Abstract
Germ cells in XY male mice establish site-specific methylation on imprinted genes during spermatogenesis, whereas germ cells in XX females establish their imprints in growing oocytes. We showed previously that in vitro, sex-specific methylation patterns of pluripotent stem cell lines derived from germ cells were influenced more by the sex chromosome constitution of the cells themselves than by the gender of the embryo from which they had been derived. To see whether the same situation would prevail in vivo, we have now determined the methylation status of H19 expressed from the maternal allele, and the expression and methylation status of a paternally expressed gene Peg3, in germ cells from sex-reversed and control embryos. For these imprinted genes, we conclude that the female imprint is a response of the germ cells to undergoing oogenesis, rather than to their XX chromosome constitution. Similarly, both our XY and our sex-reversed XX male germ cells clearly showed a male rather than a female pattern of DNA methylation; here, however, the sex chromosome constitution had a significant effect, with XX male germ cells less methylated than the XY controls.
Keywords: mouse, methylation, spermatogenesis, oogenesis, embryonic germ cells
In mouse germ cells, the erasure and reestablishment of imprints at differentially methylated sites involves epigenetic changes that begin even before entry of the germ cells into the genital ridge (1, 2). Maternal and paternal alleles are subsequently marked during gametogenesis by differential DNA methylation. In chromosomally normal XX female mice, different imprinted genes have been shown to acquire differential methylation postnatally, at different stages of oocyte growth (3). However, XY germ cells in male mice acquire differential methylation prenatally, as prospermatogonia, before entry into meiosis (4, 5).
To address the question of whether differential methylation of imprinted genes directly or indirectly reflects the tissue environment of the germ cells, rather than their chromosomal sex (XX versus XY), we initially looked at embryonic germ (EG) cells, where methylation status can readily be ascertained by Southern analysis. Differentially methylated H19 and Igf2 sites were known to be hypermethylated in EG cell lines derived from germ cells in male but not female embryos 11.5–12.5 days post coitum (dpc) (6). This hypermethylation was not due to a failure of imprint erasure (7) but probably represents the precocious establishment of a genomic imprint, because the methylation imprints of H19, at least, are established in male germ cells from 14.5 dpc onwards (8, 5).
To examine the effect of chromosomal sex of the germ cells, we used XYTdym1 males carrying a Y chromosome deleted for Sry to create sex-reversed XY females, complemented by an Sry-coding autosomal transgene to create XX males (see Materials and Methods for details). In the ovaries of XY females, germ cells enter meiosis before birth, but many degenerate during oogenesis so that the mice are subfertile. Germ cells in XX male testes undergo the early stages of spermatogenesis, entering mitotic arrest before birth, but are all lost shortly after birth. When sex reversal is incomplete, ovotestes may develop, sometimes manifesting as a transient hermaphroditism such that some of the germ cells form growing oocytes in an otherwise normal testis (9). A similar situation may occur in XX↔XY chimeras (10). Isotani et al. (11) reported that some XX oocytes in XX↔XY chimeric testes showed a maternal pattern of differential methylation in three of four imprinted genes, whereas the more numerous XX spermatogonia showed a paternal pattern in two genes. No observations were made on chimeric ovaries.
Our finding, that EG cell lines derived from XY females as well as from XY males showed the same H19 methylation pattern that Tada et al. (6) had reported for XY males, whereas both XX female and XX male embryos were hypomethylated (12), implied that the sex-chromosome constitution of the germ cells was responsible for the sex difference in methylation. The result was clear-cut, but we already knew that the methylation status of EG cells did not necessarily represent that of the germ cells from which they were derived (13). We therefore needed to extend our investigation from EG cells to germ cells, in both the male and the female germ cell lineage.
Results
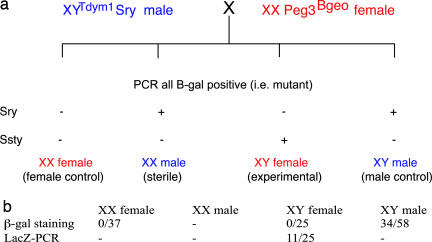
For the female germ line, we looked at gene expression, using a βgeo cassette that had been inserted into the 5’ exon of an imprinted gene, Peg3 (14), which is methylated on the maternal allele and expressed from the paternal allele only. We mated Peg3βgeo females to XYTdym1 males, to generate XX (control) and XYTdym1 (sex-reversed) females, and XX Sry (sex-reversed) and XYTdym1 Sry (control) males, all heterozygous for Peg3βgeo (Fig. 1a). From this F1 progeny, the XX males were not mated, because XX males are sterile. After discarding those that were not carrying Peg3βgeo, the two control groups (XX females and XY males) and the experimentals (XY females) were bred with wild-type partners. The (F2) newborn young were then tested for β-gal expression (Fig. 1b). From the XY male controls, we obtained the expected 50% (34/58) expressing β-gal. From the XX female controls, none of the 37 young expressed β-gal, again as expected, because Peg3 is paternally expressed. From the XY female group, we obtained 25 young (all XY females are subfertile), none expressing β-gal. Of these 25 young, PCR analysis showed that 11 were carrying, but not expressing, the mutant Peg3-βgeo gene. Moreover, the imprint at the DNA methylation level seems to have been established correctly in the XY mothers (Fig. 2), because ≈50% of the alleles were methylated.
Fig. 1.
Scheme for generating the experimental and the two control parental groups used for testing the effect of sex chromosome constitution on Peg3 expression. (a) Males carrying an Sry transgene on an autosome, compensating for their Sry deleted Y chromosome, were mated to females carrying one copy of the imprinted Peg3 gene, marked by insertion of a β-geo cassette into the 5’ exon. Because Peg3 is methylated on the maternal allele and expressed only from the paternal allele, none of the progeny expressed Peg3, but 50% were positive for β-gal (PCR). These β-gal positive progeny were analyzed by PCR for the autosomal XY transgene (Sry) and for the deleted Y chromosome (Ssty), to distinguish the control and sex-reversed male and female young (−, no PCR product was detected; +, PCR product was detected). (b) The XX males were sterile, but the other three groups were mated to wild-type partners. Because Peg3 is paternally transmitted, the expectation was that the female and male controls should have 0% and 50% β-gal positive progeny, respectively. The experimental XY females clearly resembled the XX female rather than the XY male controls. To check that some of their β-gal negative progeny had inherited the Peg3-LacZ-carrying allele from their mother, all 25 young were analyzed by PCR for the LacZ gene. Eleven proved positive.
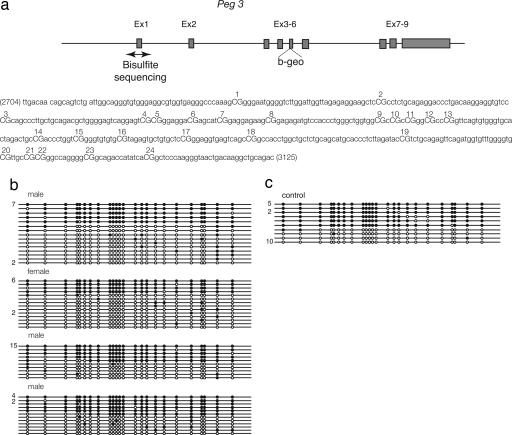
Fig. 2.
Methylation status of the Peg3 differentially methylated domain analyzed in progeny of XY females mated with normal males. (a) Schematic representation of Peg3. The β-geo cassette was knocked-in to the 5 exon (14). The double-headed arrow indicates the region in which methylation was analyzed. (b and c) Each line corresponds to a single strand of DNA, and each circle represents a CpG dinucleotide on that strand. The number of strands observed with a given methylation profile (if greater then one) is indicated to the left of each line. Twenty-four CpGs were analyzed by bisulphite mutagenesis and sequencing. A filled circle represents a methylated and an open circle an unmethylated cytosine. (b) Analyses were performed on three male and one female progeny of XY females. (c) Control sample.
Thus, when Peg3-βgeo allele was transmitted from either the XY females or the XX females, its expression was suppressed, just as Peg3 is suppressed when maternally transmitted. In contrast, the XYTdym1 Sry expressed the Peg3-βgeo allele, just as XY males express Peg3. Based on this result, we concluded that the establishment of the Peg3 imprint is a response of the germ cells to undergoing oogenesis and is in no way dependent on an XX chromosome constitution. This finding is in striking contrast to our earlier findings on EG cells, where cells derived from XX males as well as XX female primordial germ cells (PGCs) were hypomethylated, and those derived from XY female as well as XY male PGCs were methylated.
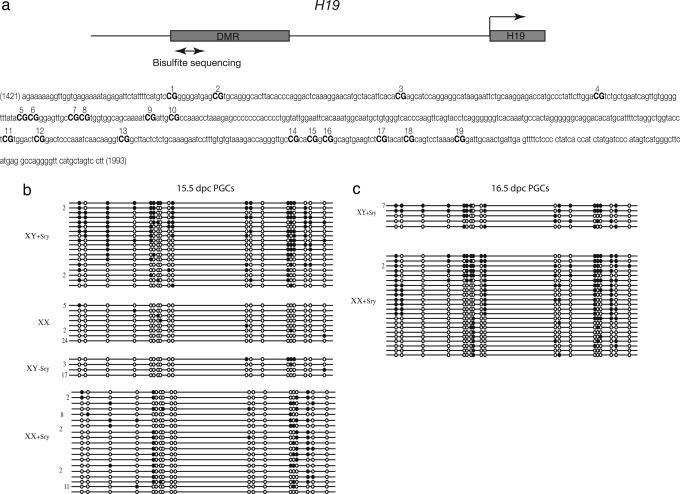
For the male germ line, we mated XYTdym1 Sry males to females carrying a GFP-tagged Oct-4 gene. After FACS-sorting germ cells from gonads at 15.5 and 16.5 dpc, bisulphite sequencing was carried out on control and sex-reversed germ cells, at differentially methylated H19 sites (Table 1 and Fig. 3). The DNA methylation pattern in the XX male germ cells is strikingly different both from that in the control XX female germ cells and in the virtually unmethylated XY female germ cells, so that again we can conclude that the establishment of the imprint is at least in part a response of the germ cell to undergoing spermatogenesis rather than oogenesis. On the other hand, the XX male germ cells are significantly less methylated than the equivalent sites in the XY controls at both 15.5 dpc and 16.5 dpc. This result suggests that, in the male germ line, the sex chromosome constitution of the germ cells has an influence on the establishment of the imprint.
Table 1.
The proportion of all methylated cytosines in the differentially methylated H19 domain analyzed by bisulphite sequencing (Fig. 3)
| Mice | 15.5 dpc |
16.5 dpc |
||||
|---|---|---|---|---|---|---|
| No. of sites | No. methylated | % methylated | No. of sites | No. methylated | % methylated | |
| XX females | 684 | 12 | 1.8 | ND | — | — |
| XY females | 418 | 8 | 2.0 | ND | — | — |
| XX males | 741 | 106 | 16.7a | 437 | 143 | 32.7b |
| XY males | 380 | 132 | 34.7b | 209 | 122 | 58.4c |
Values with different superscript letters differ from one another at a significance level of P < 0.001. ND, not determined.
Fig. 3.
Methylation status of the H19 differentially methylated domain in control and sex-reversed male and female germ cells. (a) Schematic representation of sequences upstream of the H19 promoter. The double-headed arrow indicate the region in which methylation was analyzed. (b and c) Each line corresponds to a single strand of DNA, and each circle represents a CpG dinucleotide on that strand. The number of strands observed with a given methylation profile (if greater then one) is indicated to the left of each line. Nineteen CpGs were analyzed by bisulphite mutagenesis and sequencing. A filled circle represents a methylated and an open circle an unmethylated cytosine. Analyses were performed on male and female germ cells at 15.5 dpc (b) and 16.5 dpc (c). XY + Sry, male control group; XX, female control group; XY-Sry, experimental XY female group; XX + Sry, XX male group, sterile.
Discussion
We have shown previously (12) that, in vitro, the sex chromosome constitution has a striking effect on the establishment of imprints. In this article, we provide evidence of the situation in vivo. In the female germ line, pups carried by XY as well as XX mothers showed virtual absence of site-specific methylation of H19 (a maternally expressed gene) whereas, for Peg3 (a paternally expressed gene), the imprint at the DNA methylation level seemed to have been established correctly and expression was silenced. We were unfortunately unable to test Peg3 transmission by sex-reversed males, because XX males are sterile. We conclude that imprint establishment during oogenesis is wholly independent of the germ cell's own sex chromosome constitution, unlike our earlier findings on EG cells.
In the male germ line, the differentially methylated H19 sites (Fig. 2a) that we analyzed by bisulphite sequencing covered the same region as those used by Davis et al. (8). Our results on control XY male germ cells were largely similar to theirs in respect of the establishment of the new methylation imprint, although our mice (129 genetic background) seemed to establish the new imprint more slowly than theirs (C57BL/6 genetic background). In both XY and XX prospermatogonia, the level of methylation increased significantly from 15.5 to 16.5 dpc, with the level at 16.5 dpc in XY cells being similar to that of Davis et al. (8) at 15.5 dpc.
Both our XY and our sex-reversed XX male germ cells clearly showed a male rather than a female pattern of DNA methylation (Table 1). However, the XX male germ cells were significantly less methylated at both 15.5 and 16.5 dpc than the XY male controls. Unlike in somatic cells, both X chromosomes in XX germ cells in the genital ridge are active, even when the XX germ cell is in a testis (15). The level of site-specific methylation on H19 could perhaps be affected by some transacting demethylating influence because of X-linked genes on the X chromosome. In vitro, Zvetkova et al. (16) have reported that DNA methylation, both globally and in differentially methylated regions of imprinted genes, is reduced in XX ES cell lines relative to XY or XO lines. They show that the hypomethylation effect is due to reduced levels of Dnmt3a and -3b, and they speculate that the X chromosome encodes a modifier locus whose product represses de novo methyltransferases. In addition, the X-coded protein ATRX is known to be involved in chromatin modification, and is dosage-sensitive (17, 18).
Can we reconcile our present in vivo results on germ cells, with our earlier in vitro findings on EG cells? Table 2 suggests that we can, if we assume three sources of influence on site-specific methylation of H19, namely:
There is an effect of the tissue environment, exerted presumably by Sertoli cells in the male genital ridge, follicle cells in the female. Experiments in which germ cells were exchanged between male and female genital ridges at different times (19) have shown that exposure to the environment of a male genital ridge for 1–2 days is sufficient to commit germ cells to spermatogenesis. In the male, the effect would be positive, favoring methylation, but in the female it would be strongly negative. EG cells would of course be free of any such effect.
There is a dose-dependent demethylating effect exerted by X chromosomes (one in XY, two in XX germ cells, with both X chromosomes active).
There is a cell-autonomous methylating activity dependent on the presence of a Y chromosome (independent of Sry expression, because Sry was deleted from the Y chromosome used in these experiments). It is assumed to act in germ cells at 15.5 and 16.5 dpc, and also in EG cells. Although these EG cells were derived from earlier (11.5 dpc) germ cells, we have evidence to suggest that germ cell differentiation continues for a few days after the PGCs are placed in culture (7).
Table 2.
Three hypothetical sources of influence, reconciling the H19 methylation data on EG cells derived from 11.5 dpc PGCs (4) with the data on 15.5 and 16.5 dpc PGCs from the present article (Table 1)
| Cells | Phenotype | Chromosome constitution | Tissue environment | X chromosome | Y chromosome | Resultant |
|---|---|---|---|---|---|---|
| PGCs | Males | XX | ↑ | ↓ | ↑ | Methylation |
| XY | ↑ | ↓↓ | Less methylation | |||
| Females | XY | ↓ | ↓ | ↑ | Hypomethylation | |
| XX | ↓ | ↓↓ | Hypomethylation | |||
| EG cells | Males | XY | ↓ | ↑ | Methylation | |
| XX | ↓↓ | Hypomethylation | ||||
| Females | XY | ↓ | ↑ | Methylation | ||
| XX | ↓↓ | Hypomethylation |
Upward and downward arrows indicate positive and negative influences, respectively, on site-specific methylation.
XY EG cells, whether male or female, show methylation, so the Y chromosome effect dominates, outweighing the effect of the single X chromosome. In XX EG cells, the presumed demethylating effect of the two X chromosomes is unopposed. In germ cells, on the other hand, the strong demethylating effect of the female genital ridge dominates, so that female germ cells, whether XX or XY, are hypomethylated. In male XY germ cells, the methylating effect of the tissue environment combines with the cell-autonomous effect to overcome the influence of the single X chromosome, and full methylation is achieved. In male XX germ cells, the effect of the tissue environment is sufficient to give a characteristically male methylation pattern, but, in the absence of the cell-autonomous influence and the presence of two demethylating X chromosomes, the level of methylation reached is significantly less than for male XY germ cells (Table 1).
Many questions remain. What is the nature of the cell-autonomous effect, and from which part of the Y chromosome does it emanate? Can the relevant gene on the X chromosome be identified? Is it the same as the methyltransferase-repressing modifier locus postulated by Zvetkova et al. (16)? The effect of the tissue environment will of course vary from one imprinted gene to another. Because H19 is paternally methylated, it is not unexpected that the male genital ridge should favor methylation. Spermatogenic cells derived from XY ES cells have been reported to show a normal methylation imprint pattern (20), although the germ cell/Sertoli cell interaction must have been far from normal. The striking difference between the methylation of H19 in sex-reversed XY female germ cells versus EG cells reflects the lack of somatic cell signaling in vitro, and therefore the different epigenetic modifications occurring in vivo and in vitro. It is, however, reassuring that the lack of methylation that we previously found in EG cells derived from sex-reversed XX male PGCs seems to be causally related to the lower level of site-specific methylation seen in XX as compared with XY male germ cells in the present article.
Materials and Methods
Mice.
For the experiment on oogenesis, we used the paternally expressed imprinted gene Peg3 (14, 21). Females homozygous for the Peg3βgeo construct (14) on a strain 129 background were mated to strain 129 males. The embryos were transferred to (CBA/H × C57BL10) F1 recipient females, and the resulting heterozygous females were mated to XYTdym1 Sry males. After classifying the progeny by PCR for Peg3 and sex chromosome constitution, all young not carrying Peg3βgeo were excluded from further analysis [as well as all XX males (sterile)]. XX and XY females were mated to strain 129 males, and XY males were mated to 129 females. After birth, the progeny were classified by PCR analysis for Peg3 and tested for the expression of β-gal by dipping an excised tail tip into Blue-Gal stain.
For the experiment on spermatogenesis, XYTdym1 Sry males were mated to females carrying a GFP-tagged Oct4 gene. Pregnant mice were killed at 15.5 or 16.5 dpc. The gonads were dissected out, disaggregated, and FACS-sorted for green germ cells, which were then subjected to bisulphite sequencing. The head of each fetus was stored at −20°C, so that its sex chromosome constitution could be ascertained by PCR analysis.
PCR Analysis.
For sex chromosome constitution, two PCRs were carried out (22), one detecting YMT2/B-related members of the multiple copy Ssty gene family from the Y long arm, and the other the Sry transgene, in both cases duplexed with the autosomal gene myogenin as an amplification control. For Peg3 detection, fetuses were genotyped by PCR as described (14).
Isolation and FACS Sorting of 15.5–16.5 dpc Germ Cells.
Genital ridges were collected in cold PB1 medium supplemented with 0.5 mM EGTA (Sigma). Immediately before FACS sorting, ridges were washed in PBS and disaggregated in trypsin/EDTA solution (Gibco) by pipetting. Disaggregated cells were resuspended in PB1 supplemented with 300–500 units of hyaluronidase (Sigma) per ml. After 1-min incubation, cells were centrifuged again and the pellet was resuspended in PBS supplemented with 1 mM EGTA before being passed through a 50-μm filter. GFP-positive PGCs were FACS sorted as described (1). More than 95% purity of sorted PGCs was confirmed by alkaline phosphatase staining.
Bisulphite Treatment.
The FACS-sorted germ cells (500–1,000) were embedded in agarose and lysed, and the chromosomal DNA was subjected to bisulphite treatment (1), followed by H19 DMR PCR amplifications using a nested approach (23). The PCR products were gel purified by using QuiaexII (Quiagen), ligated into pGEM T/A cloning vector (Promega), and transformed into TOP10 (Invitrogen) ultra-competent Escherichia coli cells. Positive clones were verified by using colony PCRs, and the products were sequenced by an external sequencing service. The data were collected from at least two independent FACS-sorted samples. We examined a total of 19 CpG sites in a 553-bp fragment of H19 (GenBank accession no. AF049091, pos. 1421–2002). The primers and conditions used for Peg3 (GenBank accession no. AF105262, pos. 2597–3125) have been described (1).
Acknowledgments
G.D.-H. is a Biotechnology and Biological Sciences Research Council (BBSRC)-funded Medical Research Council Career Stem Cells Fellow. This work was funded by grants from the Wellcome Trust (to M.A.S.) and by the Leverhulme Trust, Isaac Newton Trust, and BBSRC (to A.M.).
Abbreviations
- EG
embryonic germ
- PGC
primordial germ cell
- dpc
days post coitum
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Hajkova P., Erhardt S., Lane N., Haaf T., El-Maarri O., Reik W., Walter J., Surani M. A. Mech. Dev. 2002;117:15–23. doi: 10.1016/s0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- 2.Yamazaki Y., Mann M. R. W., Lee S. S., Marh J., McCarrey J. R., Yanagimachi R., Bartolomei M. S. Proc. Natl. Acad. Sci. USA. 2003;100:12207–12212. doi: 10.1073/pnas.2035119100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Obata Y., Kono T. J. Biol. Chem. 2002;277:5285–5289. doi: 10.1074/jbc.M108586200. [DOI] [PubMed] [Google Scholar]
- 4.Davis T. L., Trasler J. M., Moss S. B., Yang G. J., Bartolomei M. S. Genomics. 1999;58:18–28. doi: 10.1006/geno.1999.5813. [DOI] [PubMed] [Google Scholar]
- 5.Ueda T., Abe K., Miura A., Yuzuriha M., Zubair M., Noguchi M., Niwa K., Kawase Y., Kono T., Matsuda Y., et al. Genes Cells. 2000;5:649–659. doi: 10.1046/j.1365-2443.2000.00351.x. [DOI] [PubMed] [Google Scholar]
- 6.Tada T., Tada M., Hilton K., Barton S. C., Sado T., Takagi N., Surani M. A. Dev. Genes Evol. 1998;207:551–561. doi: 10.1007/s004270050146. [DOI] [PubMed] [Google Scholar]
- 7.Durcova-Hills G., Ainscough J. E., McLaren A. Differentiation (Berlin) 2001;68:220–226. doi: 10.1046/j.1432-0436.2001.680409.x. [DOI] [PubMed] [Google Scholar]
- 8.Davis T. L., Yang G. J., McCarrey J. R., Bartolomei M. S. Hum. Mol. Genet. 2000;9:2885–2894. doi: 10.1093/hmg/9.19.2885. [DOI] [PubMed] [Google Scholar]
- 9.McLaren A. Nature. 1980;283:688–689. doi: 10.1038/283688a0. [DOI] [PubMed] [Google Scholar]
- 10.Mystkowska E. T., Tarkowski A. K. J. Embryol. Exp. Morphol. 1968;20:33–52. [PubMed] [Google Scholar]
- 11.Isotani A., Nakanishi T., Kobayashi S., Lee J., Chuma S., Nakatsuji N., Ishino F., Okabe M. Proc. Natl. Acad. Sci. USA. 2005;102:4039–4044. doi: 10.1073/pnas.0406769102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durcova-Hills G., Burgoyne P., McLaren A. Dev. Biol. 2004;268:105–110. doi: 10.1016/j.ydbio.2003.12.018. [DOI] [PubMed] [Google Scholar]
- 13.McLaren A. Dev. Biol. 2003;262:1–15. doi: 10.1016/s0012-1606(03)00214-8. [DOI] [PubMed] [Google Scholar]
- 14.Li L.-L., Keverne E. B., Aparicio S. A., Ishino F., Barton S. C., Surani M. A. Science. 1999;284:330–333. doi: 10.1126/science.284.5412.330. [DOI] [PubMed] [Google Scholar]
- 15.McLaren A., Monk M. J. Reprod. Fertil. 1981;63:533–537. doi: 10.1530/jrf.0.0630533. [DOI] [PubMed] [Google Scholar]
- 16.Zvetkova I., Apedaile A., Ramsahoye B., Mermoud J. E., Crompton L. A., John R., Feil R., Brockdorff N. Nat. Genet. 2005;37:1274–1279. doi: 10.1038/ng1663. [DOI] [PubMed] [Google Scholar]
- 17.Gibbons R. J., McDowell T. L., Raman S., O’Rourke D. M., Garrick D., Ayyub H., Higgs D. R. Nat. Genet. 2000;24:368–371. doi: 10.1038/74191. [DOI] [PubMed] [Google Scholar]
- 18.Xue Y., Gibbons R., Yan Z., Yang D., McDowell T. L., Sechi S., Qin J., Zhou S., Higgs D., Wang W. Proc. Natl. Acad. Sci. USA. 2003;100:10635–10640. doi: 10.1073/pnas.1937626100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adams I. R., McLaren A. Development (Cambridge, U.K.) 2002;129:1155–1164. doi: 10.1242/dev.129.5.1155. [DOI] [PubMed] [Google Scholar]
- 20.Geijsen N., Horoschak M., Kim K., Gribnau J., Eggan K., Daley G. Q. Nature. 2004;427:148–154. doi: 10.1038/nature02247. [DOI] [PubMed] [Google Scholar]
- 21.Kuroiwa Y., Kaneko-Ishino T., Kagitani F., Kohda T., Li L. L., Tada M., Suzuki R., Yokoyama M., Shiroishi T., Wakana S., et al. Nat. Genet. 1996;12:186–190. doi: 10.1038/ng0296-186. [DOI] [PubMed] [Google Scholar]
- 22.Mahadevaiah S. K., Odorisio K., Elliott D. J., Rattigan A., Szot M., Laval S. H., Washburn L. L., McCarrey J. R., Cattanach B. M., Lovell-Badge R., et al. Hum. Mol. Genet. 1998;7:715–727. doi: 10.1093/hmg/7.4.715. [DOI] [PubMed] [Google Scholar]
- 23.Olek A., Walter J. Nat. Genet. 1997;17:275–276. doi: 10.1038/ng1197-275. [DOI] [PubMed] [Google Scholar]