Abstract
Although the exact etiology of Alzheimer's disease (AD) is a topic of debate, the consensus is that the accumulation of β-amyloid (Aβ) peptides in the senile plaques is one of the hallmarks of the progression of the disease. The Aβ peptide is formed by the amyloidogenic cleavage of the amyloid precursor protein (APP) by β- and γ-secretases. The endocytic system has been implicated in the cleavages leading to the formation of Aβ. However, the identity of the intracellular compartment where the amyloidogenic secretases cleave and the mechanism by which the intracellularly generated Aβ is released into the extracellular milieu are not clear. Here, we show that β-cleavage occurs in early endosomes followed by routing of Aβ to multivesicular bodies (MVBs) in HeLa and N2a cells. Subsequently, a minute fraction of Aβ peptides can be secreted from the cells in association with exosomes, intraluminal vesicles of MVBs that are released into the extracellular space as a result of fusion of MVBs with the plasma membrane. Exosomal proteins were found to accumulate in the plaques of AD patient brains, suggesting a role in the pathogenesis of AD.
Keywords: multivesicular bodies, rafts, amyloid precursor protein, β-secretase, endocytosis
Alzheimer's disease (AD) is a late-onset neurological disorder with progressive loss of memory and cognitive abilities as a result of excessive neurodegeneration (1). AD is characterized by extracellular aggregates of β-amyloid (Aβ) peptides known as amyloid plaques (2). The Aβ peptide is derived from the sequential processing of the amyloid precursor protein (APP) by β- and γ-secretases. β-secretase [(β-APP cleaving enzyme (BACE)] is a type-1 transmembrane aspartyl protease and is mainly localized to endosomes, lysosomes and the trans-Golgi network (3). γ-Secretase is a multicomponent complex that is composed of presenilin-1/presenilin-2, nicastrin, Aph-1, and PEN-2 (4) and is localized to the early secretory (5, 6) and the endocytic compartments (7, 8). Nonamyloidogenic processing of APP involves α-secretase that cleaves APP inside the Aβ region, giving rise to the α-cleaved ectodomain, thus precluding the formation of Aβ (9). Hence, the availability of APP to either α- or β-secretase determines whether Aβ peptide will be generated. Lateral organization of membranes (10) and subcellular localization (11, 12) of the substrate and the secretases have been documented to regulate Aβ generation. Recent work suggests that β-secretase associates with lipid rafts, liquid-ordered domains in the membrane (13, 14), and that integrity of raft domains is required for β-cleavage of APP to occur (ref. 10; see, however, ref. 15). α-Cleavage, in contrast, occurs outside raft domains (10). The γ-secretase complex is also raft-associated (16); hence, amyloidogenic processing of APP could occur in clustered raft domains to generate Aβ (10). Inhibition of endocytosis reduces β-cleavage but not α-cleavage, suggesting that β-cleavage mainly occurs in endosomes (10, 11, 17–19). Accumulation of Aβ peptides in extracellular plaques requires the release of Aβ peptides from the cell. An intriguing question is how the intracellularly generated, fairly hydrophobic Aβ peptide is released into the extracellular space. Here we show that β-cleavage occurs in a specific subset of endosomes and that a fraction of Aβ peptides is found in multivesicular bodies (MVBs) and is released in association with exosomes. Upon fusion of MVBs with the plasma membrane, the intraluminal vesicles of MVBs are released into the extracellular milieu as exosomes. These vesicles are enriched in raft lipids and proteins and are implicated in various functions, such as scavenging of archaic proteins, signaling, and transmission of pathogens (20). In this study, we also show that an exosome-associated protein, Alix, is specifically enriched in amyloid plaques of AD brain sections, suggesting a novel role for exosomes in AD pathogenesis.
Results
β-Cleavage of APP Occurs in Early Endosomes.
To identify the intracellular compartment within the endocytic pathway where β-cleavage occurs, we performed immunofluorescence experiments in HeLa cells expressing the Swedish mutant of APP (swAPP) (2) by using an antibody that specifically recognizes the β-cleaved ectodomain of swAPP (sAPPβ) (Fig. 6, which is published as supporting information on the PNAS web site) along with markers for different endosome populations (Fig. 1b). Colocalization of sAPPβ was observed with the early endosomal markers, rab5 (47% of colocalization) and early endosomal antigen-1 (EEA-1), suggesting early endosome to be a station of β-cleavage (Fig. 1 a and b). Colocalization of sAPPβ was also observed with the late endosomal marker, rab7 (58% of colocalization), but little colocalization was observed with rab11 (29% of colocalization) (Fig. 1b) or with transferrin, 20 min after internalization (data not shown). Similar results were also obtained with the neuroblastoma cells, N2a (Fig. 7, which is published as supporting information on the PNAS web site).
Fig. 1.
Involvement of early endosomes in β-cleavage of APP. (a) HeLa cells expressing swAPP were stained for endogenous early endosomal antigen-1 (EEA-1) (green) and the β-cleaved ectodomain (sAPPβ) (red). (b) HeLa cells expressing swAPP were transfected with rab5, rab7, dominant active rab5 mutant (Q79L), or rab11 GFP fusion proteins fixed and stained for the β-cleaved ectodomain (sAPPβ) (red) with ANJJ antibody. (c) HeLa cells expressing the rabGFP constructs, APP, and BACE were crosslinked at 4°C with anti-APP (red) and anti-BACE (blue) antibodies and endocytosed at 37°C. After 5 min, APP and BACE are internalized to rab5–GFP-positive endosomes. For a detailed version, see Fig. 9.
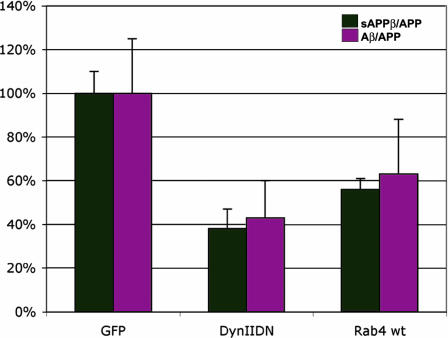
Because the β-cleaved ectodomain was found both in early and late endosomes under these steady-state conditions, it is possible that β-cleavage either occurs in early endosomes, and the cleaved ectodomain is then transported to late endosomes or that the cleavage occurs both in early and late endosomes. By overexpressing the GTPase mutant of rab5 (rab5Q79L), which inhibits cargo flow from early to late endosomes (21), we could freeze almost all of the cellular β-cleaved ectodomain in these enlarged endosomes consistent with the idea that β-cleavage occurs either in or upstream of early endosomes (Fig. 1b). Overexpression of this dominant active version of rab5, however, neither enhanced β-cleavage nor increased the cellular levels of β-cleaved ectodomain (Fig. 8, which is published as supporting information on the PNAS web site), suggesting that the secretion of sAPPβ was not affected under these conditions. The involvement of early endosomes in β-cleavage was confirmed by induced endocytosis experiments in which APP and BACE were crosslinked with antibodies at the cell surface and internalized. After 5 min of internalization, APP and BACE colocalized in rab5-positive early endosomes (Fig. 1c; see also Fig. 9, which is published as supporting information on the PNAS web site). Fluorescence resonance energy transfer measurements also confirmed that maximum intermolecular interaction between APP and BACE occurred in early endosomes [our unpublished results and ref. 22). Interestingly, expression of the dominant negative mutant of rab7 did not affect β-cleavage and Aβ secretion (data not shown), but expression of the dominant negative mutant of dynamin inhibited β-cleavage and Aβ secretion by ≈60%. The absence of a complete inhibition in β-cleavage of swAPP under endocytosis-inhibited conditions could be explained by the observations that swAPP also could undergo β-cleavage during its biosynthetic transport (12, 23). Stimulation of the recycling of cargo from the early endosome back to the plasma membrane by overexpression of wild-type rab4 (24) decreased β-cleavage and Aβ secretion (Fig. 2), showing that reducing the residence time of APP and/or BACE in early endosomes reduces β-cleavage, suggesting early endosomes to be a major site of β-cleavage.
Fig. 2.
The effect of endocytic effectors on β-cleavage and Aβ secretion. N2a cells expressing CFP-swAPP and the dynaminK44A mutant (DynIIDN) or rab4wt constructs were assayed for sAPPβ, Aβ, and full-length APP as described in Materials and Methods. The y axis represents the amount of sAPPβ as a fraction of full-length APP (sAPPβ/APP) (black) or Aβ as a fraction of full-length APP (Aβ/APP) (magenta). The GFP control was normalized to 100%.
A Fraction of Aβ Peptides Is Localized to MVBs and Is Released in Association with Exosomes.
Most of the extracellular Aβ is known to be soluble (23, 25, 26), but we were interested in knowing whether a fraction of the extracellular Aβ remained membrane-bound as Aβ is generated in the membrane (27, 28). Therefore, we performed differential centrifugation of the culture supernatants and analyzed for Aβ peptides in the pelleted membrane fractions. Indeed, a minute fraction pelleted with membranes (Fig. 10, which is published as supporting information on the PNAS web site). Moreover, immunogold labeling of N2a cells with an anti-Aβ antibody showed that Aβ peptides also were localized to MVBs (Fig. 3), consistent with previous reports (29, 30). Proteins that are destined for lysosomal degradation are sorted from the early endosomes to intraluminal vesicles of the MVBs or late endosomes. These multivesicular endosomes can then either fuse with lysosomes and degrade the cargo or fuse with the plasma membrane to release these vesicles as exosomes (31).
Fig. 3.
Aβ peptides are localized in MVBs in N2a cells. Immunoelectron microscopy on anti-Aβ-stained cryosections of N2a cells shows that Aβ localizes to MVBs.
To test whether Aβ could be released in association with exosomes, we isolated exosomes from culture supernatants of N2a cells stably transfected with swAPP through differential centrifugation and density gradient centrifugation as described in Materials and Methods (32, 33). Alix and flotillin-1 were used as marker proteins (34, 35) to identify exosome-containing fractions (Fig. 4a). These fractions were devoid of nonexosomal proteins, such as transferrin receptor and full-length APP (Fig. 4a). Only Aβ peptides but none of the other APP-derived fragments were detected in the exosomal fractions by Western blotting using 6E10 antibody (Figs. 4a and 6a).
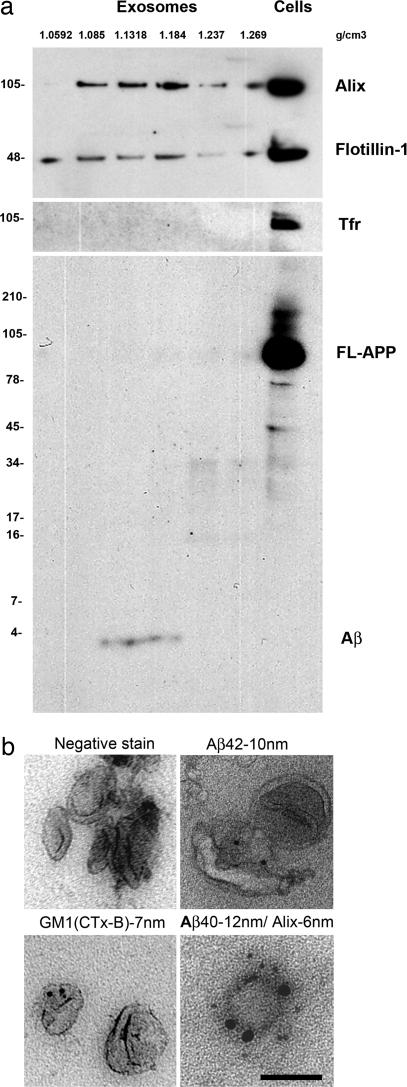
Fig. 4.
Exosomes released from N2a cells contain Aβ peptides. (a) Sucrose gradient fractions of an exosomal preparation from N2a-swAPP cell culture supernatants were immunoblotted with several antibodies. Alix and flotillin-1 mark the exosome-positive fractions (fractions 2–4). Transferrin receptor is excluded from the exosomal fractions and could be detected only in the whole-cell lysates (Cells). Immunoblotting with 6E10 that recognizes the Aβ peptides, full-length APP (FL-APP), β-cleaved C-terminal fragment, and α-cleaved ectodomain reveals that only Aβ peptides fractionate in the exosomal fractions and that full-length APP and other fragments are excluded from the exosomes and could be detected only in the cell lysate (Cells). Note that, in N2a cells, Aβ peptides are efficiently secreted into the medium and, hence, are not detectable at the concentrations of the cell lysates used for blotting. Molecular masses in kilodaltons are indicated to the left of the blots. The densities of the fractions as measured with a refractometer are indicated by the values labeled g/cm3. (b) Exosomes from fractions 3 and 4 of the sucrose gradient were negatively stained with 1% uranyl acetate and immunolabeled with antibodies for the exosomal marker Alix. Exosomes also were immunolabeled for Aβ40 or Aβ42 and cholera toxin B subunit (CTx-B), which binds to the ganglioside GM1.
Electron microscopy of exosome-enriched fractions showed that these vesicles were typically 60–100 nm in size (Fig. 4b). To confirm that Aβ peptides were released in association with exosomes, we performed double immunogold-labeling of these vesicles with specific antibodies against Aβ42/Aβ40 and the exosome markers, Alix or flotillin-1. In agreement with our biochemical data, Aβ peptides were indeed found on the Alix (Fig. 4b) or flotillin-1-positive vesicles (data not shown). The ganglioside GM1 has been shown to be present in exosomal vesicles from prion protein-expressing epithelial and neuroglial cells (35). Cholera-toxin-crosslinked GM1 is found in raft clusters (14). Immunogold labeling with the B subunit of cholera toxin confirmed that these exosomal vesicles contained GM1 (Fig. 4b). These results show that Aβ40 and Aβ42 could be secreted by an exosomal pathway in neuroblastoma cells.
Exosomal Proteins Are Enriched in Amyloid Plaques.
To investigate whether these findings have any bearing on Alzheimer pathology, we performed immunohistochemistry analysis with antibodies against exosomal proteins on autopsied brain sections from three AD patients, two Parkinson's disease patients, and two age-matched control subjects. Enrichment of Alix, a marker for exosomes, was seen around the small neuritic plaques, and a moderate signal was detected in large diffuse plaques (Fig. 5) in brain sections from all AD patients tested. Alix staining was largely absent in brain sections of control subjects and of Parkinson's disease patients. Interestingly, previous work has shown that another exosomal protein, flotillin-1, is associated with part of Aβ within the senile plaques of the Tg2756 mouse brain (36) and suggested flotillin-1 to be an indicator of the progression of the pathology (37). Our results now provide a plausible mechanistic explanation for these findings. Accumulation and enrichment of exosomal proteins around amyloid plaques in AD patients are in agreement with our findings that Aβ peptides can be released complexed with exosomes from MVBs. These findings suggest that exosome-associated Aβ could be involved in plaque formation.
Fig. 5.
Exosomal proteins are enriched in human amyloid plaques. Paraffin-embedded human autopsy tissues from patients with AD or Parkinson's disease or from controls were studied. We performed either Gallyas silver stain and subsequent immunochemistry for Alix on hippocampal sections (d–f) or immunochemistry with hematoxylin counterstain only (a–c). Alix was stained by indirect alkaline phosphatase method with new fuchsin as the chromogen. (Magnification, ×63.) (a and d) Hippocampus of a normal control brain without amyloid plaques or detection of Alix. (b and e) Hippocampus of a patient with Parkinson's disease displaying slight accumulation of pathological tau-filaments but no amyloid plaques and no Alix. (c and f) Hippocampus of a patient with moderate to advanced Alzheimer's pathology [CERAD (Consortium to Establish a Registry for Alzheimer's Disease) score B] with discrete deposits of Alix within typical neuritic plaques (f) containing amyloid, hyperphosphorylated tau-fibrils, and cellular elements (astrocytes and microglia).
Discussion
The data of the present study substantiate that the endocytic pathway is a major site for Aβ generation. β-cleavage occurs in early endosomes, and γ-cleavage can take place there (38). How the bulk of Aβ is released from its membrane-bound state into the extracellular space is currently not understood. We show here that a minute fraction (<1%) of Aβ is secreted into the extracellular medium bound to exosomes. Because only a minor component of Aβ is associated with exosomes, it raises a concern about whether extracellular soluble Aβ simply “sticks” to the extracellular exosomes or a small fraction of Aβ peptides are indeed released via exosomes. Several observations support the idea that some of the Aβ is released via exosomes: (i) Aβ peptides are seen in MVBs (Fig. 3), and these MVBs fuse with the plasma membrane and release the intraluminal vesicles as exosomes that bear Aβ. (ii) After several stringent washes during the cryoelectron microscopy specimen preparation, Aβ was still seen on the exosomes, which supports that Aβ must be inserted in the membrane. However, it is important to note that additional Aβ also could be carried by multivesicular endosomes and be released in a non-membrane-bound form after fusion with the plasma membrane. Initial reports on characterization of extracellular Aβ used immunoprecipitation and Western blotting analysis and, hence, did not identify the membrane bound, extracellular Aβ (26). Amyloidogenic processing of APP also occurs in the biosynthetic route, and this pool of Aβ could be a component of the nonexosomal release (5). Macroautophagy, a process by which both the biosynthetic and endocytic organelles are degraded, has been recently shown to be involved in Aβ generation (39). Aβ generated by this process also could contribute to the exosomal fraction of Aβ via fusion with late endosomes (39).
Whether exosome-associated Aβ could play a role in the pathogenesis of AD is yet to be studied. The observations that exosome-associated proteins, such as Alix and flotillins (data not shown and ref. 37), are enriched in the amyloid plaques suggest that exosome-associated Aβ may be involved in plaque formation. We have recently shown that raft lipids (neutral sphingolipids and cholesterol) activate β-secretase in proteoliposomes (40). Because exosomes are enriched in glycolipids, raft lipids, and raft-associated proteins, such as glycosylphosphatidylinositol-anchored proteins (34, 35), raft-bound Aβ could be incorporated into exosomes. Aβ-associated GM1, a ganglioside, has been shown to act as an amyloid seed for Aβ fibrillation (41), and the presence of GM1 and other raft lipids (42) in exosomes bolsters the idea that exosomes could act as nucleation centers for plaque formation. Another amyloid fibril-forming protein, Pmel1, involved in melanosome biogenesis and melanin polymerization (43) has been shown to be routed to MVBs and to be released via exosomes (44). These studies hypothesized that the intraluminal vesicles of MVBs or exosomes could provide a conducive environment for amyloidogenic fibril formation (43). Because AD is a slow and progressive disorder, the release of Aβ associated with exosomes, although minute, could still be involved in the progression of the disease. Aβ could also hitch on exosomes to mediate its intercellular transfer (45). HIV uses the exosome pathway for its assembly and release (46), and prion proteins mediate their intercellular transfer via exosomes (35). Exosomes are thus emerging as Trojan horses in pathogenesis. Further work will be required to assess the significance of the exosome route for the extracellular release of both soluble and membrane-bound Aβ.
Materials and Methods
Constructs and Generation of Recombinant Adenoviruses.
The cyan fluorescent protein (CFP)-swAPP construct was similar to the yellow fluorescent protein-APP construct described previously (10). The BACE1A–vesicular stomatitis virus glycoprotein construct also was described previously (10). Plasmids encoding rab proteins fused to GFP were obtained from Marino Zerial (Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany).
Cells.
Mouse neuroblastoma N2a cells and HeLa cells were cultured at 37°C in DMEM supplemented with 10% FCS/100 units/ml penicillin/100 μg/ml streptomycin. Gang Yu (University of Texas Southwestern Medical Center, Dallas) kindly provided HeLa cells stably expressing swAPP. N2a cells stably expressing the wild-type APP (N2a-wtAPP) and the Swedish mutant (N2a-swAPP) were kind gifts from Gopal Thinakaran (University of Chicago, Chicago) and were cultured as described in ref. 23. Adenovirus expressing either CFP-swAPP or swAPP-CFP and BACE–vesicular stomatitis virus glycoprotein were used to infect both N2a and HeLa cells. In some cases, plasmids encoding for CFP-swAPP or swAPP-CFP were also used.
Antibodies.
Rabbit polyclonal antibody 7523 against the N-terminal end of BACE1 was a kind gift from Christian Haass (Ludwig-Maximilians-University, Munich, Germany) (47). Mouse monoclonal antibodies against Alix/AIP1, Flotillin-2, Flotillin-1, and Transferrin receptor were from BD Biosciences (Heidelberg, Germany). The fluorophore-conjugated (Cy2, Cy3, Cy5, or FITC) secondary antibodies were purchased from The Jackson Laboratory. Monoclonal antibodies against Aβ40 and Aβ42 (G2–10 and G2–13, respectively) were from The Genetics Company (Zurich, Switzerland). Monoclonal antibodies 6E10, which recognizes the 1–12 region of Aβ peptide, and 4G8, which is directed against the 17–24 region of Aβ, were purchased from Biocat (Heidelberg, Germany). Rabbit polyclonal antibodies IP60 (10), which is directed against the C terminus of APP, 70JE (10), which is directed against amino acids 1–11 of Aβ, and ANJJ, which is directed against the C terminus of the sAPPβ, were developed in our laboratory. ANJJ recognizes sAPPβ of swAPP but no other APP-processing products.
Reagents.
Alexa Fluor 546-conjugated Transferrin and the unconjugated holotransferrin were purchased from Molecular Probes. Cholera toxin B and other routinely used chemicals were purchased from Sigma. All reagents for the Meso Scale Discovery (MSD) electrochemiluminesence assays were purchased from Meso Scale Discovery (Gaithersburg, MD).
Transfection, Viral Infection, and Biochemical Assays.
At 24 h after the seeding of N2a cells into 3.5-cm dishes, transient transfections were performed with 1–3 μg of each expression plasmid either using calcium phosphate precipitation as described by Chen and Okayama (48) or Lipofectamine 2000 (Invitrogen). Additionally, at 24 h after seeding, the N2a cells were infected with recombinant adenoviruses for 1 h at 37°C in complete medium. After a change of medium, the cells were incubated for 16–20 h at 37°C and then used for biochemical assays.
Immunoprecipitation and Quantification.
After metabolic labeling for 1 h, the cell culture medium was collected, and cell extracts were prepared in PBS containing 2% Nonidet P-40, 0.2% SDS, and chymostatin, leupeptin, antipain, and pepstatin A each at 25 μg/ml. Immunoprecipitates were recovered on protein A-Sepharose CL4B beads (Amersham Pharmacia Biosciences) preincubated with antibodies IP60, ANJJ, and 70JE, respectively, and separated on 10–20% Tris-tricine (Invitrogen) gels. PhosphorImager plates were exposed to the fixed and dried gels, and bands were quantified by using the Fuji BAS 1800II image plate reader and Science Lab 99 image gauge 3.3 software (Raytest Isotopenmessgeraete, Straubenhardt, Germany).
Immunofluorescence Microscopy.
Coverslip-grown cells were transfected with various rab-GFP constructs and infected with adenoviruses expressing the swAPP construct. In the case of N2a cells, cells were grown on polylysine/laminin-coated coverslips. After 6–8 h of transfection/infection, the cells were fixed with 3.7% paraformaldehyde, washed with ammonium chloride, permeabilized with methanol at 20°C for 5 min, washed with PBS, and blocked for 1 h with 0.2%BSA/0.2% fish skin gelatin in PBS (blocking buffer). Cells were then incubated with primary antibodies in blocking buffer for 1 h, subsequently washed thoroughly with PBS, and the primary signal was detected with various fluorochrome-conjugated (FITC, Cy2, Cy3, or Cy5) anti-mouse or anti-rabbit antibodies.
Antibody Crosslinking and Induced Endocytosis.
Coverslip-grown cells were transfected with either rab-GFP or control GFP constructs and infected with adenoviruses expressing swAPP and BACE. After 6–8 h of transfection/infection, cells were washed twice in CO2-independent medium containing 2% BSA (BCM). Cells were then incubated in the cold (4°C) with anti-APP (6E10) and anti-BACE (7523) in BCM for 1 h and then washed and further incubated with donkey anti-mouse Cy3 and donkey anti-rabbit Cy5 antibodies in BCM. The cells were then washed and incubated at 37°C for 5, 10, and 20 min. After these times, the cells were immediately fixed in paraformaldehyde and processed as described in Immunofluorescence Microscopy.
Preparation of Exosome-Depleted Medium.
The medium was depleted of exosomes from bovine serum by essentially following an established protocol (35). DMEM/glutamine containing 20% FCS and penicillin/streptomycin (Invitrogen) was centrifuged overnight at 4°C and 100,000 × g using a Ti45 rotor in a Beckmann ultracentrifuge. The supernatant was carefully removed with a pipette and passed through a vacuum-connected 0.22-μm filter. This medium was then added to an equal volume of DMEM/glutamine containing penicillin/streptomycin and used for the purification of exosomes as described below.
Purification of Exosomes.
Exosomes from untransfected or APP stably transfected N2a cells were prepared as described (35). Briefly, cells from 40–60 T175 flasks were cultured in DMEM with 10% FCS. A day before the exosome preparation, culture medium was replaced with exosome-depleted medium. Culture supernatants of cells grown for 24 h were collected and spun at 300 × g for 10 min to remove cells. The supernatants were then sequentially centrifuged at 1000 × g, 10,000 × g, and 100,000 × g. The 100,000 × g pellet was resuspended in 2.5 M sucrose in 20 mM Hepes (pH 7.4), and a step gradient of sucrose (2.25, 2.0, 1.75, 1.5, 1.25, 1.0, 0.75, 0.5, and 0.25 M) was layered over the exosome-containing, 2.5 M sucrose solution. The gradient was spun at 200,000 × g for at least 16 h using an SW55 rotor. Fractions were collected from the top of the gradient, diluted with PBS, and spun at 100,000 × g with a TLA-100.3 rotor. The pelleted fractions were then used either for immunoblotting or for electron microscopy.
Immunoelectron Microscopy and MSD Electrochemiluminescence Assays.
The isolated exosome fractions (fractions 3 and 4 from the gradient) and the cells were immunolabeled as described in refs. 35 and 49. MSD assays for Aβ1–40, sAPPα, sAPPβ, and full-length APP were performed on a SECTOR Imager 6000 (Meso Scale Discovery). This platform is based on MSD's MultiArray technology, which is a proprietary combination of patterned arrays and electrochemiluminescence detection enabling large numbers of measurements with exceptional sensitivity, wide dynamic range, and convenience. Individual wells of a High-Bind MultiArray 384-well-plate were coated with capture antibodies for Aβ1–40 (6E10), sAPPα (6E10), sAPPβ (ANJJ), and full-length APP (IP60). After a blocking step with 3% MSD Blocker A in MSD Tris wash buffer, 25-μl aliquots of the cell lysate, the 100,000 × g supernatant, and the 100,000 × g pellet were added to individual wells for 1 h at room temperature with shaking. After four washes with 40 μl of MSD Tris wash buffer, the SULFO-TAG-labeled detection antibodies were added in 25 μl of 1% MSD Blocker A in MSD Tris wash buffer. They were prepared by incubating antibody G2–10 (for Aβ1–40) or an antibody binding close to the N terminus of APP (for sAPPα, sAPPβ, and full-length APP) with SULFO-TAG-NHS-ester [ruthenium(II) tris-bipyridine, N-hydroxysuccinimide; Meso Scale Discovery] as suggested by the manufacturer. Note that the latter antibody binds human, but not mouse, APP. After an incubation for 1 h at room temperature with shaking, the wells were washed again, 35 μl of MSD read buffer T with surfactant was added, and the plates were read immediately.
Histology and Immunochemistry on Human Autopsy Tissues.
Sections (5 μm thick) of formalin-fixed and paraffin-embedded autopsy tissues of the left hippocampus (obtained from the Department of Pathology, University Clinic, Technical University, Dresden) were deparaffinized and then received Gallyas silver stain (reagents from Merck, Darmstadt, Germany) (50) and subsequent immunochemistry for Alix or immunochemistry only. The slides were first heated in a vapor cooking apparatus (Multigourmet; Braun, Kronberg, Germany) for 20 min in 10 mM sodium citrate with 0.01% Tween, pH 6.5. The primary antibody was applied at a dilution of 1/100 overnight at 4°C. Development followed with the use of the ABC indirect alkaline phosphatase kit mouse IgG (Vector Laboratories, Burlingame, CA) and a fuchsin derivative (new fuchsin; Sigma) as a chromogen (51).
Supplementary Material
Acknowledgments
We thank Marino Zerial, Christian Haass, Bart de Strooper, Graça Raposo, Mikael Simons, Uwe Koenietzko, and the members of the Abnormal Proteins in the Pathogenesis of Neurodegenerative Disorders Consortium for critical input to the study; Aki Manninen for critical comments on the manuscript; Benoit Fevrier, Aude de Gassart, and Uenal Coskun for advice on gradient centrifugation; Dr. Gopal Thinakaran and Prof. Gang Yu for providing APP stable cell lines; and Jana Mantler for expert technical support. L.R. thanks Stephanie Dienel, Vineeth Surendranath, Aparna Katoch, Indrani Sarkar, Jonas Ries, and Manonmani Arunachalam for help with cell culture. K.D.G. thanks Marcus Domula and Brigitte Hamaan for technical assistance and the Alzheimer Research Foundation of Germany for funding. This work was supported by Abnormal Proteins in the Pathogenesis of Neurodegenerative Disorders Grant LSHM-CT-2003-503330 and Deutsche Forschungs Gemeinschaft Transregio Grant DFG SFB TR13-04 SFB-TR.
Abbreviations
- Aβ
β-amyloid
- AD
Alzheimer's disease
- APP
amyloid precursor protein
- swAPP
Swedish APP
- sAPPβ
β-cleaved ectodomain of swAPP
- BACE
Aβ cleaving enzyme
- MVBs
multivesicular bodies
- CFP
cyan fluorescent protein
- MSD
Meso Scale Discovery
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Ross C. A., Poirier M. A. Nat. Med. 2004;10:S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 2.Selkoe D. J. Physiol. Rev. 2001;81:741–766. doi: 10.1152/physrev.2001.81.2.741. [DOI] [PubMed] [Google Scholar]
- 3.Capell A., Steiner H., Willem M., Kaiser H., Meyer C., Walter J., Lammich S., Multhaup G., Haass C. J. Biol. Chem. 2000;275:30849–30854. doi: 10.1074/jbc.M003202200. [DOI] [PubMed] [Google Scholar]
- 4.Edbauer D., Winkler E., Regula J. T., Pesold B., Steiner H., Haass C. Nat. Cell Biol. 2003;5:486–488. doi: 10.1038/ncb960. [DOI] [PubMed] [Google Scholar]
- 5.Hartmann T., Bieger S. C., Bruhl B., Tienari P. J., Ida N., Allsop D., Roberts G. W., Masters C. L., Dotti C. G., Unsicker K., Beyreuther K. Nat. Med. 1997;3:1016–1020. doi: 10.1038/nm0997-1016. [DOI] [PubMed] [Google Scholar]
- 6.Capell A., Beher D., Prokop S., Steiner H., Kaether C., Shearman M. S., Haass C. J. Biol. Chem. 2005;280:6471–6478. doi: 10.1074/jbc.M409106200. [DOI] [PubMed] [Google Scholar]
- 7.Runz H., Rietdorf J., Tomic I., de Bernard M., Beyreuther K., Pepperkok R., Hartmann T. J. Neurosci. 2002;22:1679–1689. doi: 10.1523/JNEUROSCI.22-05-01679.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rechards M., Xia W., Oorschot V. M., Selkoe D. J., Klumperman J. Traffic. 2003;4:553–565. doi: 10.1034/j.1600-0854.2003.t01-1-00114.x. [DOI] [PubMed] [Google Scholar]
- 9.Kojro E., Gimpl G., Lammich S., Marz W., Fahrenholz F. Proc. Natl. Acad. Sci. USA. 2001;98:5815–5820. doi: 10.1073/pnas.081612998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ehehalt R., Keller P., Haass C., Thiele C., Simons K. J. Cell Biol. 2003;160:113–123. doi: 10.1083/jcb.200207113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haass C., Koo E. H., Mellon A., Hung A. Y., Selkoe D. J. Nature. 1992;357:500–503. doi: 10.1038/357500a0. [DOI] [PubMed] [Google Scholar]
- 12.Haass C., Lemere C. A., Capell A., Citron M., Seubert P., Schenk D., Lannfelt L., Selkoe D. J. Nat. Med. 1995;1:1291–1296. doi: 10.1038/nm1295-1291. [DOI] [PubMed] [Google Scholar]
- 13.Simons K., Vaz W. L. Annu. Rev. Biophys. Biomol. Struct. 2004;33:269–295. doi: 10.1146/annurev.biophys.32.110601.141803. [DOI] [PubMed] [Google Scholar]
- 14.Rajendran L., Simons K. J. Cell Sci. 2005;118:1099–1102. doi: 10.1242/jcs.01681. [DOI] [PubMed] [Google Scholar]
- 15.Abad-Rodriguez J., Ledesma M. D., Craessaerts K., Perga S., Medina M., Delacourte A., Dingwall C., De Strooper B., Dotti C. G. J. Cell Biol. 2004;167:953–960. doi: 10.1083/jcb.200404149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vetrivel K. S., Cheng H., Lin W., Sakurai T., Li T., Nukina N., Wong P. C., Xu H., Thinakaran G. J. Biol. Chem. 2004;279:44945–44954. doi: 10.1074/jbc.M407986200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Selkoe D. J., Yamazaki T., Citron M., Podlisny M. B., Koo E. H., Teplow D. B., Haass C. Ann. N.Y. Acad. Sci. 1996;777:57–64. doi: 10.1111/j.1749-6632.1996.tb34401.x. [DOI] [PubMed] [Google Scholar]
- 18.Grbovic O. M., Mathews P. M., Jiang Y., Schmidt S. D., Dinakar R., Summers-Terio N. B., Ceresa B. P., Nixon R. A., Cataldo A. M. J. Biol. Chem. 2003;278:31261–31268. doi: 10.1074/jbc.M304122200. [DOI] [PubMed] [Google Scholar]
- 19.Refolo L. M., Sambamurti K., Efthimiopoulos S., Pappolla M. A., Robakis N. K. J. Neurosci. Res. 1995;40:694–706. doi: 10.1002/jnr.490400515. [DOI] [PubMed] [Google Scholar]
- 20.Fevrier B., Raposo G. Curr. Opin. Cell Biol. 2004;16:415–421. doi: 10.1016/j.ceb.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 21.Rink J., Ghigo E., Kalaidzidis Y., Zerial M. Cell. 2005;122:735–749. doi: 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
- 22.Kinoshita A., Fukumoto H., Shah T., Whelan C. M., Irizarry M. C., Hyman B. T. J. Cell Sci. 2003;116:3339–3346. doi: 10.1242/jcs.00643. [DOI] [PubMed] [Google Scholar]
- 23.Thinakaran G., Teplow D. B., Siman R., Greenberg B., Sisodia S. S. J. Biol. Chem. 1996;271:9390–9397. doi: 10.1074/jbc.271.16.9390. [DOI] [PubMed] [Google Scholar]
- 24.de Renzis S., Sonnichsen B., Zerial M. Nat. Cell Biol. 2002;4:124–133. doi: 10.1038/ncb744. [DOI] [PubMed] [Google Scholar]
- 25.Seubert P., Vigo-Pelfrey C., Esch F., Lee M., Dovey H., Davis D., Sinha S., Schlossmacher M., Whaley J., Swindlehurst C., et al. Nature. 1992;359:325–327. doi: 10.1038/359325a0. [DOI] [PubMed] [Google Scholar]
- 26.Haass C., Schlossmacher M. G., Hung A. Y., Vigo-Pelfrey C., Mellon A., Ostaszewski B. L., Lieberburg I., Koo E. H., Schenk D., Teplow D. B., et al. Nature. 1992;359:322–325. doi: 10.1038/359322a0. [DOI] [PubMed] [Google Scholar]
- 27.Haass C., De Strooper B. Science. 1999;286:916–919. doi: 10.1126/science.286.5441.916. [DOI] [PubMed] [Google Scholar]
- 28.Hartmann T. Trends Neurosci. 2001;24:S45–S48. doi: 10.1016/s0166-2236(00)01990-1. [DOI] [PubMed] [Google Scholar]
- 29.Langui D., Girardot N., El Hachimi K. H., Allinquant B., Blanchard V., Pradier L., Duyckaerts C. Am. J. Pathol. 2004;165:1465–1477. doi: 10.1016/s0002-9440(10)63405-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takahashi R. H., Milner T. A., Li F., Nam E. E., Edgar M. A., Yamaguchi H., Beal M. F., Xu H., Greengard P., Gouras G. K. Am. J. Pathol. 2002;161:1869–1879. doi: 10.1016/s0002-9440(10)64463-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pelchen-Matthews A., Raposo G., Marsh M. Trends Microbiol. 2004;12:310–316. doi: 10.1016/j.tim.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 32.Raposo G., Nijman H. W., Stoorvogel W., Liejendekker R., Harding C. V., Melief C. J., Geuze H. J. J. Exp. Med. 1996;183:1161–1172. doi: 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zitvogel L., Regnault A., Lozier A., Wolfers J., Flament C., Tenza D., Ricciardi-Castagnoli P., Raposo G., Amigorena S. Nat. Med. 1998;4:594–600. doi: 10.1038/nm0598-594. [DOI] [PubMed] [Google Scholar]
- 34.Thery C., Boussac M., Veron P., Ricciardi-Castagnoli P., Raposo G., Garin J., Amigorena S. J. Immunol. 2001;166:7309–7318. doi: 10.4049/jimmunol.166.12.7309. [DOI] [PubMed] [Google Scholar]
- 35.Fevrier B., Vilette D., Archer F., Loew D., Faigle W., Vidal M., Laude H., Raposo G. Proc. Natl. Acad. Sci. USA. 2004;101:9683–9688. doi: 10.1073/pnas.0308413101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kokubo H., Saido T. C., Iwata N., Helms J. B., Shinohara R., Yamaguchi H. Neurobiol. Aging. 2005;26:409–418. doi: 10.1016/j.neurobiolaging.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 37.Kokubo H., Lemere C. A., Yamaguchi H. Neurosci. Lett. 2000;290:93–96. doi: 10.1016/s0304-3940(00)01334-3. [DOI] [PubMed] [Google Scholar]
- 38.Kaether C., Schmitt S., Willem M., Haass C. Traffic. 2006;7:408–415. doi: 10.1111/j.1600-0854.2006.00396.x. [DOI] [PubMed] [Google Scholar]
- 39.Yu W. H., Cuervo A. M., Kumar A., Peterhoff C. M., Schmidt S. D., Lee J. H., Mohan P. S., Mercken M., Farmery M. R., Tjernberg L. O., et al. J. Cell Biol. 2005;171:87–98. doi: 10.1083/jcb.200505082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kalvodova L., Kahya N., Schwille P., Ehehalt R., Verkade P., Drechsel D., Simons K. J. Biol. Chem. 2005;280:36815–36823. doi: 10.1074/jbc.M504484200. [DOI] [PubMed] [Google Scholar]
- 41.Hayashi H., Kimura N., Yamaguchi H., Hasegawa K., Yokoseki T., Shibata M., Yamamoto N., Michikawa M., Yoshikawa Y., Terao K., et al. J. Neurosci. 2004;24:4894–4902. doi: 10.1523/JNEUROSCI.0861-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Gassart A., Geminard C., Hoekstra D., Vidal M. Traffic. 2004;5:896–903. doi: 10.1111/j.1600-0854.2004.00223.x. [DOI] [PubMed] [Google Scholar]
- 43.Theos A. C., Truschel S. T., Raposo G., Marks M. S. Pigment Cell Res. 2005;18:322–336. doi: 10.1111/j.1600-0749.2005.00269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wolfers J., Lozier A., Raposo G., Regnault A., Thery C., Masurier C., Flament C., Pouzieux S., Faure F., Tursz T., et al. Nat. Med. 2001;7:297–303. doi: 10.1038/85438. [DOI] [PubMed] [Google Scholar]
- 45.Morelli A. E., Larregina A. T., Shufesky W. J., Sullivan M. L., Stolz D. B., Papworth G. D., Zahorchak A. F., Logar A. J., Wang Z., Watkins S. C., et al. Blood. 2004;104:3257–3266. doi: 10.1182/blood-2004-03-0824. [DOI] [PubMed] [Google Scholar]
- 46.von Schwedler U. K., Stuchell M., Muller B., Ward D. M., Chung H. Y., Morita E., Wang H. E., Davis T., He G. P., Cimbora D. M., et al. Cell. 2003;114:701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- 47.Capell A., Meyn L., Fluhrer R., Teplow D. B., Walter J., Haass C. J. Biol. Chem. 2002;277:5637–5643. doi: 10.1074/jbc.M109119200. [DOI] [PubMed] [Google Scholar]
- 48.Chen C., Okayama H. Mol. Cell. Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scheiffele P., Verkade P., Fra A. M., Virta H., Simons K., Ikonen E. J. Cell Biol. 1998;140:795–806. doi: 10.1083/jcb.140.4.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gallyas F. Acta Morphol. Acad. Sci. Hung. 1971;19:1–8. [PubMed] [Google Scholar]
- 51.Malik N. J., Daymon M. E. J. Clin. Pathol. 1982;35:1092–1094. doi: 10.1136/jcp.35.10.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.