). This indicates recombination between Ec1a genes. The C1 and C2 regions of H1, H45, and H12 provide another example (Fig. 2). It seems clear that, while there has been little if any recombination between Ec1 and Ec2 genes within either C1 or C2, there has been some reassortment of these regions that must have involved recombination within each group.
Another example of varying association between fliC regions is H24 and H48, which are in Ec1 for the C regions but group together at the base of Se2 for the V region. Also, H10 and H43 are at the base of Ec1b in the V region but in Ec1a in the C regions; it is interesting that these two sequences cluster together in the V and C1 regions but not in the C2 region.
Application of the Stephens test for nonrandom clustering of polymorphic nucleotide sites (37) revealed many intragenic recombination events in E. coli fliC genes (for polymorphic sites see Supplementary Table 8 at http://www.mmb.usyd.edu.au/archives/). For example, regions from positions 465 to 537 of H14, H31, and H28 are almost identical and are distinguished from the other Ec1a sequences at 11 to 13 sites. The overall picture is one of recombination between and within the major groups but in general one of stability of the major groups, with the C1, V, and C2 regions of most fliC genes in the same major group.
Mutations under selection pressure as a source of variation in the central region.
Joys (10) suggested that random mutations accumulated in the central region of the fliC region due to absence of functional constraint. The alternative view is that selection pressure and lateral gene transfer provide the source of variation (13, 29). It is difficult to obtain from sequence data alone evidence for the V region being free of functional constraint. In bacteria the frequency of KA rarely exceeds that of KS, which would give clear evidence for selection, but a decrease in KS/KA is usually taken as evidence for change due to selection, although a decrease could arise from a lower level of functional constraint or from counterselection. The genes studied in general encode proteins of known function; hence, relative lack of constraints is not considered an explanation for cases of a lower-than-usual value for KS/KA, because function has to be maintained. In 1988 the V region was not known to have any specific function, as it codes for the exposed part of the flagellin that is not greatly involved in the interactions that give the flagellum its shape and structural properties. It was on this basis that Joys put forward his proposal that the V region lacks functional constraint.
However, we now know that the V region has a major functional role. Flagellin monomer moves through the central pore of the growing flagellum before being incorporated at its distal end, and it is now known that in flagellin monomer only the V domain is ordered, the other domains being disordered until incorporation into the growing flagellum occurs (8). The V region therefore has the important function of being folded first and keeping the two components of the C domain in proximity such that they can fold together when they contact the tip of the growing flagellum. It has also been shown that a D313Y substitution in the D2 domain affects filament structure and that the nearby residue Asn315 makes a hydrogen bond with Gly133 in D1 (31), showing that D2 does play a role in maintaining integrity of the flagellum.
The V region is also typical of globular protein domains in many respects. It is almost entirely comprised of secondary structures, mostly β structures with one α helix. It has a calculated partial specific volume of 0.79 cm3/g, within the range of 0.75 to 0.83 for four well-documented globular proteins (Materials and Methods), and very close to the normal experimental range of 0.72 to 0.75 (5) (We thank Charles Collyer for this analysis.) The enthalpy and entropy changes from native state to fully folded state (ΔH and ΔS) for flagellin monomer are also within the range for other globular proteins, if applied to only the V domain to allow for the finding that the C domain does not contribute to these changes (8). The active site of an enzyme comprises a relatively small part of the protein and involves only a few residues, the major constraint to amino acid substitution overall being maintenance of tertiary structure. It is this that keeps the thermodynamics of the folded state and the partial specific volume within the typical range, and the fact that the V domain of flagellin is within this range implies comparable constraints on amino acid substitution. The V region is typical of globular protein domains, and we see no reason to treat overall levels of sequence variation in flagellin differently from how those in other proteins are treated.
Since 1988 there has also been direct support for selection contributing to the variation. With knowledge of the clonal structure of bacterial populations has come recognition that surface antigen genes are subject to higher levels of lateral transfer than are most other genes, with the implication that there is selection for the diversity in these genes. We will therefore proceed on the assumption that variation in fliC can be interpreted in the same manner as variation in other genes.
The full set of E. coli fliC genes provides the opportunity to test hypotheses on the source of variation by using either pairs of related sequences or defined groups of sequences. We selected three pairs of closely related fliC genes, H1/H12, H30/H32, and H5/H56, with no indels and DNA identity levels of 97.4, 93.99, and 92.6%, respectively. The KA and KS values for C1, C2, and V regions are presented in Table 2. The atypical value for KA and the KS/KA ratio (<5.4) for the V regions of all three pairs, much lower than those found for housekeeping genes of E. coli and S. enterica, indicate selection pressure for amino acid substitutions. The C1 and C2 regions have a KS/KA ratio of more than 35 for the three pairs (Table 2), consistent with substitutions being due to genetic drift, but one should note that the level of variation here is too low for the values to be very accurate.
We also obtained KA and KS values for C1 and C2 for all pairs of sequences within Ec1, where diversity is high enough to give reliable values for KA and KS but not so high as to be greatly affected by the built-in correction for reverse mutations. The average KS/KA ratios are 23.78 and 26.93 for C1a and C2b (Table 2), close to those for comparisons of E. coli and S. enterica genes, indicating that there is not significant selection pressure for change. However, C1b and C2a of Ec2 and Se1 give higher KA values, and C1b also has a higher KS. We suggest that C1b and C2a, which are adjacent to the V region, have either undergone mutation under selection pressure or were transferred from another species into the E. coli fliC locus together with the V region (to give higher values for both KA and KS). There could be selection for mutational change in these subregions to adapt in some way to changes in the adjacent V region, while cotransfer with the V region from another species could account for the higher KS of C1b.
Potential for molecular typing.
The H antigen has been used for the detection and identification of bacteria in many species (40). In S. enterica and E. coli, the combination of H and O antigens in general defines a clone (6, 23). In this study we showed that 43 of the 53 E. coli H antigens are encoded by genes in the fliC locus and identified specific primers for each allele (Supplementary Table 2 at http://www.mmb.usyd.edu.au/archives/). These primers can be used to develop PCR- or microarray-based methods for identification of E. coli strains to replace serotyping methods.
The genes for 10 H antigens remain to be sequenced. All map at loci other than fliC, and it is unlikely that their inclusion will reduce the ability to find antigen-specific primers. Indeed, one might expect that, as at least three loci are involved, the additional sequences will increase the overall level of diversity. Nevertheless, trials need to be carried out for each primer pair before routine use. The H7 primers were tested by PCR against all 53 H type strains and a panel of H7 strains; it was found that they amplify only DNA from H7 strains (38), confirming that the primers chosen are specific in this case.
Concluding remarks.
The full set of fliC genes of E. coli was sequenced. The data confirm earlier observations of a central highly variable region and flanking conserved regions. Surprisingly, the sequences fell into two divergent groups. Comparison with the S. enterica sequences available showed that only part of each of the two so-called conserved regions is completely free of selection for diversity, but these segments allowed us to show that one group of E. coli fliC genes (Ec2) is consistent with derivation from the fliC gene of the common ancestor of E. coli and S. enterica. Variation in the V region was extremely high and also high in the contiguous parts of the C regions as generally recognized. Comparison of closely related forms revealed lower KS/KA ratios for these regions characteristic of genes under directional mutation pressure, but it was also apparent that recombination was a major contributor to generation of H-antigen polymorphism. Both H and O antigens are on the cell surface and appear to be the major targets of the immune system, which must apply intense selection and contribute to the origin and maintenance of the high level of variation. The variation is thought to allow the various clones of a species to each present a surface that offers a selective advantage in the niche occupied by that clone. It is difficult to demonstrate directly the selective advantage of specific antigenic variants, but we have estimated that a selective advantage of only 0.1% for one antigen over another in a given niche is more than sufficient to maintain different alleles in different clones (28). However, the data presented provide strong indirect support for selection for change of H-antigen specificity.
The morphological variation observed in the 1970s (12a) correlates well with the fliC sequence variation that we observe, but studies on flagellin genes of other species will be needed to determine the extent to which this variation arose within E. coli and reflects long-standing polymorphism or lateral gene transfer.
Acknowledgments
We thank all the people and institutes listed in Materials and Methods for kindly supplying strains and Charles Collyer for help with protein partial specific volumes.
The work was supported by the Australian Research Council.
REFERENCES
- 1.Al Mamun, A. A. M., A. Tominaga, and M. Enomoto. 1997. Cloning and characterization of the region III flagellar operons of the four Shigella subgroups: genetic defects that cause loss of flagella of Shigella boydii and Shigella sonnei. J. Bacteriol. 179:4493-4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey, S. 1994. The CCP4 Suite: programs for protein crystallography. Acta Crystallogr. Sect. D 50:760-763. [DOI] [PubMed] [Google Scholar]
- 3.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 1999. Laboratory methods for the diagnosis of epidemic dysentery and cholera. Centers for Disease Control and Prevention, Atlanta, Ga.
- 5.Creighton, T. E. 1993. Proteins: structures and molecular properties. W. H. Freeman, New York, N.Y.
- 6.Ewing, W. H.1986. Edwards and Ewing's identification of the Enterobacteriaceae. Elsevier Science Publishers, Amsterdam, The Netherlands.
- 7.Gordon, D., C. Abajian, and P. Green. 1998. CONSED—a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 8.Honda, S., H. Uedaira, F. Vonderviszt, S. Kidokoro, and K. Namba. 1999. Folding energetics of a multidomain protein, flagellin. J. Mol. Biol. 293:719-732. [DOI] [PubMed] [Google Scholar]
- 9.Joys, T. M. 1985. The covalent structure of the phase-1 flagellar filament protein of Salmonella typhimurium and its comparison with other flagellins. J. Biol. Chem. 260:15758-15761. [PubMed] [Google Scholar]
- 10.Joys, T. M. 1988. The flagellar filament protein. Can. J. Microbiol. 34:452-458. [DOI] [PubMed] [Google Scholar]
- 11.Kawagishi, I., V. Muller, A. W. Williams, V. M. Irikura, and R. M. Macnab. 1992. Subdivision of flagellar region III of the Escherichia coli and Salmonella typhimurium chromosomes and identification of two additional flagellar genes. J. Gen. Microbiol. 138:1051-1065. [DOI] [PubMed] [Google Scholar]
- 12.Kimura, M. 1980. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 116:111-120. [DOI] [PubMed] [Google Scholar]
- 12a.Lawn, A. M., I. Orskov, and F. Orskov. 1977. Morphological distinction between different H serotypes of Escherichia coli J. Gen. Microbiol. 101:111-119. [DOI] [PubMed] [Google Scholar]
- 13.Li, J., K. Nelson, A. C. McWhorter, T. S. Whittam, and R. K. Selander. 1994. Recombinational basis of serovar diversity in Salmonella enterica. Proc. Natl. Acad. Sci. USA 91:2552-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li, W. H. 1993. Unbiased estimation of the rates of synonymous and nonsynonymous substitution. J. Mol. Evol. 36:96-99. [DOI] [PubMed] [Google Scholar]
- 15.Lior, H. 1994. Classification of Escherichia coli, p. 31-72. In C. L. Gyles (ed.), Escherichia coli in domestic animals and humans. CAB International, Wallingford, United Kingdom.
- 16.Mabeck, C. E., F. Ørskov, and I. Ørskov. 1971. Escherichia coli serotypes and renal involvement in urinary-tract infections. Lancet i:1312-1314. [DOI] [PubMed]
- 17.Macnab, R. M. 1996. Flagella and motility, p. 123-145. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. American Society for Microbiology, Washington, D.C.
- 18.Mytelka, D. S., and M. J. Chamberlin. 1996. Escherichia coli fliAZY operon. J. Bacteriol. 178:24-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nei, M., and J. C. Miller. 1990. A simple method for estimating average number of nucleotide substitutions within and between populations from restriction data. Genetics 125:873-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okazaki, N., S. Matsuo, K. Saito, A. Tominaga, and M. Enomoto. 1993. Conversion of the Salmonella phase 1 fliC gene to the phase 2 gene fljB on the Escherichia coli K-12 chromosome. J. Bacteriol. 175:758-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ørskov, I., F. Ørskov, K. A. Bettelheim, and M. E. Chandler. 1975. Two new Escherichia coli O antigens, O162 and O163, and one new H antigen, H56. Withdrawal of H antigen H50. Acta Pathol. Microbiol. Scand. 83:121-124. [DOI] [PubMed] [Google Scholar]
- 22.Parish, C. R., R. Wistar, and G. L. Ada. 1969. Cleavage of bacterial flagellin with cyanogen bromide: antigenic properties of the protein fragments. Biochem. J. 113:501-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Popoff, M. Y., and L. L. Minor. 1997. Antigenic formulas of the Salmonella serovars, 7th revision. WHO Collaborating Centre for Reference and Research on Salmonella. Institut Pasteur, Paris, France.
- 24.Pupo, G. M., D. K. R. Karaolis, R. Lan, and P. R. Reeves. 1997. Evolutionary relationships among pathogenic and nonpathogenic Escherichia coli strains inferred from multilocus enzyme electrophoresis and mdh sequence studies. Infect. Immun. 65:2685-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pupo, G. M., R. Lan, and P. R. Reeves. 2000. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc. Natl. Acad. Sci. USA 97:10567-10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ratiner, Y. A. 1985. Two genetic arrangements determining flagellar antigen specificities in two diphasic Escherichia coli strains. FEMS Microbiol. Lett. 29:317-323. [Google Scholar]
- 27.Ratiner, Y. A. 1998. New flagellin-specifying genes in some Escherichia coli strains. J. Bacteriol. 180:979-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reeves, P. R. 1992. Variation in O antigens, niche specific selection and bacterial populations. FEMS Microbiol. Lett. 100:509-516. [DOI] [PubMed] [Google Scholar]
- 29.Reid, S. D., R. K. Selander, and T. S. Whittam. 1999. Sequence diversity of flagellin (fliC) alleles in pathogenic Escherichia coli. J. Bacteriol. 181:153-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 31.Samatey, F. A., I. Katsumji, S. Nagashima, F. Vonderviszt, T. Kumasaka, M. Yamamoto, and K. Namba. 2001. Structure of the bacterial flagellar protofilament and implications for a switch in supercoilling. Nature 410:331-337. [DOI] [PubMed] [Google Scholar]
- 32.Sanderson, K. E., A. Hessel, and K. E. Rudd. 1995. Genetic map of Salmonella typhimurium, edition VIII. Microbiol. Rev. 59:241-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schoenhals, G., and C. Whitfield. 1993. Comparative analysis of flagellin sequences from Escherichia coli strains possessing serologically distinct flagellar filaments with a shared complex surface pattern. J. Bacteriol. 175:5395-5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharp, P. M. 1991. Determinants of DNA sequence divergence between Escherichia coli and Salmonella typhimurium: codon usage, map position, and concerted evolution. J. Mol. Evol. 33:23-33. [DOI] [PubMed] [Google Scholar]
- 35.Silverman, M., and M. Simon. 1980. Phase variation: genetic analysis of switching mutants. Cell 19:845-854. [DOI] [PubMed] [Google Scholar]
- 36.Silverman, M., J. Zieg, M. Hilmen, and M. Simon. 1979. Phase variation in Salmonella: genetic analysis of a recombination switch. Proc. Natl. Acad. Sci. USA 76:391-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stephens, J. C. 1985. Statistical methods of DNA sequence analysis: detection of intragenic recombination or gene conversion. Mol. Biol. Evol. 2:539-556. [DOI] [PubMed] [Google Scholar]
- 38.Wang, L., D. Rothemund, H. Curd, and P. R. Reeves. 2000. Sequence diversity of the Escherichia coli H7 fliC genes: implication for a DNA-based typing scheme for E. coli O157:H7. J. Clin. Microbiol. 38:1786-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei, L., and T. M. Joys. 1985. Covalent structure of three phase-1 flagellar filament proteins of Salmonella. J. Mol. Biol. 186:791-803. [DOI] [PubMed] [Google Scholar]
- 40.Winstanley, C., and A. W. Morgan. 1997. The bacterial flagellin gene as a biomarker for detection, population genetics and epidemiological analysis. Microbiology 143:3071-3084. [DOI] [PubMed] [Google Scholar]