Abstract
Genomic amplifications and deletions, the consequence of somatic variation, are a hallmark of human cancer. Such variation has also been observed between “normal” individuals, as well as in individuals with congenital disorders. Thus, copy number measurement is likely to be an important tool for the analysis of genetic variation, genetic disease, and cancer. We developed representational oligonucleotide microarray analysis, a high-resolution comparative genomic hybridization methodology, with this aim in mind, and reported its use in the study of humans. Here we report the development of a representational oligonucleotide microarray analysis microarray for the genomic analysis of the mouse, an important model system for many genetic diseases and cancer. This microarray was designed based on the sequence assembly MM3, and contains ≈84,000 probes randomly distributed throughout the mouse genome. We demonstrate the use of this array to identify copy number changes in mouse cancers, as well to determine copy number variation between inbred strains of mice. Because restriction endonuclease digestion of genomic DNA is an integral component of our method, differences due to polymorphisms at the restriction enzyme cleavage sites are also observed between strains, and these can be useful to follow the inheritance of loci between crosses of different strains.
Keywords: mouse genome, polymorphism, segmentation, CGH, probe selection
Copy number variation is increasingly being recognized as important to understanding pathophysiology and genetics in humans. For example, in cancer, deletions are used to map tumor suppressors and amplifications to map oncogenes. Some genetic diseases, both inherited and spontaneous, can be attributed to mutations altering copy number. Moreover, wide variation in copy number is present in the normal human population, and although not yet proven, this variation is expected to alter normal physiology.
Very large scale deletions and duplications can be observed cytogenetically by chromosome banding and comparative genomic hybridization (CGH) on chromosomes. More recently, higher resolution has been achieved by array CGH, first performed with BACs (1). With the advent of the completion of the human genome sequence, methods with even higher resolution are now available, all based on using oligonucleotide probes mapped to the genome. These methods include whole genome hybridization (2, 3) and representational approaches (4) that we pioneered. High-density SNP arrays that use representational technology (5) can also be used to detect copy number changes.
The success of representational oligonucleotide microarray analysis (ROMA) depends in part on the simplification of the genome that results from PCR amplification of genomic DNA, restriction endonuclease cleavage, and adapting fragments by ligation to primers (6). Algorithms based on the human genome assembly (7) make possible the design of probes that hybridize to representations. Fundamentally, ROMA depends on the development of high-density oligonucleotide arrays, and in particular on methodology that allowed for the flexible design and rapid fabrication of arrays, enabling us to optimize probe selection. Such technology is available from NimbleGen Systems, that allows up to 800,000 oligonucleotide features to be fabricated by using mirror-directed laser photochemistry (8). The NimbleGen fabrication system allows the rapid redesign of an array, from assay to assay, at no extra cost.
Our initial design for a ROMA array was based on the human genome sequence assembly. The array was designed with ≈84,000 probes, designed from short (200–1,200 bp) candidate fragments based on BglII representations, and then further empirically selected based on performance parameters we established. We demonstrated its use in detecting amplifications and deletions in cancers, the widespread variation of copy number present in the human population, and de novo mutations found in some children with genetic disorders (4, 9). Here, we describe the design and implementation of a ROMA array based on BglII representations of the mouse genome sequence assembly.
Mouse models have become increasingly important for the study of human diseases, such as cancer and genetics. Many cancer models revolve around the study and understanding of one particular gene, but others are more global models, focused on the recapitulation of cancer etiology and gene discovery. Mouse models have also been widely used to understand the known human disease genes (10) and to model known genetic alterations (11), but also to discover genes that are involved in pathophysiology, such as obesity (12) and diabetes (13). In these efforts, particularly the latter, we see a role for measuring copy number, both as a means for gene discovery, and as a method to build appropriate models of human disease.
Both BAC and cDNA fragment arrays have already been used to measure gene copy number in numerous mouse neoplasias (14, 15). We have developed a mouse ROMA array because such arrays are more available to us, but also because they are better defined, more accurately reproduced, and can be of higher resolution. Moreover, because representations are generated by restriction endonuclease digestion, the ROMA is sensitive to single nucleotide polymorphisms between strains, and this can be used to advantage in certain circumstances.
We show the utility of these arrays in detecting changes in copy number in cancers, and assessing differences between inbred strains. The method for probe selection is provided online at our web site (http://roma.cshl.org). Although the method for making the mouse array follows closely the method we used for making the human array, there is much greater genetic variation within the murine species than in Homo sapiens, and hence application of ROMA to the laboratory mouse requires greater care in experimental design.
Results
Representations.
A representation is a sampling of the genome that reduces complexity. This effectively increases signal to noise during hybridization, because there is less “extraneous” labeled genome contributing to nonspecific hybridization. In addition, representations can be prepared from very small amounts of DNA, i.e., very few cells, a common practice when working with tumors, and some other biological samples. The disadvantage of making representations is the extra step, one that must be conducted with great care to achieve reproducibility. We generally use a two-color hybridization to compare a test sample to a reference sample, each prepared in parallel to minimize variation caused by representation. A representation is prepared by digestion of the genome with a restriction endonuclease, ligation of oligonucleotide adaptors and PCR amplification with the same adaptors. Taq polymerase generally does not amplify large fragments as efficiently as small fragments, resulting in the preferential amplification of fragments in the size range of ≈200–1,200 base pairs, thereby reducing the complexity. The degree of complexity reduction depends on the choice of restriction endonuclease. We have based the mouse array on representations prepared by using the restriction endonuclease BglII, the same enzyme we used for the human array. We estimate complexity is reduced to ≈3% of the original genome.
Probe and Array Design.
Probes were designed based on Build 30 (updated to Build 33) of the mouse genome sequence, which is an assembly of reads predominantly from C57BL/6. The genome was digested “in silico” with BglII (AGATCT), and ≈200,000 fragments (100–1,200 bp long) were culled. Multiple distinct and minimally overlapping 50-bp oligonucleotide sequences, the candidate probes, were generated for each fragment. Each 50-mer sequence was annotated for exact matches in the genome by using a Burroughs–Wheeler transformation and algorithm as described (7). Two critical counts were calculated for each candidate probe, corresponding to the number of exact matches of 21- and 15-bp substrings in the mouse genome. These are measures of repetitiveness. Based on humans, we have identified empirically acceptable counts for probes (7). Limits were set on consecutive runs of single nucleotides, and GC content for further probe selection. After taking these criteria into account, up to four probes were picked per fragment. Up to 25 bp of overlap between probes was allowed. A final set of 650,000 probes was chosen to be as unique as possible within the genome, and satisfying the above conditions. The resulting probe set was synthesized on our arrays with overlapping subsets so that the resulting data could be normalized across arrays to more accurately pick a final set of 84,000 probes.
Probe Selection.
As described in ref. 4, the probes were validated by using depletion experiments. A depletion experiment compares a standard BglII representation to a BglII representation that is depleted of a predictable subset of fragments. A depletion representation is commonly prepared by digesting the representation just before its amplification with a second restriction endonuclease. By doing so, all representational fragments containing a cleavage site for the second restriction endonuclease will not be amplified in the final representation, the equivalent of being depleted from the representation. We then compare a representation to a depleted representation. Assuming the mouse genome is reliable we should be able to predict which probes will have elevated ratios.
We do not perform a set of depletion studies that covers all probes. Rather, we determined during the development of the human ROMA array, and confirmed this for the mouse array, that the probes with the highest ratio also had the highest intensities in the nondepleted representation. For an example, see Fig. 1A where the probe ratios are graphed as a function of intensity in the (undepleted) representation channel. Probes predicted to be depleted due to the presence of an internal cleavage site are in red, and all others are in green. The depleted probes have ratios correlated to the intensity. The intensity measurements for all probes for all 650,000 probes from four different hybridizations were measured, and as expected a fraction, of the probes do not perform as well as the others. The top 84,000 probes were picked based on intensity and a new NimbleGen array designed and synthesized. Having optimized for probe performance on the array, the final array design could now be used to analyze biological samples such as tumor material or DNAs from specific mouse strains.
Fig. 1.
The distribution of measured probe ratios from a depletion experiment as a function of intensity of the nondepleted representation channel, for the 84,000 probes in the final selection. The y axis is the ratio measurements and the x axis is the intensity, both plotted in log scale. Probes that derive from fragments with an internal HindIII cleavage site are red, and those that derive from fragments without an internal site are shown in blue.
Tumor Genomes.
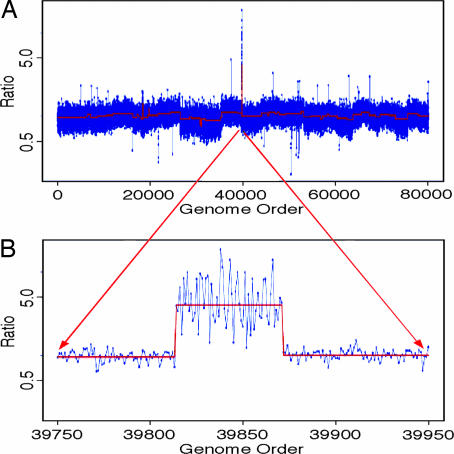
As an illustration of mouse ROMA, Fig. 2A shows the analysis of a tumor obtained from a mouse model of liver cancer (23). Hepatoblasts from a p53−/− mouse were transduced with a murine stem cell virus retrovirus coexpressing the Myc oncogene, and transplanted into mice. The tumors were excised from the liver and cultured, and DNA was prepared. These samples were compared to a DNA sample from the parent animal’s tail by ROMA.
Fig. 2.
The genomic profile of a DNA sample derived from a tumor generated in a mouse model of liver cancer. Tumor representations were compared to representations prepared from the parent DNA. (A) The profile of the entire genome. The x axis is the genome order concatenating the entire genome from chromosome 1 to chromosomes Y. The y axis is the ratio measurements obtained, graphed in log scale. The blue dots connected by a blue line are the normalized ratio measurements for each probe. The red line is the segmented data for the same experiments. Deletions are in the downward direction, and amplifications are in the upward direction. (B) Magnified view of a region on chromosome 9 that is genomically amplified and contains the oncogene candidates IAP1, IAP2, or YAP1.
It is clear that there is an amplicon in this tumor. The probe data are annotated for probe genomic position so that it is easy to obtain the chromosome location of any lesion observed. We can zoom in on an amplicon that is found in the beginning of the q arm of chromosome 9 (Fig. 2B). The genome coordinates of the probes lying outside of the amplicon can be used as break points to identify the gene content for the regions of interest by using publicly available gene browsers such as the University of California, Santa Cruz, Genome Browser (www.genome.ucsc.edu). If we select the amplicon (coordinates chr9:6,896,942–8,515,171, build 33) and identify the gene content, we can easily determine that there are 15 full-length gene candidates within this region. This region is seen amplified in several types of cancer such as liver and ovarian cancer (23). These data were used along with that from other samples mouse and human to determine the most likely candidate genes. Gene candidate validation was used to determine that the genes IAP1, IAP2, and YAP1 within this region are amongst the likely target genes important to the development and progression of cancer (see ref. 23 for a more detailed analysis of these gene candidates).
To make the identification of lesions, we use more automated algorithms to perform what is commonly referred to as “segmentation,” to partition the log ratios of the intensities into segments of uniform distributions. The ratios are arranged in genome order and are divided into blocks of 100 data points with arbitrary boundaries. Then, the boundaries of the segments are iteratively moved by minimizing the variance, and further refined by using a two-distribution Kolmogorov–Smirnov null hypothesis test (16). Only boundaries that give a P value statistic of <10−5 are accepted. This method is excellent for large lesions comprising of six or more probes. A Hidden Markov Model may detect smaller lesions.
This technique has utility to identify copy number variations in tumor samples from mouse models or human cancer. These regions can be used to map cancer gene candidates, and this information can be cross referenced to the human location based on synteny. Using these methods, we have identified a gene that is involved in human cancer, demonstrating the utility of identification of copy number fluctuations in mouse cancer models to identify genes that are mutated in human cancer.
Strain Polymorphism and Implications for Analysis.
ROMA performed on two different humans reveals two major types of polymorphisms (4, 9). SNPs are detected when the two individuals differ at a BglII cleavage site that causes a restriction fragment length polymorphism deleting a fragment in the representation. Copy number polymorphisms (CNPs) are detected as altered intensity ratios of clusters of nearby probes. In humans, CNPs can be found as more than, or less than, the expected copies per genome.
Much like human ROMA, the mouse ROMA chip will also identify both of these types of polymorphisms, much more striking between inbred laboratory strains than anything seen in humans. Fig. 3B shows a comparison of BALB/cByJ to C57BL/6J (hereafter referred to as BALB/c and C57BL/6). We observe an exceedingly asymmetric distribution of differences, with a large number of clustered single probe events, indicating low hybridization from BALB/c. This finding is in stark comparison to a self-to-self hybridization where the samples compared were liver and tail from a C57BL/6 mouse (Fig. 3A). This difference is readily explainable as SNPs distinguishing the strains. In the present case, the mouse array is designed from the sequence assembled mainly from C57BL/6, with the expectation of BglII fragment lengths predicted from that strain. Moreover, probes were selected by hybridization to DNA from that strain. Interestingly, the SNPs between the two strains cluster into regions as shown in Fig. 3D surrounded by genomic regions of much higher similarity between the two strains.
Fig. 3.
The genomic profile comparison of differing mouse strains. (A) The entire genome profile of a comparison of one strain to itself. (B) The entire genome profile of the mouse strain BALB/c compared to C57BL/6. (C) Magnified view of a region containing a CNP, with fewer copies in BALB/c as compared to C57BL/6. (D) Magnified view of a region from the comparison of C57BL/6 to BALB/c, where there is a region dense with SNPs between the two strains. On all graphs, the x axis is the genomic order as described above, and the y axis is the ratio measurements obtained, graphed in log scale. The red line is the segmented data.
This interpretation was confirmed by PCR analysis (Fig. 4). PCR primers were designed 100–300 bp on either side of the BglII cleavage sites of the expected representational fragment, and genomic DNA from C57BL/6 and BALB/c were used as template. The amplified fragments were digested with BglII. If both BglII cleavage sites are present, the expected fragment length will be obtained. If one of the BglII cleavage sites is altered by the presence of a SNP, the digested fragment will be larger. Three proposed SNPs were selected from a cluster and tested by this method, and the expected results were obtained. For each fragment, both BglII cleavage sites in C57BL/6 were retained, and one cleavage site in BALB/c was lost. This analysis shows that the isolated probes with large negative log ratios are likely to reflect SNPs within BglII restriction sites, although in theory such single probe differences could also occur as a result of large internal insertions that alter the size of a BglII fragment, significant alteration within the probe sequence itself, or small deletions that encompass only a single probe.
Fig. 4.
Validation by PCR of several probes containing possible SNPs identified from the comparison of C57BL/6 to BALB/c. Fragments were amplified such that they were larger than the genomic BglII representational fragment and digested with BglII to detect the presence of SNPs in one or both BglII cleavage sites. C57BL/6, fragment amplified from C57BL/6; C57BL/6-D, fragment amplified from C57BL/6 digested with BglII; BALB/c, fragment amplified from BALB/c; and BALB/c-D, fragment amplified from BALB/c digested with BglII. The arrows point to the fragment being queried. The ratio is plotted on the y axis in log scale, and the x axis is a genome order. The markers on the gel picture (M) are a 100-bp ladder.
Most of these polymorphisms, although not in adjacent probes, are regionally clustered, indicating that different regions of highly diverged ancestral genomes became fixed during inbreeding, whereas much of the remaining genome between C57BL/6 and BALB/c is nearly identical. Different strains show different patterns of fixation (I.M.H., C.E., B.L., Srinath Srindar, Deepa Pai, and M.W., unpublished data).
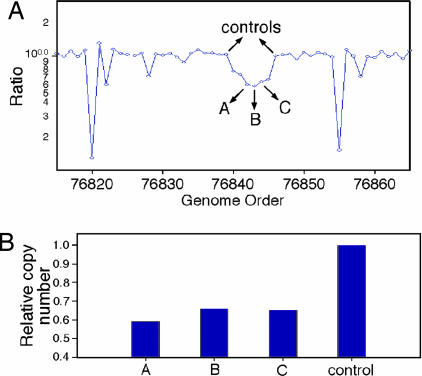
As expected, CNPs are also identified in the comparison of the two strains. In Fig. 3C, a CNP was identified, where fewer copies are present in BALB/c. Therefore, this region is likely to be present as two copies in BALB/c as compared to four copies in C57BL/6 because these are inbred strains. Another CNP, shown in Fig. 5A, was one of several CNPs detected with the lower limit of probes for the segmentor to identify a lesion, and copy number was validated by quantitative PCR. Three probes within the CNP region were tested and normalized to the proximal sequences flanking the CNP. The results show roughly half the copy number for the probes within the CNP (Fig. 5B) in BALB/c as compared to C57BL/6 (similar to the CNP discussed earlier).
Fig. 5.
Quantitative PCR validation of a CNP identified in the comparison of the two mouse strains analyzed. (A) The probe ratio data for the CNP region graphically represented and arrows identifying the probes whose sequences were used to generate Q-PCR primers. The x axis is the genome order, and the y axis is the probe ratio plotted in log scale. (B) The results of quantitative PCR graphed as a histogram. The x axis is the three probes and the average of the controls. The y axis is the calculated copy number in relation to the control probes, being set to a value of 1.
Discussion
Measurement of gene copy number in humans is now widely recognized as a valuable tool for the analysis of genetic variation in cancer and genetic disease. The laboratory mouse is the premier animal model for human disease, and we therefore sought to develop a similar tool for mice. We used our experience with ROMA as a guide. The resulting genome array is built on the same principles, and functions similarly to the human version. The array can detect duplications, amplifications and deletions in the mouse genome, with applications in mouse cancer models and to discriminating the genetic differences between mouse strains. Experimental design and application in mice must differ somewhat from humans because of the existence of inbred strains of mice, because there is greater genetic variation in mouse than in humans, and because these genetic differences are distributed in distinct clusters between strains.
We detect two types of genetic variation between mice, most prominently between inbred strains. These are SNPs that result in restriction fragment-length polymorphisms, which become apparent because ROMA is based on restriction endonuclease cleavage and CNPs (see Fig. 3). Both types of variation are highly nonuniform, clustering in specific locations along the genome. The variation within these regions is far greater than anything we observe in humans, and more resembles differences we see between humans and primates (data not shown). Between these regions are regions with almost no variation, far less than we observe between different individual humans. In this report, we show a comparison of only two inbred strains of mice, BALB/c and C57BL/6, but the phenomenon is present between any two strains, with different regions of clustering. These types of studies have been undertaken by others (17, 18) using arrays that cannot detect SNPs. The extreme variation we observe is undoubtedly a consequence of the mosaic nature of the inbred mouse genome, with different strains containing unique combinations of genomic segments derived from genetically diverged subspecies (19). The marked differences between strains can be exploited during genetic crosses of strains in the mapping of complex traits, but the corollary is that, in tumor studies, one must take great care to match genomes.
When we examine tumor genomes in humans, we are often forced to compare a cancer from one patient to the normal DNA of another individual. In humans, where there are few polymorphisms, we can still interpret our data by making a compendium of the common and rare CNPs, and masking differences at these sites. Between inbred strains of mice, because there are so many differences dispersed through out the genomes, this would be an extreme nuisance. However, in the controlled setting of the laboratory, and without the need for institutional review board approval, comparing a tumor to normal DNA from the tumor-bearing animal is, fortunately, not an issue. It might seem that it suffices to compare tumors to DNA from the same inbred strain, but we have found that even this has perils. Inbred strains do have genetic variation that we detect, so the comparison should be between the tumor and the tumor-bearing animal whenever possible. (This is all of the more imperative if the strain has been out-crossed.) When the experiment is carefully designed, we can clearly see genomic alterations that are tumor specific, as demonstrated in Fig. 2. The alterations can be easily mapped to the mouse genome to identify oncogene candidates and the region can be compared to the syntenic region in the human genome. In the particular case that we illustrate, this method was used to find a strong candidate oncogene in mouse liver cancer that is also amplified in the syntenic region in human cancer.
We have used ROMA for measuring copy number, but there are several other methods that have been used for measurements with humans that can be adapted to the mouse. BAC arrays have already been used in mice for this purpose (17). Our preference for ROMA, aside from the obvious one of familiarity (to us), is its flexibility, reproducibility, and even cost. Our arrays, which are manufactured by NimbleGen Systems, are made to our design, and the arrays are reusable. In this report, we use arrays with 84,000 probes, and this is sufficient for most applications, but we have recently designed arrays with 390,000 probes for humans, and these work well and give higher resolution. Because of the density of restriction fragment-length polymorphisms in mice, it would not be difficult to design a specialty ROMA array for detecting strain heterogeneity at high resolution. Recently, we have adapted the ROMA methodology to detect differences in methylation patterns in two tissues, a method we call MOMA (methylation detection representational oligonucleotide microarray analysis). MOMA should be especially valuable when applied to mouse because any tissue from the same individual animal is readily available. The probe selection procedures, current probe coordinates, and protocols for our mouse ROMA BglII array are available from the authors upon request.
Materials and Methods
Supplies.
Cot-1 DNA (15279-011) and yeast tRNA (15401-029) were supplied by Invitrogen. Restriction enzymes, ligase, and Klenow fragments (M0212M) were supplied by New England Biolabs. The Megaprime labeling kit, Cy3-conjugated dCTP, and Cy5-conjugated dCTP were supplied by Amersham Pharmacia. Taq polymerase was supplied by Eppendorf. Centricon YM-30 filters were supplied by Amicon (42410), and formamide was supplied by Amresco (0606-500). Phenol–chloroform was supplied by Sigma (P2069).
Array Design.
Oligonucleotide probes are designed in silico from the mouse genome, build 30. The genome was digested in silico by the restriction enzyme, and ≈200,000 fragments were selected that are 100–1,200 bp long. For every fragment, constituent 50-bp probes were annotated by using a genome dictionary (7), a previously developed algorithm. We allowed the probes to overlap by 25 bp. Annotation of each probe includes the frequency of occurrence of consecutive 21-mer, frequency of occurrence of consecutive 15-mer in the genome, consecutive runs of single nucleotides, and its GC content. A final set of 650,000 probes was chosen to be as unique as possible within the genome. All probes that satisfied the following criteria were selected for validation: frequency of 21-mer < 2; runs of A/Ts < 6; runs of C/Gs < 4, and GC percentage within 30–60% and 15-mer frequency as low as possible to give us four probes per fragment. We tested all 650,000 probes, predicted to be complementary to short BglII fragments, arrayed on four chips.
Representation.
BglII representations, in general, were prepared as described (20). A major change is that amplification was carried out in an MJ Research Tetrad. Eight 250-μl tubes were used for amplification. The cycle conditions were 95°C for 1 min, 72°C for 3 min, for 20 cycles, followed by a 10-min extension at 72°C. Representations depleted of specific fragments by restriction enzyme were prepared in the same manner with the following modification. After ligation of adaptor, the mixture was cleaned by phenol–chloroform extraction, precipitated, and resuspended. The ligated fragments were split, half being digested with the second enzyme (HindIII in this case) and the other half mock digested. This material was then used as template in the PCR as described above.
Labeling and Hybridization.
Two DNAs for comparison were labeled as described (21) using reagents from the Amersham Pharmacia Megaprime labeling kit with 10 μl of label (Cy3-dCTP or Cy5-dCTP). In addition, each experiment was hybridized in duplicate, where in one replicate, the Cy5 and Cy3 dyes were swapped (i.e., “color reversal”). Hybridizations, washing, and scanning were also performed as described (4, 21). Samples were denatured in an MJ Research Tetrad at 95°C for 5 min and then incubated at 37°C for 30 min. Samples were spun down and pipetted onto a slide prepared with lifter slip and incubated in a hybridization oven such as the Boekel InSlide Out oven set at 42°C for 14–16 h. Slides were washed and then scanned immediately with an Axon GenePix 4000B scanner. genepix pro 4.0 software was used for quantitation of intensity for the arrays. Array data were imported into s-plus for further analysis. Measured intensities without background subtraction were used to calculate ratios. Data were normalized by using an intensity-based lowess curve fitting algorithm similar to that described in ref. 22. Data obtained from color-reversal experiments were averaged and displayed as presented in the figures
Quantitative PCR.
Three BglII fragments (A, B, C) were chosen for verification. For each fragment, primers were designed to be in the 75- to 150-bp range. For each primer pair, a group of three reactions is set up. The three reactions correspond to the reference DNA (C57BL/6), BALB/c, and no template. This group is duplicated on the plate, and a similar plate was repeated three times to achieve statistical significance. The reactions were formulated by using Applied Biosystems SYBR Green PCR core reagents kit, and the PCR was performed on an ABI Prism 7700 sequence detection system.
Acknowledgments
We thank Scott Powers for critical comments on the manuscript, Linda Rodgers for technical assistance, Jim Duffy for artwork, and Patricia Bird for secretarial assistance. This work was supported by Department of Defense Grant W81XWH-05-1-0068 (to R.L.); National Cancer Institute Grant K01 CA93634-04 (to R.L.); National Cancer Institute Grant 5R01-CA078544-07 (to M.W.); and grants from the Miracle Foundation, Breast Cancer Research Foundation, Long Islanders Against Breast Cancer, West Islip Breast Cancer Foundation, Long Island Breast Cancer (1 in 9), Elizabeth McFarland Breast Cancer Research Grant, and Breast Cancer Help, Inc. (all to M.W.). M.W. is an American Cancer Society Research Professor.
Abbreviations
- ROMA
representational oligonucleotide microarray analysis
- CNP
copy number polymorphism.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Pinkel D., Segraves R., Sudar D., Clark S., Poole I., Kowbel D., Collins C., Kuo W. L., Chen C., Zhai Y., et al. Nat. Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- 2.Barrett M. T., Scheffer A., Ben-Dor A., Sampas N., Lipson D., Kincaid R., Tsang P., Curry B., Baird K., Meltzer P. S., et al. Proc. Natl. Acad. Sci. USA. 2004;101:17765–17770. doi: 10.1073/pnas.0407979101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Selzer R. R., Richmond T. A., Pofahl N. J., Green R. D., Eis P. S., Nair P., Brothman A. R., Stallings R. L. Genes Chromosomes Cancer. 2005;44:305–319. doi: 10.1002/gcc.20243. [DOI] [PubMed] [Google Scholar]
- 4.Lucito R., Healy J., Alexander J., Reiner A., Esposito D., Chi M., Rodgers L., Brady A., Sebat J., Troge J., et al. Genome Res. 2003;13:2291–2305. doi: 10.1101/gr.1349003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang J., Wei W., Zhang J., Liu G., Bignell G. R., Stratton M. R., Futreal P. A., Wooster R., Jones K. W., Shapero M. H. Hum. Genomics. 2004;1:287–299. doi: 10.1186/1479-7364-1-4-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lisitsyn N., Lisitsyn N., Wigler M. Science. 1993;259:946–951. doi: 10.1126/science.8438152. [DOI] [PubMed] [Google Scholar]
- 7.Healy J., Thomas E. E., Schwartz J. T., Wigler M. Genome Res. 2003;13:2306–2315. doi: 10.1101/gr.1350803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nuwaysir E. F., Huang W., Albert T. J., Singh J., Nuwaysir K., Pitas A., Richmond T., Gorski T., Berg J. P., Ballin J., et al. Genome Res. 2002;12:1749–1755. doi: 10.1101/gr.362402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sebat J., Lakshmi B., Troge J., Alexander J., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., et al. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 10.Ikehara S., Ohtsuki H., Good R. A., Asamoto H., Nakamura T., Sekita K., Muso E., Tochino Y., Ida T., Kuzuya H., et al. Proc. Natl. Acad. Sci. USA. 1985;82:7743–7747. doi: 10.1073/pnas.82.22.7743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daley G. Q., Van Etten R. A., Baltimore D. Science. 1990;247:824–830. doi: 10.1126/science.2406902. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y., Proenca R., Maffei M., Barone M., Leopold L., Friedman J. M. Nature. 1994;372:425–432. doi: 10.1038/372425a0. [DOI] [PubMed] [Google Scholar]
- 13.Chen H., Charlat O., Tartaglia L. A., Woolf E. A., Weng X., Ellis S. J., Lakey N. D., Culpepper J., Moore K. J., Breitbart R. E., et al. Cell. 1996;84:491–495. doi: 10.1016/s0092-8674(00)81294-5. [DOI] [PubMed] [Google Scholar]
- 14.Sander S., Bullinger L., Karlsson A., Giuriato S., Hernandez-Boussard T., Felsher D. W., Pollack J. R. Oncogene. 2005;24:6101–6107. doi: 10.1038/sj.onc.1208751. [DOI] [PubMed] [Google Scholar]
- 15.Urzua U., Frankenberger C., Gangi L., Mayer S., Burkett S., Munroe D. J. Tumour Biol. 2005;26:236–244. doi: 10.1159/000087378. [DOI] [PubMed] [Google Scholar]
- 16.Conover W. J. Practical Nonparametric Statistics. New York: Wiley; 1980. [Google Scholar]
- 17.Snijders A. M., Nowak N. J., Huey B., Fridlyand J., Law S., Conroy J., Tokuyasu T., Demir K., Chiu R., Mao J. H., et al. Genome Res. 2005;15:302–311. doi: 10.1101/gr.2902505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J., Jiang T., Mao J. H., Balmain A., Peterson L., Harris C., Rao P. H., Havlak P., Gibbs R., Cai W. W. Nat. Genet. 2004;36:952–954. doi: 10.1038/ng1417. [DOI] [PubMed] [Google Scholar]
- 19.Wade C. M., Kulbokas E. J., III, Kirby A. W., Zody M. C., Mullikin J. C., Lander E. S., Lindblad-Toh K., Daly M. J. Nature. 2002;420:574–578. doi: 10.1038/nature01252. [DOI] [PubMed] [Google Scholar]
- 20.Lucito R., Wigler M. Preparation of Target DNA. Plainview, NY: Cold Spring Harbor Lab. Press; 2003. [Google Scholar]
- 21.Lucito R., Wigler M. Preparation of Slides and Hybridization. Plainview, NY: Cold Spring Harbor Lab. Press; 2003. [Google Scholar]
- 22.Yang Y. H., Dudoit S., Luu P., Lin D. M., Peng V., Ngai J., Speed T. P. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zender L., Spector M. S., Xue W., Flemming P., Cordon-Cardo C., Sike J., Fan S.-T., Luk J. M., Wigler M., Hannon G. J., et al. Cell. 2006;125:1–15. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]