Abstract
The zinc-finger transcription factors GATA4 and GATA6 play critical roles in embryonic development. Mouse embryos lacking GATA4 die at embryonic day (E) 8.5 because of failure of ventral foregut closure and cardiac bifida, whereas GATA6 is essential for development of the visceral endoderm. Although mice that are heterozygous for either a GATA4 or GATA6 null allele are normal, we show that compound heterozygosity of GATA4 and GATA6 results in embryonic lethality by E13.5 accompanied by a spectrum of cardiovascular defects, including thin-walled myocardium, ventricular and aortopulmonary septal defects, and abnormal smooth muscle development. Myocardial hypoplasia in GATA4/GATA6 double heterozygous mutant embryos is associated with reduced proliferation of cardiomyocytes, diminished expression of the myogenic transcription factor MEF2C (myocyte enhancer factor 2C), and down-regulation of β-myosin heavy chain expression, a key determinant of cardiac contractility. These findings reveal a threshold of GATA4 and GATA6 activity that is required for gene expression in the developing cardiovascular system and underscore the potential of recessive mutations to perturb the delicate regulation of cardiovascular development.
Keywords: GATA factors, heart development, ventricular septal defect, heart defects, cardiogenesis
The GATA family of transcription factors plays an important role in differentiation, growth, and survival of diverse cell types (1, 2). Six GATA family members have been identified in vertebrates, all of which contain two zinc-finger domains that bind the consensus site (A/T)GATA(A/G) and mediate cofactor interactions. GATA1, 2, and 3 are primarily expressed in hematopoietic lineages (2), and GATA4, 5, and 6 are expressed in mesoderm- and endoderm-derived tissues such as the heart, liver, lung, and gut (1).
GATA4 regulates the expression of genes that are critical for cardiac contraction, as well as the expression of cardiac transcription factors such as Nkx2.5, Hand2, and MEF2C (myocyte enhancer factor 2C) (3–11). GATA4 null mice display defects in heart morphogenesis and ventral foregut closure (12), resulting in embryonic lethality by embryonic day (E) 8.5. Tetraploid rescue experiments with GATA4 null embryonic stem cells give rise to embryos with abnormal looping of the heart tube and thin-walled myocardium (13). Early cardiac-specific deletion of GATA4 also results in myocardial thinning, abnormal endocardial cushion development, and right ventricular hypoplasia (14), whereas cardiac-specific deletion at later time points results in reduced cardiac function and an inability to undergo hypertrophy after pressure overload or exercise (15). Mice that are homozygous for a hypomorphic GATA4 mutation display a variety of heart defects, including double outlet right ventricle and hypoplasia of the compact myocardium (16). Heterozygous mutations in GATA4 are also associated with congenital heart defects in humans (17).
GATA6 null mice die after implantation because of defects in visceral endoderm function and extraembryonic development (18). Tetraploid rescue experiments have implicated GATA6 in liver differentiation and growth and suggest that GATA4 provides functional redundancy in liver specification (19). Tissue-specific deletion of GATA6 in smooth muscle or neural crest suggests a role for this factor in patterning the cardiac outflow tract and the aortic arch (20). To date, defects in myocardial development have not been observed in GATA6 mutant mice.
Mice that are heterozygous for GATA4 or GATA6 null mutations are viable and without obvious cardiovascular phenotypes. However, given the similarities in protein structure and expression pattern of GATA4 and 6, and their ability to physically interact and synergistically enhance gene transcription (21), we postulated that GATA4 and 6 might act cooperatively to regulate cardiovascular development. Here, we show that GATA4/6 compound heterozygous mice die by E13.5 with 100% penetrance. These mutant mice display a spectrum of cardiovascular defects, including ventricular septal defects (VSDs), a persistent truncus arteriosis (pta), myocardial hypoplasia, reduced myocardial proliferation, and impaired differentiation of vascular smooth muscle cells (SMCs). Our findings reveal an exquisite sensitivity of the developing cardiovascular system to the levels of GATA4 and GATA6 and suggest that these GATA factors act cooperatively to regulate downstream target genes in cardiac cells and SMCs in vivo.
Results
Generation of GATA6 Mutant Mice.
The mouse GATA6 gene was disrupted by replacing exon 2 with a neomycin resistance cassette (see Fig. 6A, which is published as supporting information on the PNAS web site), resulting in deletion of the first coding exon, which encodes amino acids 1–372. The targeted locus lacks the translation initiation codon, and the majority of the transcriptional activation domain and should therefore function as a null allele. Mice that were heterozygous for the targeted allele were viable and fertile and were intercrossed to obtain GATA6 null offspring. Consistent with previous studies (18), no GATA6 null embryos were observed after E7.5 because of failure in visceral endoderm differentiation (data not shown).
Compound Heterozygosity of GATA4/6 Results in Embryonic Lethality by E13.5.
To investigate whether a threshold of GATA4 and GATA6 activity might be required for normal embryonic development, we intercrossed GATA4+/− and GATA6+/− mice. No GATA4/6 compound heterozygous mice were observed at birth (Table 1, which is published as supporting information on the PNAS web site), suggesting that the combined heterozygous mutations resulted in embryonic lethality. Analysis of embryos from timed matings revealed that compound heterozygous offspring were viable up to E13.5. However, no viable GATA4/6 compound heterozygous animals were observed at E14.5 or later, indicating that this genotype results in embryonic lethality with complete penetrance by E13.5. Embryonic lethality of GATA4/6 compound heterozygous embryos was observed in C57BL6/129 mixed genetic backgrounds, suggesting its independence of possible strain variability.
Compound Heterozygosity of GATA4/6 Results in Abnormal Vascular Development.
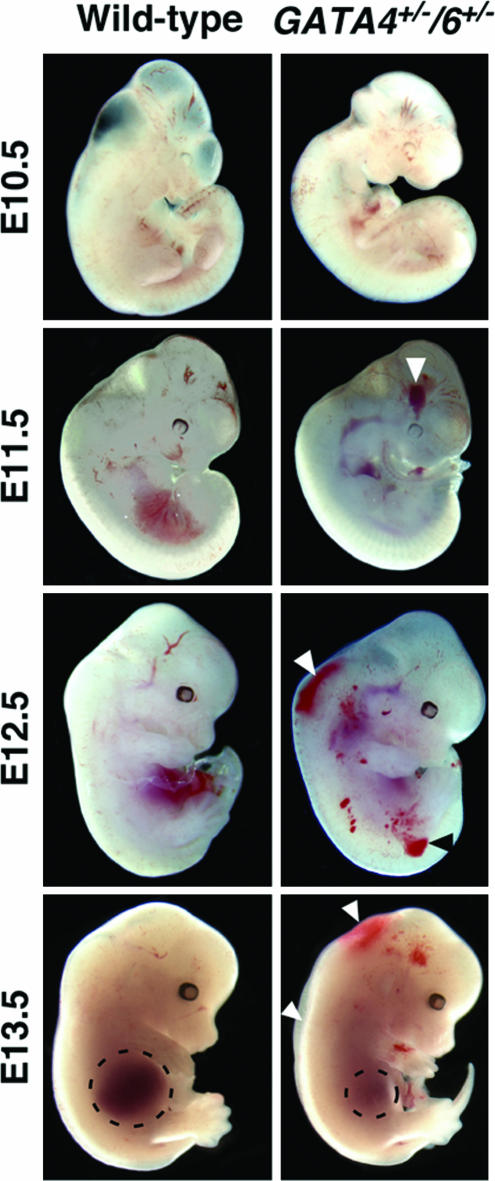
GATA4/6 compound heterozygous embryos displayed widespread hemorrhages by E11.5 and edema at E13.5, as well as reduced liver size compared with control littermates (Fig. 1 and data not shown). We did not observe a difference in hepatic gene expression in the GATA4/6 mutants, although we did observe a reduction in the number of mature erythrocytes in the peripheral blood (data not shown). These results are consistent with GATA4/6 loss-of-function studies in zebrafish, which demonstrate the genes' redundant roles in growth of the specified liver bud (22).
Fig. 1.
Edema and hemorrhage in GATA4+/−/6+/− embryos at E13.5. WT and GATA4+/−/6+/− mutant embryos at E10.5, E11.5, E12.5, and E13.5 are shown. Arrowheads denote hemorrhage and edema. Dashed lines outline the liver in both control and GATA4/6 mutants.
The heartbeat of GATA4/6 mutants at E11.75 was sluggish and irregular compared with that of control littermate embryos (data not shown), suggesting possible abnormalities in cardiac function. To visualize potential vascular abnormalities in GATA4/6 mutant embryos, we stained the vasculature of E10.5 embryos for platelet/endothelial cell adhesion molecule. As shown in Fig. 2A, the cranial and intersomitic vasculature in the GATA4/6 double heterozygous embryos was enlarged and disorganized compared with WT embryos, indicative of possible defects in blood circulation and/or vessel development and remodeling.
Fig. 2.
Vascular defects in GATA4+/−/6+/− embryos. (A) Whole-mount staining of WT and mutant embryos at E10.5 by using anti-platelet/endothelial cell adhesion molecule antibodies. Arrowheads denote dilated and less-developed vessels within the cranial and intersomitic vasculature in the GATA4+/−/6+/− embryos. (B) β-Gal staining of GATA4+/−, GATA6+/−, or GATA4+/−/6+/− embryos at E12 reveals reduced expression of the SM22-LacZ transgene (arrows) and the presence of pta (see arrowhead). jv and phv denote expression of the SM22-LacZ transgene in venous smooth muscle of GATA4+/−/6+/− mutants. Boxed areas in Upper were rephotographed and enlarged in Lower. (C) Transverse sections of β-gal-stained embryos in B demonstrate LacZ staining within the primary head (arrow) and jugular veins (arrowhead). (D) H&E staining (Upper) of E12.5 embryos shows a hypoplastic and dilated aorta in the GATA4/6 mutants. Smooth muscle actin staining (Lower) shows reduced smooth muscle differentiation in the GATA4+/−/6+/− mutants. ba, branchial arch artery; va, vertebral artery; jv, jugular vein; phv, primary head vein; da, dorsal aorta; aao, ascending aorta; pt, pulmonary trunk.
To better visualize the defects in vascular patterning in mutant mice, we intercrossed GATA4 and GATA6 heterozygous mice with mice that were transgenic for SM22-LacZ, a cardiac and arterial smooth muscle-specific reporter (23). LacZ staining at E12 revealed patterning defects of the outflow tract in GATA4/6 mutants, as evidenced by the presence of pta, a defect resulting from incomplete septation of the conotruncus into the aorta and pulmonary artery (see arrowhead in Fig. 2B and Fig. 3). Although the process of aortopulmonary septation occurs between E10.5 and E13.5 (24), the pta observed in the GATA4/6 mutants was completely penetrant (n > 10) when compared with control littermates, suggesting either a delay in the mutants or an inability to completely septate. Because the GATA4/6 compound heterozygotes die by E13.5, we cannot rule out the possibility that this component of the phenotype is a result of general delay in cardiovascular development beginning at E11.5. LacZ staining in the descending aorta (da) and vertebral artery (va) of compound mutant embryos was also diminished compared with littermate controls (Fig. 2B arrows). Because the SM22 promoter is not regulated directly by GATA factors (25), its down-regulation suggests a general defect in smooth muscle differentiation. Notably, the SM22-LacZ reporter, which is normally expressed specifically in arterial SMCs, was activated in venous SMCs of the GATA4/6 mutants [see Fig. 2B, jugular vein (jv) and primary head vein (phv)]. Transverse sections of stained embryos showed that the primary head vein (Fig. 2C, arrow) and jugular veins (Fig. 2C, arrowhead) of mutant embryos were positive for LacZ expression, suggesting an abnormality in SMC identity within the developing vasculature. A large arterial-venous malformation can also be seen in the embryo shown in Fig. 2B. Similar defects were seen in multiple embryos, although their severity varied. Whether GATA4 and 6 participate directly in the establishment of vascular identity, or whether the SM22 promoter becomes inappropriately activated in venous SMCs in response to pathological signals, such as hypoxia, requires further study.
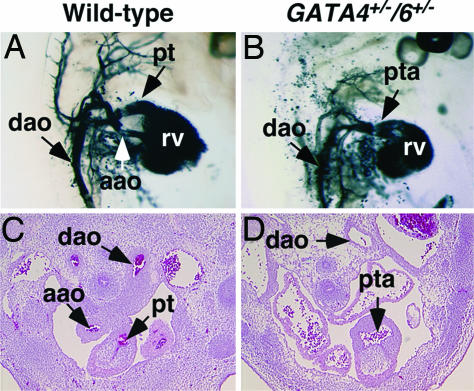
Fig. 3.
Defects in the cardiac outflow tract of GATA4+/−/6+/− embryos. (A and B) India ink was injected into the left ventricle of beating hearts of WT (A) and GATA4+/−/6+/− (B) embryos at E12.5. The atria were removed, and the right lateral views of the heart and great vessels are shown. (C and D) Transverse sections of WT and GATA4+/−/6+/− mutants at E12.5 displaying incomplete septation between the outflow tract and the pulmonary trunk. aao, ascending aorta, dao, descending aorta; pt, pulmonary trunk; pta, persistent truncus arteriosis; rv, right ventricle.
Histological sections also revealed thin, dilated vessels in GATA4/6 mutants compared with littermate controls. Immunostaining for smooth muscle α-actin at E12.5 showed that there was less smooth muscle in the medial layer of the aorta (Fig. 2D) and supported our observation of a reduction in arterial smooth muscle. These data suggest that GATA4 and 6 are required for proper development of vascular SMCs and suggest a role for these factors in maintaining overall vessel integrity.
GATA4/6 Compound Heterozygotes Display Outflow Tract Defects.
To further examine the pta and abnormalities in the great vessels and cardiac outflow tract in the GATA4/6 mutants, we performed ventricular India ink injections into the beating heart. As shown in Fig. 3 (A and C), WT animals display complete septation (or division) of the cardiac outflow tract by the aortopulmonary septum by E12.5. In the mutants, only a single outlet was visible, indicating a failure of septation between the aorta and pulmonary trunk and confirming the abnormalities seen in transverse sections of E12.5 embryos (Fig. 3 B and D). We also observed a hypoplastic transcending aortic arch in the GATA4/6 double heterozygous embryos, further suggesting patterning defects of the great vessels.
GATA4/6 Mutants Display Myocardial Thinning and VSDs as a Result of a Reduction in Myocyte Proliferation.
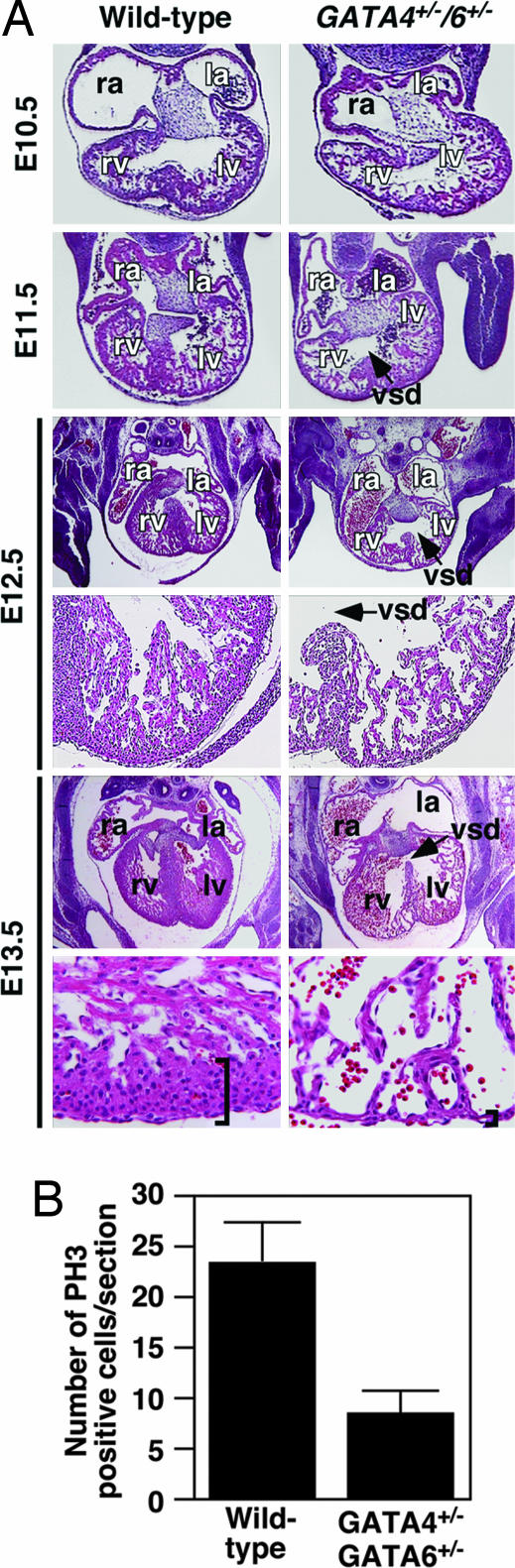
Histological sections of E10.5–13.5 embryos revealed a narrow temporal window for the onset of cardiac defects in GATA4/6 mutants (Fig. 4A). There was a modest delay in the formation of the ventricular septum beginning at E11.5. Myocardial thinning first became apparent at E12.5. Hearts of mutant embryos at E13.5 contained only two myocardial cell layers within the compact zone compared with a five-cell layer in WT embryos. In addition, GATA4/6 mutant embryos displayed VSDs that persisted until the time of death at E13.5, a finding that was consistent with the phenotype resulting from hypomorphic GATA4 alleles (16). Because defects in cardiovascular development have been shown to be secondary to placental defects (26), we examined GATA4/6 mutant embryos for perturbations in placental structure and found no obvious morphological abnormalities, suggesting that the cardiovascular defects observed are not secondary to defects in early embryonic development.
Fig. 4.
Cardiac defects in GATA4+/−/6+/− embryos. (A) H&E staining of transverse sections of WT and GATA4+/−/6+/− mutants. E12.5 and E13.5 embryo upper images show the entire heart, and lower images show ×10 magnification of the left ventricle of each heart. Arrows point to VSDs. Higher magnification (×40) of E13.5 hearts reveals thinning due to a decrease in myocardial cell layers in mutant embryos. Brackets show the thickness of the myocardial layer of the left ventricle (lv). ra, right atrium; la, left atrium; rv, right ventricle; vsd, ventricular septal defect. (B) Phosphohistone H3 (PH3) staining of sections of hearts from WT and GATA4+/−/6+/− embryos at E10.5 revealed reduced cardiomyocyte proliferation in the double mutant embryos.
To determine the molecular mechanism underlying the thin myocardium observed in the GATA4/6 mutants, we evaluated myocardial apoptosis and proliferation. We observed no differences in TUNEL staining between WT and mutant hearts at E10.5, E11.5, E12.5, and E13.5 (data not shown), suggesting that the GATA mutants might have defects in myocyte proliferation. Indeed, GATA4/6 compound heterozygous hearts displayed reduced cellular proliferation at E10.5 as assayed by phosphohistone H3 staining (Fig. 4B), suggesting that the reduction in the myocardial cell layer resulted from reduced myocyte proliferation and not apoptosis.
Altered Cardiac Gene Expression in GATA4/6 Mutants.
Quantitative RT-PCR on RNA isolated from hearts of WT and compound heterozygous mutant mice at E11.75 showed that GATA4 and GATA6 expression was reduced to 60% in mutants compared with WT littermates (Fig. 5). In addition, expression of the β-myosin heavy chain (β-MHC) gene, a known GATA target that encodes a major protein involved in embryonic cardiac contractility (27), was also down-regulated in the GATA4/6 mutants, suggesting possible defects in cardiac function. Intriguingly, MEF2C transcripts were also markedly reduced in hearts from GATA4/6 mutant embryos at E11.75. The reduction of MEF2C expression provides a potential explanation for the paucity of cardiomyocytes and the patterning defects seen in GATA4/6 compound heterozygotes.
Fig. 5.
Modulation of myocardial gene expression in GATA4+/−/6+/− embryos. Real-time PCR was performed by using RNA isolated from hearts of E11.75 embryos. Relative expression normalized to GAPDH in WT (filled bars) and GATA4+/−/6+/− (open bars) embryos is shown. Note the down-regulation of GATA4, GATA6, β-MHC, and MEF2C in GATA4+/−/6+/− embryos.
Discussion
Mice that are heterozygous for a null mutation in either GATA4 or GATA6 are viable, whereas we show in the present study that compound heterozygosity of GATA4 and 6 null alleles results in a spectrum of lethal cardiovascular phenotypes, including VSDs, myocardial hypoplasia, pta, and abnormalities in vascular smooth muscle development. The defects observed in GATA4/6 heterozygous embryos are distinct from those in embryos that are homozygous for either a GATA4 or GATA6 null allele, suggesting that these GATA factors play a cooperative role in cardiovascular development. Dosage sensitivity in development has not been previously observed with other combinations of heterozygous GATA mutations.
Cardiovascular Phenotypes of GATA4/6 Mutant Mice.
The VSDs and thin ventricular myocardium accompanied by reduced myocyte proliferation in GATA4/6 mutant embryos resemble the phenotype resulting from conditional deletion of GATA4 by using an Nkx2–5-Cre transgene (14, 16). The fact that vascular defects were not observed upon cardiac-specific deletion of GATA4 (14, 16) suggests that such abnormalities in GATA4/6 compound heterozygous embryos reflect a cell-autonomous function of GATA4 and 6 in the vascular system rather than a secondary response to cardiac demise. It is also interesting to note that down-regulation of β-MHC and MEF2C expression was not observed after cardiac-specific deletion of GATA4, suggesting a combinatorial role for both GATA4 and GATA6 in maintaining adequate levels of these transcripts for proper cardiogenesis.
GATA6 has been implicated in maturation of cardiac mesoderm in Xenopus and zebrafish embryos (28), but, to our knowledge, there has been no prior evidence for a specific role of GATA6 in heart development based on phenotypes of GATA6 mutant mice. In fact, conditional deletion of GATA6 by using either Wnt1-Cre or SM22-Cre transgenes resulted in perinatal lethality from defects in septation of the cardiac outflow tract and patterning of the aortic arch arteries without apparent myocardial abnormalities (20). Moreover, rescue of GATA6 null embryos by tetraploid embryo complementation revealed no specific requirement for GATA6 in cardiac development (19). Such results are intriguing and suggest that GATA4 and GATA6 may still function independently of one another in select tissues or at certain times in development, despite the functional redundancy suggested by the compound heterozygous phenotype described here.
Although we presume that GATA4 and 6 act synergistically in the heart and vasculature such that deletion of one copy of each gene diminishes GATA activity below a threshold required for activation of essential genes in these muscle cell types, we cannot be certain from the present studies whether GATA4 and 6 are required cell autonomously in both of these tissues or, alternatively, whether developmental abnormalities in the heart or vasculature cause abnormalities in the other tissue secondarily. It is also conceivable, but probably unlikely, that GATA4 and 6 are required in different cell types such that deletion of one copy of both genes compromises development.
GATA5 has also been shown to be required for production of normal numbers of myocardial precursors and for expression of multiple cardiac genes in zebrafish (29). In contrast, mice lacking GATA5 do not show cardiac abnormalities (30). Moreover, GATA4/5 or GATA5/6 compound heterozygous mutant mice are normal (J.D.M. and E.N.O., unpublished data), demonstrating the specificity in cardiac functions of GATA4 and 6.
Functions of GATA Factors.
GATA 4 and 6 heterodimerize and synergistically activate the ANF (atrial natriuretic factor) and BNP (brain natriuretic peptide) genes in cardiomyocytes (21). However, neither of these genes was down-regulated in GATA4/6 mutants, indicating that the phenotype of these mutants does not result from a general diminution in expression of GATA target genes in the cardiovascular system. Instead, it seems more likely that specific GATA target genes, such as β-MHC and MEF2C or others yet to be identified, are highly sensitive to the combined level of GATA4 and 6, perhaps reflecting preferential responsiveness to GATA4/6 heterodimers compared with GATA homodimers. Because a subset of transcriptional coactivators has been shown to interact with GATA4 and not GATA6 (31), it is tempting to speculate that the heterodimerization of these factors is required to mediate full transcriptional competency.
In light of the down-regulation of MEF2C in GATA4/6 mutant mice and the synergistic interactions between MEF2 and GATA4 (32), we analyzed the expression of several MEF2 target genes, including Srpk23 (33) and Bop (34), and found no change in their expression patterns. Thus, we cannot attribute the GATA4/6 mutant phenotype to a general diminution in expression of MEF2 targets. It is possible, however, that genes regulated cooperatively by GATA4/6 and MEF2C might be especially sensitive to the reduced expression of these factors in GATA4/6 mutant embryos.
The transcriptional activities of GATA4 and GATA6 are influenced by corepressors and non-DNA-bound transcriptional activators (35–37). In concert with Tbx20, GATA4 synergistically activates the expression of MEF2C in the anterior heart field (38). In addition, GATA4 has also been shown to interact with NFAT3 (nuclear factor of activated T cells 3) (39), MEF2C (32), Nkx2.5 (40, 41), SRF (serum response factor) (42), Hand2 (43), and myocardin (44) to regulate cardiac gene expression. Full transcriptional activation by both GATA4 and GATA6 also requires interaction with the histone acetyltransferase p300 (36, 37), and this interaction is lost in the presence of the competing corepressors FOG-2 (friend of GATA2) (45) and Hey-2 (46, 47). It will be of interest to determine whether mutations in the genes encoding any of these GATA cofactors, when combined with GATA4 or 6 mutations, evoke unique phenotypes not seen with the single heterozygous mutations.
Implications for Human Congenital Heart Disease.
A variety of GATA4 mutations have been linked to cardiac septal defects in humans (17, 48). The results of the present study point to the potential involvement of GATA6 mutations in human congenital heart disease. It is unclear why heterozygous mutations in GATA4 or GATA6 alone do not result in cardiac defects in mice, although it is well established that human heart development is more sensitive to subtle genetic abnormalities (49).
It is not uncommon for congenital heart disease to appear in human pedigrees with incomplete penetrance and variable expressivity, which has suggested the existence of modifier genes that influence cardiac phenotypes (49). The finding that heterozygous mutations in GATA4 and GATA6 cause no observable phenotype alone in mice, whereas together they result in complete embryonic lethality, illustrates the power of recessive genetic interactions to influence heart development. Moreover, the realization that cardiovascular development is exquisitely sensitive to the threshold of GATA4/6 activity suggests that therapeutic strategies to augment the activity of these transcription factors, even subtly, might overcome certain congenital cardiac abnormalities. In light of the repressive influence of corepressors and histone deacetylases (36, 37) on GATA factors, we are currently investigating whether partial inhibition of such negative regulators or augmentation of positive effectors of GATA activity in vivo might restore cardiac function in GATA4/6 mutant mice.
Materials and Methods
Gene Targeting.
A GATA6 targeting vector was constructed to replace exon 2, which contains the translational start site and the majority of the transcriptional activation domain, with a neomycin cassette from pNeoTK (see Supporting Materials and Methods, which is published as supporting information on the PNAS web site). Targeting of the GATA4 locus and the generation of GATA4 mutant mice are described in ref. 12.
Histology and Immunostaining.
Histology and immunostaining were performed by standard procedures as described in Supporting Materials and Methods.
India Ink Injections.
E12.5 embryos were harvested and subjected to intracardial injection of India ink by using custom-made glass pipettes. After injection, embryos were fixed in 4% paraformaldehyde for 12 h, dehydrated through graded methanol, and cleared in benzyl benzoate:benzyl alcohol (2:1) as described in ref. 50.
RNA Isolation, Real-Time PCR, and Microarray Analyses.
Total RNA was isolated from embryonic hearts collected at E11.75 by using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. For RT-PCR, 1 μg of total RNA was used as a template for reverse transcription with random hexamer primers. cDNA (25 ng) was amplified in each real-time PCR by using the TaqMan Universal PCR Master Mix kit (Applied Biosystems, Foster City, CA). Mean relative gene expression was calculated by using standard curves from serial dilutions of cDNA from WT hearts and normalized to GAPDH (n = 3 per group).
Supplementary Material
Acknowledgments
We thank Jiyeon Oh for helpful discussions, Alisha Tizenor for graphics, Jennifer Brown for editorial support, Cheryl Nolen for technical assistance, and Vidu Garg and Rhonda Bassel-Duby for helpful comments on the manuscript. This work was supported by National Institutes of Health (NIH) Training Grant F32 5 HL71450-03 (to C.A.D.), NIH grants, the Donald W. Reynolds Cardiovascular Clinical Research Center, and the Robert A. Welch Foundation (E.N.O.).
Abbreviations
- En
embryonic day n
- VSD
ventricular septal defect
- SMC
smooth muscle cell
- pta
persistent truncus arteriosis
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Molkentin J. D. J. Biol. Chem. 2000;275:38949–38952. doi: 10.1074/jbc.R000029200. [DOI] [PubMed] [Google Scholar]
- 2.Weiss M. J., Orkin S. H. Exp. Hematol. 1995;23:99–107. [PubMed] [Google Scholar]
- 3.McFadden D. G., Charite J., Richardson J. A., Srivastava D., Firulli A. B., Olson E. N. Development (Cambridge, U.K.) 2000;127:5331–5341. doi: 10.1242/dev.127.24.5331. [DOI] [PubMed] [Google Scholar]
- 4.Reecy J. M., Li X., Yamada M., Demayo F. J., Newman C. S., Harvey R. P., Schwartz R. J. Development (Cambridge, U.K.) 1999;126:839–849. doi: 10.1242/dev.126.4.839. [DOI] [PubMed] [Google Scholar]
- 5.Pikkarainen S., Tokola H., Kerkela R., Ruskoaho H. Cardiovasc. Res. 2004;63:196–207. doi: 10.1016/j.cardiores.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 6.Nemer G., Nemer M. Dev. Biol. 2003;254:131–148. doi: 10.1016/s0012-1606(02)00026-x. [DOI] [PubMed] [Google Scholar]
- 7.Nemer G., Nemer M. Ann. Med. 2001;33:604–610. doi: 10.3109/07853890109002106. [DOI] [PubMed] [Google Scholar]
- 8.Molkentin J. D., Kalvakolanu D. V., Markham B. E. Mol. Cell. Biol. 1994;14:4947–4957. doi: 10.1128/mcb.14.7.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lien C. L., Wu C., Mercer B., Webb R., Richardson J. A., Olson E. N. Development (Cambridge, U.K.) 1999;126:75–84. doi: 10.1242/dev.126.1.75. [DOI] [PubMed] [Google Scholar]
- 10.Jiang Y., Drysdale T. A., Evans T. Dev. Biol. 1999;216:57–71. doi: 10.1006/dbio.1999.9469. [DOI] [PubMed] [Google Scholar]
- 11.Dodou E., Verzi M. P., Anderson J. P., Xu S. M., Black B. L. Development (Cambridge, U.K.) 2004;131:3931–3942. doi: 10.1242/dev.01256. [DOI] [PubMed] [Google Scholar]
- 12.Molkentin J. D., Lin Q., Duncan S. A., Olson E. N. Genes Dev. 1997;11:1061–1072. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- 13.Watt A. J., Battle M. A., Li J., Duncan S. A. Proc. Natl. Acad. Sci. USA. 2004;101:12573–12578. doi: 10.1073/pnas.0400752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeisberg E. M., Ma Q., Juraszek A. L., Moses K., Schwartz R. J., Izumo S., Pu W. T. J. Clin. Invest. 2005;115:1522–1531. doi: 10.1172/JCI23769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oka T., Maillet M., Watt A. J., Schwartz R. J., Aronow B. J., Duncan S. A., Molkentin J. D. Circ. Res. 2006;98:837–845. doi: 10.1161/01.RES.0000215985.18538.c4. [DOI] [PubMed] [Google Scholar]
- 16.Pu W. T., Ishiwata T., Juraszek A. L., Ma Q., Izumo S. Dev. Biol. 2004;275:235–244. doi: 10.1016/j.ydbio.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 17.Garg V., Kathiriya I. S., Barnes R., Schluterman M. K., King I. N., Butler C. A., Rothrock C. R., Eapen R. S., Hirayama-Yamada K., Joo K., et al. Nature. 2003;424:443–447. doi: 10.1038/nature01827. [DOI] [PubMed] [Google Scholar]
- 18.Morrisey E. E., Tang Z., Sigrist K., Lu M. M., Jiang F., Ip H. S., Parmacek M. S. Genes Dev. 1998;12:3579–3590. doi: 10.1101/gad.12.22.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao R., Watt A. J., Li J., Luebke-Wheeler J., Morrisey E. E., Duncan S. A. Mol. Cell. Biol. 2005;25:2622–2631. doi: 10.1128/MCB.25.7.2622-2631.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lepore J. J., Mericko P. A., Cheng L., Lu M. M., Morrisey E. E., Parmacek M. S. J. Clin. Invest. 2006;116:929–939. doi: 10.1172/JCI27363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Charron F., Paradis P., Bronchain O., Nemer G., Nemer M. Mol. Cell. Biol. 1999;19:4355–4365. doi: 10.1128/mcb.19.6.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holtzinger A., Evans T. Development (Cambridge, U.K.) 2005;132:4005–4014. doi: 10.1242/dev.01978. [DOI] [PubMed] [Google Scholar]
- 23.Li L., Miano J. M., Mercer B., Olson E. N. J. Cell Biol. 1996;132:849–859. doi: 10.1083/jcb.132.5.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma P. R., Anderson R. H., Copp A. J., Henderson D. J. Anat. Rec. A Discov. Mol. Cell. Evol. Biol. 2004;277:355–369. doi: 10.1002/ar.a.20006. [DOI] [PubMed] [Google Scholar]
- 25.Li L., Liu Z., Mercer B., Overbeek P., Olson E. N. Dev. Biol. 1997;187:311–321. doi: 10.1006/dbio.1997.8621. [DOI] [PubMed] [Google Scholar]
- 26.Sapin V., Dolle P., Hindelang C., Kastner P., Chambon P. Dev. Biol. 1997;191:29–41. doi: 10.1006/dbio.1997.8687. [DOI] [PubMed] [Google Scholar]
- 27.Hasegawa K., Lee S. J., Jobe S. M., Markham B. E., Kitsis R. N. Circulation. 1997;96:3943–3953. doi: 10.1161/01.cir.96.11.3943. [DOI] [PubMed] [Google Scholar]
- 28.Peterkin T., Gibson A., Patient R. EMBO J. 2003;22:4260–4273. doi: 10.1093/emboj/cdg400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reiter J. F., Kikuchi Y., Stainier D. Y. Development (Cambridge, U.K.) 2001;128:125–135. doi: 10.1242/dev.128.1.125. [DOI] [PubMed] [Google Scholar]
- 30.Molkentin J. D., Tymitz K. M., Richardson J. A., Olson E. N. Mol. Cell. Biol. 2000;20:5256–5260. doi: 10.1128/mcb.20.14.5256-5260.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Durocher D., Charron F., Warren R., Schwartz R. J., Nemer M. EMBO J. 1997;16:5687–5696. doi: 10.1093/emboj/16.18.5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morin S., Charron F., Robitaille L., Nemer M. EMBO J. 2000;19:2046–2055. doi: 10.1093/emboj/19.9.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakagawa O., Arnold M., Nakagawa M., Hamada H., Shelton J. M., Kusano H., Harris T. M., Childs G., Campbell K. P., Richardson J. A., et al. Genes Dev. 2005;19:2066–2077. doi: 10.1101/gad.1338705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phan D., Rasmussen T. L., Nakagawa O., McAnally J., Gottlieb P. D., Tucker P. W., Richardson J. A., Bassel-Duby R., Olson E. N. Development (Cambridge, U.K.) 2005;132:2669–2678. doi: 10.1242/dev.01849. [DOI] [PubMed] [Google Scholar]
- 35.Yanazume T., Hasegawa K., Morimoto T., Kawamura T., Wada H., Matsumori A., Kawase Y., Hirai M., Kita T. Mol. Cell. Biol. 2003;23:3593–3606. doi: 10.1128/MCB.23.10.3593-3606.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wada H., Hasegawa K., Morimoto T., Kakita T., Yanazume T., Sasayama S. J. Biol. Chem. 2000;275:25330–25335. doi: 10.1074/jbc.M000828200. [DOI] [PubMed] [Google Scholar]
- 37.Dai Y. S., Markham B. E. J. Biol. Chem. 2001;276:37178–37185. doi: 10.1074/jbc.M103731200. [DOI] [PubMed] [Google Scholar]
- 38.Takeuchi J. K., Mileikovskaia M., Koshiba-Takeuchi K., Heidt A. B., Mori A. D., Arruda E. P., Gertsenstein M., Georges R., Davidson L., Mo R., et al. Development (Cambridge, U.K.) 2005;132:2463–2474. doi: 10.1242/dev.01827. [DOI] [PubMed] [Google Scholar]
- 39.Molkentin J. D., Lu J. R., Antos C. L., Markham B., Richardson J., Robbins J., Grant S. R., Olson E. N. Cell. 1998;93:215–228. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sepulveda J. L., Belaguli N., Nigam V., Chen C. Y., Nemer M., Schwartz R. J. Mol. Cell. Biol. 1998;18:3405–3415. doi: 10.1128/mcb.18.6.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee Y., Shioi T., Kasahara H., Jobe S. M., Wiese R. J., Markham B. E., Izumo S. Mol. Cell. Biol. 1998;18:3120–3129. doi: 10.1128/mcb.18.6.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Belaguli N. S., Sepulveda J. L., Nigam V., Charron F., Nemer M., Schwartz R. J. Mol. Cell. Biol. 2000;20:7550–7558. doi: 10.1128/mcb.20.20.7550-7558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dai Y. S., Cserjesi P., Markham B. E., Molkentin J. D. J. Biol. Chem. 2002;277:24390–24398. doi: 10.1074/jbc.M202490200. [DOI] [PubMed] [Google Scholar]
- 44.Oh J., Wang Z., Wang D. Z., Lien C. L., Xing W., Olson E. N. Mol. Cell. Biol. 2004;24:8519–8528. doi: 10.1128/MCB.24.19.8519-8528.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hirai M., Ono K., Morimoto T., Kawamura T., Wada H., Kita T., Hasegawa K. J. Biol. Chem. 2004;279:37640–37650. doi: 10.1074/jbc.M401737200. [DOI] [PubMed] [Google Scholar]
- 46.Fischer A., Klattig J., Kneitz B., Diez H., Maier M., Holtmann B., Englert C., Gessler M. Mol. Cell. Biol. 2005;25:8960–8970. doi: 10.1128/MCB.25.20.8960-8970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Okubo A., Miyoshi O., Baba K., Takagi M., Tsukamoto K., Kinoshita A., Yoshiura K., Kishino T., Ohta T., Niikawa N., et al. J. Med. Genet. 2004;41:e97. doi: 10.1136/jmg.2004.018895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kathiriya I. S., King I. N., Murakami M., Nakagawa M., Astle J. M., Gardner K. A., Gerard R. D., Olson E. N., Srivastava D., Nakagawa O. J. Biol. Chem. 2004;279:54937–54943. doi: 10.1074/jbc.M409879200. [DOI] [PubMed] [Google Scholar]
- 49.Srivastava D., Olson E. N. Nature. 2000;407:221–226. doi: 10.1038/35025190. [DOI] [PubMed] [Google Scholar]
- 50.Oh J., Richardson J. A., Olson E. N. Proc. Natl. Acad. Sci. USA. 2005;102:15122–15127. doi: 10.1073/pnas.0507346102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.