Abstract
Since its introduction a decade ago, whole-genome shotgun sequencing (WGS) has been the main approach for producing cost-effective and high-quality genome sequence data. Until now, the Sanger sequencing technology that has served as a platform for WGS has not been truly challenged by emerging technologies. The recent introduction of the pyrosequencing-based 454 sequencing platform (454 Life Sciences, Branford, CT) offers a very promising sequencing technology alternative for incorporation in WGS. In this study, we evaluated the utility and cost-effectiveness of a hybrid sequencing approach using 3730xl Sanger data and 454 data to generate higher-quality lower-cost assemblies of microbial genomes compared to current Sanger sequencing strategies alone.
The existing Sanger sequencing (1) method has served as the cornerstone for genome sequencing, including microbial sequencing, for over a decade. Continued improvements in DNA sequencing techniques, bioinformatics, and data analysis over the past few years have helped reduce the cost and time associated with sequencing a genome. However, it still costs an estimated $8,000-$10,000 per megabase pair to produce a high-quality microbial genome draft sequence. There is a need for a more efficient and cost-effective approach for genome sequencing that can maintain the high quality of data produced by conventional Sanger sequencing. One of the most promising new sequencing technologies is the 454 GS20 sequencing platform (454 Life Sciences, Branford, CT; ref. 2). It is a highly parallel noncloning pyrosequencing-based (3, 4) system capable of sequencing 100 times faster than current state-of-the-art Sanger sequencing and capillary electrophoresis platforms. However, our experience with this new technology suggests that it is not yet ready to replace current sequencing methods for de novo assembly of genomes. The key issues are short read lengths (100 bp on average), a lack of paired end reads, and the accuracy of individual reads, particularly in regions where homopolymers are observed. Short read lengths make it impossible to span repetitive genomic elements. In addition, the current lack of paired end reads information for each DNA fragment limits assembly to contigs separated by coverage gaps or repetitive elements, such that larger scaffolds required for high-quality draft sequence and gap closure are difficult to achieve. Given these limitations, we have found that, for de novo genome sequencing, the 454 platform is better used as a complement to, rather than a replacement of, existing sequencing methods.
In this paper, we explore the utility and cost-effectiveness of a hybrid sequencing approach that incorporates 454 pyrosequencing data with conventionally generated 3730xl Sanger sequencing data to produce high-quality low-cost assemblies of small marine microbial genomes. It is our belief that the evaluation of this sequencing approach and the results obtained in this study will make a substantial difference in how microbial sequencing is performed. By combining the advantages of two sequencing technologies, we are able to produce better-quality microbial genome assemblies than with the current Sanger sequencing strategy alone.
Here we present how genomic assemblies generated with a hybrid sequencing approach using 3730xl Sanger and 454 sequencing data compare to assemblies generated using Sanger data alone. In addition, we examine the utility of such a hybrid approach to cost-effectively sequence, assemble, and close genomes. Our results indicate there are benefits as well as issues that must be considered as part of the application of the 454 technology to any large-scale sequencing project. The limitations of the 454 process hinder its usefulness in de novo sequencing where the assembly of sequencing reads cannot be aligned to a reference genome. A hybrid approach, leveraging the strengths of both conventional and 454 sequencing, appears quite promising for producing higher-quality assemblies.
In particular, the 454’s lack of cloning bias and ability to sequence through regions of the genome that exhibit strong secondary structure (“hard stops”) provide dramatic improvement over the quality of assemblies available from either sequencing technology alone. In our tests of the hybrid approach on six marine microbes, we have observed complete closure of all sequencing gaps in two of the six organisms and up to an 86% gap reduction in the remaining organisms. The improved quality of the draft assemblies and reduced cost of genome closure justify the application of the 454 sequencing technology to these genomes.
In addition to exploring the quality of assemblies from the hybrid approach, we also investigate the optimal proportions of conventional Sanger vs. 454 sequencing data that will yield high-quality yet cost-effective assemblies. Based on our results, we believe that most de novo sequencing projects should continue to rely primarily on Sanger sequencing data, while incorporating 454 sequencing data to use the complementary strengths of the 454 platform. For the small marine microbes sequenced to date, we have determined that an initial assembly of 5.3× Sanger sequencing data is sufficient to determine final genome size and map out a cost-effective plan for additional sequencing. Encountering nonclonable regions and “hard stops” at this stage would warrant 454 sequencing runs, whereas large sequencing gaps, repetitive areas, and physical gaps would warrant additional Sanger sequencing reads. Improvements in the 454 technology in the areas of increasing read lengths and addition of a paired end reads capability could very well change the coverage requirements needed from each technology.
Results
Six marine microbial genomes (Janibacter sp. HTCC2649, Erythrobacter litoralis HTCC2594, Prochlorococcus marinus str. MIT 9211, Pseudoalteromonas tunicata D2, Vibrio angustum S14, and Psychromonas sp. CNPT3) were sequenced as part of the Gordon and Betty Moore Marine Microbial Sequencing Project (https://research.venterinstitute.org/moore). All genomes were sequenced to different levels of coverage using Sanger sequencing on 3730xl sequencers and the 454 GS20 sequencing platform. These genomes were chosen for hybrid sequencing, because they provide a representative spectrum of assembly characteristics within ≈70 genomes sequenced for the Gordon and Betty Moore Marine Microbial Sequencing Project. Both MIT 9211 and HTCC2594 consisted of one scaffold with few sequencing gaps and were chosen to seek out the effectiveness of the hybrid 454/Sanger sequencing in closing a genome with little or no manual intervention. HTCC2649 consisted of one scaffold with >100 sequencing gaps, several of which were believed to be hard stop gaps, i.e., gaps in which a high number of clone end reads ended within 30 bp of the gap. The remaining three genomes, D2, S14, and CNPT3, represented genomes with a high number of both sequencing and physical gaps, as well as repetitive structures within the DNA.
Gaps and N50 Contigs.
We compared assemblies at six levels of Sanger sequencing coverage and three levels of 454 sequencing runs. A summary of these results is provided in Figs. 1 and 2 and Table 1. A general trend of decreasing gap number and increasing N50 scaffold size was seen across all six levels of Sanger sequencing with the incorporation of one and two 454 runs (one 454 run corresponds to ≈20–40 million bases).
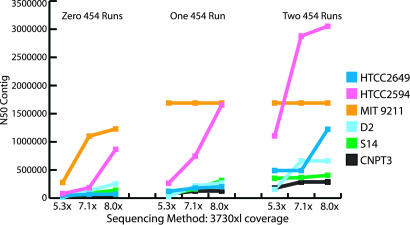
Fig. 1.
Results of mixed sequencing runs, ABI 3730xl, and 454 technology. Hybrid assembly comparison, N50 contigs. The x axis represents the amount and type of data used for an assembly. The y axis is a logarithmic scale representation of the N50 contig size (i.e., half the nucleotides in assembled contigs are in contigs of this size or greater).
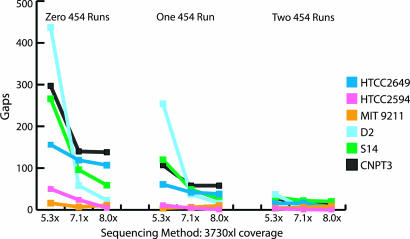
Fig. 2.
Results of mixed sequencing runs, ABI 3730xl, and 454 technology. Hybrid assembly comparison, number of sequencing gaps in assembly. The x axis represents the amount and type of data used for an assembly. The y axis represents the number of gaps remaining after assembly.
Table 1.
Hybrid data assembly
| Organism | Sequencing gaps | Hard stop gaps | Average sequence gap size | Missing sequence | Base pairs in 1× or 0× coverage | Q20 BP in Scaff | Percent Q20 in scaffold | Physical gaps | N50 contig | Coverage | Genome size | Cost difference, % |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIT9211 5.3× | 11 | 0 | 366 | 4,026 | 8,378 | 1,746,693 | 99.20 | 0 | 277,174 | 6 | 1,760,746 | −37.00 |
| MIT9211 5.3×–one 454 | 0 | 0 | 0 | 0 | 222 | 1,763,720 | 97.66 | 0 | 1,847,284 | 24 | 1,806,060 | 19.13 |
| MIT9211 5.3×–two 454 | 0 | 0 | 0 | 0 | 133 | 1,765,790 | 97.65 | 0 | 1,688,962 | 42 | 1,808,271 | 52.00 |
| MIT9211 two 454 only | 9 | 0 | 394 | 3,546 | 1,499 | 1,707,409 | 99.62 | 0 | 427,124 | 36 | 1,713,978 | 6.55 |
| MIT9211 8×–0 454 | 1 | 0 | 20 | 20 | 31 | 1,854,846 | 97.92 | 0 | 1,228,306 | 10 | 1,894,186 | 0.00 |
| HTCC2594 5.3× | 50 | 0 | 600 | 30,000 | 47,037 | 3,030,260 | 98.80 | 0 | 78,081 | 5 | 3,066,971 | −26.00 |
| HTCC2594 5.3×–one 454 | 11 | 0 | 746 | 8,206 | 5,161 | 3,048,942 | 99.61 | 0 | 265,562 | 15 | 3,060,803 | 9.42 |
| HTCC2594 5.3×–two 454 | 0 | 0 | 0 | 0 | 1,273 | 3,053,801 | 99.93 | 0 | 3,053,801 | 34 | 3,055,839 | 24.28 |
| HTCC2594 8×–one 454 | 1 | 0 | 156 | 156 | 386 | 3,053,178 | 99.98 | 0 | 1,655,186 | 18 | 3,053,827 | 26.50 |
| HTCC2594 8×–two 454 | 0 | 0 | 0 | 0 | 150 | 3,053,677 | 100.00 | 0 | 3,053,801 | 37 | 3,053,801 | 36.60 |
| HTCC2594 two 454 only | 50 | 0 | 4,863 | 243,150 | 6,019 | 3,029,260 | 92.34 | 0 | 125,688 | 29 | 3,280,646 | −14.00 |
| HTCC2594 8×–0 454 | 4 | 0 | 158 | 632 | 4,817 | 3,051,930 | 99.93 | 0 | 865,987 | 9 | 3,054,012 | 0.00 |
| HTCC2649 5.3× | 156 | 28 | 646 | 100,776 | 60,499 | 4,165,998 | 97.37 | 0 | 44,457 | 5 | 4,278,365 | −3.00 |
| HTCC2649 5.3×–1 454 | 61 | 11 | 235 | 14,335 | 29,059 | 4,216,813 | 99.40 | 0 | 121,019 | 12 | 4,242,346 | 2.00 |
| HTCC2649 5.3×–two 454 | 15 | 4 | 1,177 | 17,655 | 19,626 | 4,235,057 | 99.21 | 0 | 489,600 | 21 | 4,268,677 | 13.35 |
| HTCC2649 8×–one 454 | 38 | 11 | 248 | 9,424 | 14,016 | 4,233,589 | 99.64 | 0 | 200,552 | 15 | 4,249,087 | 22.12 |
| HTCC2649 8×–two 454 | 3 | 3 | 13 | 40 | 15,259 | 4,233,767 | 99.97 | 0 | 1,221,599 | 25 | 4,235,038 | 31.26 |
| HTCC2649 two 454 only | 114 | 20 | 6,757 | 770,298 | 12,073 | 4,117,973 | 83.57 | 1 | 54,348 | 16 | 4,927,326 | −33.00 |
| HTCC2649 8×–0 454 | 107 | 76 | 58 | 6,206 | 19,320 | 4,223,352 | 99.72 | 68,491 | 9 | 4,235,057 | 0.00 | |
| S14 5.3× | 266 | 0 | 983 | 261,478 | 149,819 | 5,072,226 | 94.76 | 10 | 29,979 | 5 | 5,352,635 | −24.00 |
| S14 5.3×–one 454 | 120 | 0 | 1,278 | 153,360 | 54,485 | 4,965,321 | 96.50 | 11 | 61,684 | 10 | 5,145,225 | 4.61 |
| S14 5.3×–two 454 | 30 | 0 | 4,500 | 135,000 | 15,594 | 5,044,842 | 97.20 | 5 | 351,723 | 17 | 5,190,011 | 17.97 |
| S14 8×–one 454 | 26 | 0 | 2,878 | 74,828 | 15,130 | 5,027,642 | 98.39 | 6 | 313,097 | 12 | 5,109,753 | 22.11 |
| S14 8×–two 454 | 18 | 0 | 4,000 | 72,000 | 8,493 | 5,095,940 | 98.35 | 5 | 404,679 | 21 | 5,181,460 | 31.25 |
| S14 two 454 only | 125 | 0 | 9,529 | 1,191,125 | 18,500 | 4,768,229 | 78.54 | 95 | 22,966 | 13 | 6,070,860 | −33.00 |
| S14 8×–0 454 | 59 | 0 | 1,979 | 116,761 | 57,071 | 5,047,120 | 98.94 | 6 | 139,234 | 8 | 5,101,447 | 0.00 |
| CNPT3 5.3× | 297 | 0 | 1,009 | 299,673 | 105,754 | 2,698,477 | 89.27 | 8 | 14,163 | 4 | 3,022,992 | −18.00 |
| CNPT3 5.3×–one 454 | 107 | 0 | 1,232 | 131,824 | 40,304 | 2,973,045 | 94.84 | 5 | 67,811 | 11 | 3,134,706 | 20.83 |
| CNPT3 5.3×–two 454 | 12 | 0 | 3,400 | 40,800 | 8,884 | 3,038,758 | 97.62 | 0 | 286,979 | 23 | 3,112,910 | 34.73 |
| CNPT3 8×–one 454 | 42 | 0 | 2,316 | 97,272 | 20,524 | 3,038,403 | 96.74 | 1 | 127,506 | 13 | 3,140,913 | 30.90 |
| CNPT3 8×–two 454 | 12 | 0 | 3,387 | 40,644 | 9,610 | 3,070,512 | 98.38 | 0 | 286,979 | 24 | 3,121,209 | 41.73 |
| CNPT3 two 454 only | 88 | 0 | 2,977 | 261,976 | 7,480 | 2,918,009 | 90.91 | 18 | 44,842 | 19 | 3,209,644 | 5.72 |
| CNPT3 8×–0 454 | 136 | 0 | 1,305 | 177,480 | 58,925 | 2,932,134 | 95.31 | 3 | 47,074 | 6 | 3,076,427 | 0.00 |
| D2 5.3× | 437 | 0 | 417 | 182,303 | 196,439 | 4,808,623 | 98.47 | 28 | 14,970 | 4 | 4,883,418 | −42.00 |
| D2 5.3×–one 454 | 254 | 0 | 380 | 96,523 | 118,977 | 4,895,192 | 99.05 | 27 | 26,468 | 8 | 4,942,189 | −16.00 |
| D2 5.3×–two 454 | 37 | 0 | 614 | 22,733 | 39,971 | 4,934,010 | 99.31 | 12 | 155,774 | 15 | 4,968,469 | −1.00 |
| D2 8×–one 454 | 16 | 0 | 1,161 | 18,570 | 40,346 | 4,944,178 | 99.46 | 12 | 269,204 | 13 | 4,971,208 | 20.56 |
| D2 8×–two 454 | 5 | 0 | 2,430 | 12,148 | 28,912 | 4,975,666 | 99.62 | 10 | 660,254 | 20 | 4,994,573 | 29.29 |
| D2 two 454 only | 77 | 0 | 9,320 | 717,640 | 16,195 | 4,593,629 | 84.54 | 130 | 21,045 | 12 | 5,433,827 | −62.94 |
| D2 8×–0 454 | 23 | 0 | 986 | 22,685 | 42,629 | 4,921,016 | 98.77 | 14 | 253,418 | 9 | 4,982,425 | 0.00 |
Comparison of gaps (type and size), base pairs in low-coverage areas and high-quality areas, as well as coverage, N50 numbers, and cost differentials. The assembly quality score is a rough indication of assembly quality, with a higher number being an assembly of lower quality based on the equation in the table.
For the two genomes containing very few sequencing gaps after an assembly of 8× Sanger-generated coverage, MIT 9211 and HTCC2594, the remaining sequencing gaps were easily closed with the addition of 454 data. By using 5.3× Sanger sequencing data plus one 454 sequencing run for MIT 9211 and 5.3× Sanger sequencing data plus two 454 sequencing runs for HTCC2594, all sequencing gaps were closed.
The 454 platform has proven to be effective in sequencing through hard stops that are often found in genomes with high GC content (>60% GC). One of the six genomes, HTCC2649, fell into this category and showed the biggest reduction in the number of gaps with the addition of 454 data. Seventy-six of the 107 gaps present at 8× Sanger sequencing were found to be hard stop gaps. The addition of two 454 runs to the 8× Sanger data closed all but three of these gaps. In addition, the N50 contig size for HTCC2649 increased by a magnitude of 5.5 times over that attained through Sanger sequencing alone.
Although the number of gaps decreased with the addition of 454 data, the total amount of missing sequence increased in many cases, leading to the theory that many small gaps were being closed, whereas larger gaps remained essentially unaffected.
The hypothesis that clonal bias is not introduced with 454 sequencing would suggest that the number of physical ends of an assembly would drastically decrease with the addition of the 454 pseudoreads (see Informatics). However, this was not found to be the case. The addition of 454 data was observed to have a much lower impact on the number of physical ends than on the number of sequencing gaps. Physical ends are created within the assembly process when no clonal information exists to link scaffolds. These ends are usually the by-product of clonal bias or large repeat structure. The addition of 454 reads did not help link physical ends in these assemblies.
Repeat Structure.
The addition of 454 data did not increase the quality of assembly in repetitive areas of any of the six genomes. After the addition of two 454 runs, there were still >100 significant repeat structures (>500 bp) found in each of D2 and VAS14 and >50 found in CNPT3. A direct correlation between the remaining gaps in the hybrid assemblies and the number of repeats in the genomes was found. This observation is supported by a comparative genomics study in which a de novo assembly of Escherichia coli was performed by using only 454-generated data (www.454.com/downloads/454_case_study_denovo.pdf), with the results showing that many of the gaps remaining after 454 sequencing corresponded to known repeats within the Sanger sequence finished genome.
Low Coverage and Q20 Values.
Many characteristics, in addition to gap number, are important in identifying the quality of a genome assembly. It is evident from our results that a significant number of sequencing gaps can be closed, or at the very least contigs extended, with the addition of 454 sequencing data to Sanger sequencing data. However, it is important to determine whether the gaps being closed, as well as the areas of low coverage being filled, are of high-quality sequence.
For the six microbes sequenced here, the addition of 454 sequencing data decreased the areas of low coverage, in some cases by two orders of magnitude. For genomes containing many sequencing gaps, this correlated with more of the genome lying in low coverage areas. These genomes also saw a large reduction of nucleotides in low coverage areas, with S14 dropping from 57,000 to 8,500 bp, as shown in Table 1. MIT9211 contained only 100–200 bp in low coverage regions after 5.3× Sanger sequencing and two 454 runs.
Another measure of assembly quality is the total number of Q20 base pairs found in scaffolded contigs. For all six microbes, Q20 values increased when two 454 runs were incorporated with 8× Sanger sequencing coverage. When comparing 5.3× Sanger sequencing plus one or two 454 runs to 8× Sanger sequencing alone, we found the number of Q20 bases within an assembly varied by only 0.5–1% in most cases.
Sequencing Strategy.
This study shows that the 454 technology is able to sequence regions of a genome for which Sanger sequencing is ineffective (i.e., hard stops). However, in other difficult areas, such as large sequencing gaps, repetitive areas, and physical ends, the addition of 454 sequencing data does not significantly improve assembly quality. For the marine microbes sequenced, we were able to determine that an initial assembly of 5.3× Sanger sequencing data is sufficient to determine final genome size and devise a cost-effective plan for additional sequencing. Encountering nonclonable regions and hard stops at this stage would warrant 454 sequencing runs, whereas large sequencing gaps, repetitive areas, and physical gaps would warrant additional Sanger sequencing reads.
To determine a sequencing strategy, we needed more informative measures of assembly quality, other than gap numbers alone. Several factors were taken into account, including genome size, the number of physical and sequencing gaps, type of sequencing gap, total missing sequence, sequence coverage, sequence coverage quality, and overall genome repeat structure. Both standard sequencing gaps and hard stops were given heavier weight in the evaluation, because the 454 technology seems to be more effective in closing these types of gaps. Coverage was taken into account, with high coverage adding significant weight to the evaluation, because a genome with high coverage and more gaps could be seen as being in poor condition and, therefore, a good candidate for 454 sequencing. After initial assembly of 5.3× Sanger sequencing data and depending on the evaluation criteria outlined in Fig. 3, genomes would either receive 454 processing consideration or continue to standard 8× Sanger sequencing coverage.
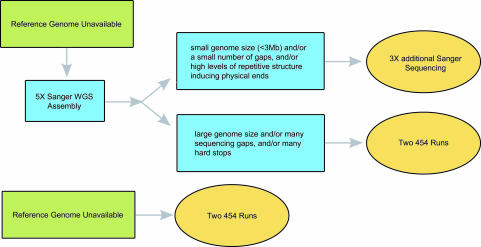
Fig. 3.
Decision tree for hybrid sequencing strategy. For organisms with a small genome size (<3 Mb) and/or a small number of gaps and/or high levels of repetitive structure inducing physical ends, we found 8× Sanger sequencing to be the most cost-effective approach. For organisms with a large genome size, many sequencing gaps, and/or hard stops, we found initial sequencing of 5.3× Sanger data followed by the addition of two 454 runs to be the most cost-effective approach.
Discussion
In this study, we evaluated the use of both 454 and 3730xl Sanger sequencing data to generate high-quality low-cost draft assemblies of small marine microbial genomes. Our goal was to determine the optimal combination of 454 and Sanger sequencing data that would produce the best possible high-quality genome assembly in the most timely and cost-effective manner for marine microbial genomes.
A severe degradation in assembly was found when using less than 5.3×** Sanger sequencing data, with or without the addition of 454 sequencing data. Therefore, 5.3× Sanger sequencing data were used as a baseline for optimal data combination studies.
By increasing the amount of 454 sequencing data at any ratio to Sanger sequencing data, we observed an improvement to the final draft genome in terms of coverage, reduction of gaps, and reduction of poorly sequenced regions that degrade the value of an assembly. Also, we observed that using two 454 runs, without the addition of Sanger sequencing data, could be a viable solution for comparative genomics if a reference genome were available (www.454.com/downloads/454_case_study_denovo.pdf). This could reduce the sequencing cost 20% on average when compared to sequencing using the Sanger-only approach, as shown in Table 1.
When using a combination of one or two 454 runs and 5.3× Sanger data, the addition of more Sanger reads did not improve the quality of the assembly enough to justify the added expense of additional Sanger sequencing. Although sequencing gap numbers were reduced, the overall gap reduction was not enough to warrant additional high-throughput sequencing.
From this study, we determined that a two-tiered sequencing strategy is ideal for cost-effective high-throughput sequencing of microbial genomes (see Fig. 3). Initial sequencing and assembly of 5.3× Sanger data are used to determine final genome size and map out a cost-effective plan for additional sequencing. For organisms with a small genome size (<3 Mb) and/or a small number of gaps and/or high levels of repetitive structure inducing physical ends, continuing to 8× Sanger coverage is the most cost-effective approach. For organisms with a large genome size, many sequencing gaps and/or hard stops, continuing with two 454 runs, is the most cost-effective approach.
The choice and justification of finishing a genome depend heavily upon the scope of a project. A working draft assembly can be produced more quickly than a finished draft assembly and can represent up to 99% of a genome’s sequence. However, gaps and low-quality regions may still remain in the data, decreasing the value of the working draft for studying DNA features that span large regions or require high accuracy. In cases where a genome is slated for finishing, the ≈10–20% increase in initial sequencing cost associated with 454 sequencing is justified by reducing the cost of closure by 25% or more. In such cases, a hybrid assembly is a sound approach for de novo sequencing of microbial genomes.
By adding 454 sequencing data to Sanger sequencing data for the six marine genomes processed in this project, we were able to obtain a higher-quality genome assembly with fewer gaps than would have been produced by using Sanger sequencing data alone. For those interested in finishing genomes, the use of 454 sequencing data as a prefinishing technique is advantageous. Traditional genome finishing is very time-consuming, labor-intensive, and costly. Using the 454 technology decreases, significantly, the work load, time, manual labor, and rearraying instrumentation needed for conventional finishing, leading to an ≈25% reduction in cost.
Conclusions
In this study, we were able to explore the utility and cost-effectiveness of a hybrid-sequencing approach that incorporates 454 sequencing data with conventionally generated Sanger sequencing data to produce high-quality cost-effective assemblies of small marine microbial genomes. We observed that, by combining low-sequence high-clone coverage of mate-pair Sanger sequencing reads with 454 sequencing data, a very good first-draft genomic sequence can be generated. More importantly, we observed that the 454 sequencing platform is capable of producing sequence coverage for microbial genomes that can be used for closing gaps in assembly projects, especially where sequencing or cloning bias exists with current technologies, without a reduction in sequence quality.
We believe that the results obtained in this study will make a substantial difference in how microbial sequencing is performed. For years, conventional Sanger sequencing has been the foundation for whole-genome shotgun microbial sequencing. By combining the advantages of two sequencing technologies in a hybrid approach, we have been able to produce better-quality draft microbial genome assemblies in the same amount of time. This hybrid sequencing strategy has the potential for being the first in years to have a major impact on the sequencing community.
Additional developmental efforts by 454 to improve their technology and explore new applications are under way. These improvements include longer read lengths (180 bp+), the generation of 100 million bases per run, paired end-read capabilities, and improvements to the 454 assembler that will allow for the assembly of larger genomes. Undoubtedly, the introduction of this system and its improvements into our microbial sequencing pipeline will allow for an increase in sequencing capacity and higher-quality genome assemblies with fewer sequencing gaps.
Materials and Methods
Laboratory.
454 processing.
The laboratory methods and protocols used in this experiment were as described by Rothberg and coworkers (2). For each of the six microbes sequenced, 5 μg of microbial DNA was used for the library preparation step. The input genomic DNA met the following requirements: (i) It was double-stranded, (ii) it was nonamplified, (iii) it was nondegraded, (iv) it had DNA fragments of >1.5 kb, (v) it had an OD 260/280 reading of ≥1.8, (vi) it had a concentration of ≥50 ng/μl, and (vii) it was suspended in TE buffer (10 mM Tris/1 mM EDTA, pH 8.0). Each microbial DNA sample was sequenced by using two 454 sequencing runs.
High-throughput AB 3730xl Sanger sequencing processing.
The high-throughput sequencing process for the production of plasmid and fosmid reads included the construction of a small insert 3- to 4-kb plasmid and a 36- to 40-kb fosmid library, library QC, clone plating and picking, template production, and sequencing. These procedures are described below.
Library construction.
Genomic DNA was randomly sheared by nebulization to produce fragments with a distribution of ≈1–25 kb, end-polished with consecutive BAL31 nuclease and T4 DNA polymerase treatments, and size-selected using gel electrophoresis on 1% low-melting-point agarose. After ligation to BstXI adapters, DNA was purified by three rounds of gel electrophoresis to remove excess adapters, after which the fragments were inserted into BstXI-linearized medium-copy pBR322 plasmid vectors. The resulting library was electroporated into GC10 cells. To ensure construction of high-quality random plasmid libraries containing few to no clones without inserts and no clones with chimeric inserts, we used a series of vectors (pHOS) containing BstXI cloning sites that include several features: (i) the sequencing primer sites immediately flank the BstXI cloning site to avoid excessive resequencing of vector DNA, (ii) elimination of strong promoters oriented toward the cloning site, and (iii) using BstXI sites for cloning facilitates the preparation of libraries with a low incidence of no-insert clones and a high frequency of single inserts. Clones were sequenced from both ends, producing pairs of linked sequences representing ≈800 bp at the end of each insert.
The fosmid libraries were constructed by using ≈1 μg of DNA that was sheared by using bead beating to generate cuts in the DNA. The staggered ends or nicks were repaired by filling with dNTPs. A size-selection process followed on a pulsed-field electrophoresis system with lambda ladder to select for 39- to 40-kb fragments. The DNA was then recovered from a gel, ligated to the blunt-ended pCC1FOS vector, packaged into lambda packaging extracts, incubated with the host cells, and plated to select for the clones containing an insert.
Clone picking and inoculation.
Libraries were transformed, and cells were plated onto large-format (16 × 16-cm) diffusion plates, prepared by layering 150 ml of fresh molten antibiotic-free agar onto a previously set 50-ml layer of agar-containing antibiotic. Colonies were picked for template preparation by using Q-bot or Q-Pix colony-picking robots (Genetix, New Milton, Hampshire, U.K.) and inoculated into 384-well blocks containing liquid media and incubated overnight with shaking.
DNA template preparation.
High-purity plasmid DNA was prepared by using the DNA purification robotic workstation custom-built by Thermo CRS and based on the alkaline lysis miniprep (5), modified for high-throughput processing in 384-well plates. Bacterial cells were lysed, cell debris was removed by centrifugation, and plasmid DNA was recovered from the cleared lysate by isopropanol precipitation. DNA precipitate was washed with 70% ethanol, dried, and resuspended in 10 mM Tris·HCl buffer containing a trace of blue dextran. The typical yield of plasmid DNA from this method is ≈600–800 ng per clone, providing sufficient DNA for at least four sequencing reactions per template.
Sequencing reactions.
Sequencing protocols were based on the dideoxy sequencing method (1). Two 384-well cycle sequencing reaction plates were prepared from each plate of plasmid template DNA for opposite-end paired-sequence reads. Sequencing reactions were completed by using Big Dye Terminator (BDT) Chemistry, Ver. 3.1, Cycle Sequencing Ready Reaction Kits (Applied Biosystems) and standard M13 forward and reverse primers. Reaction mixtures, thermal cycling profiles, and electrophoresis conditions were optimized to both reduce the volume of the BDT mix to 1/32 of that recommended by the manufacturer and extend read lengths. Sequencing reactions were set up by Biomek FX (Beckman Coulter) pipetting workstations. Robots were used to aliquot and combine templates with reaction mixes consisting of deoxy- and fluorescently labeled dideoxynucleotides, DNA polymerase, sequencing primers, and reaction buffer in a 5-μl volume. Bar coding and tracking promoted error-free template and reaction-mix transfer. After 30–40 consecutive cycles of amplification, reaction products were precipitated by isopropanol, dried at room temperature, resuspended in water, and transferred to one of the 3730xl DNA Analyzers (Applied Biosystems). Set-up times were <1 hr, and 12 runs per day were completed with average trimmed sequence read lengths of >800 bp.
Informatics.
Pseudoread creation.
A mix of 454 “pseudoreads” and complementary Sanger reads were used as input into the Celera Assembler (6). The 454 reads/flows were not input directly to the Celera Assembler, because, unlike the 454 Newbler assembler, the Celera Assembler has not been optimized to use short reads with the error characteristics inherent in the 454 platform. Conversely, Newbler cannot make use of 3730xl reads. The 454 pseudoreads were generated by assembling the 454-generated flows with 454’s Newbler assembler (www.454.com/enabling-technology/the-software.asp) and then breaking or shredding these assemblies into overlapping and evenly spaced 600-bp fragments, while maintaining the effective depth of the assembly. The length of 600 bp was chosen to emulate the read size of standard Sanger data, for which the Celera Assembler is optimized. To minimize the possibility of misassembled 454 Newbler contigs, a threshold was set to break 454 contigs apart where their underlying depth was less than two reads deep. The depth of the 454 assembly in the resultant pseudoreads was maintained by controlling the stepping between consecutive shreds. In effect, the deeper the coverage we wanted to emulate, the smaller the stepping was between shreds (see Fig. 4). Because the depth of the coverage allows the assembler to differentiate between unique and repeat regions of the genome, the depth of the contigs needed to be maintained.
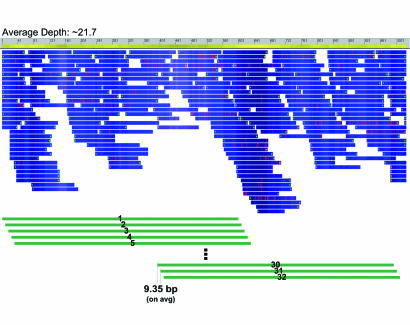
Fig. 4.
454 pseudoread creation and assembled contigs. In this example, the average depth of the 454 assembled contig was ≈21.7. The ungapped length of the contig was 890 bp. Because we used 600-bp pseudoread shreds, the number of pseudoreads we needed to generate was floor(contig_length × target_depth/pseudoread_length) or floor(890 × 21.7/600) = 32 pseudoreads. The distance between consecutive pseudoreads was approximated by (contig_length−pseudoread_length)/(num_pseudoreads−1) or (890–600)/(32−1) = 9.35 bp.
There are inherent limitations to this process, just as there are for using 454 reads directly. This methodology does not allow for the use of mate pairing from clone ends, so each pseudoread can be used for sequence coverage only. Higher-quality scores supersede lower-quality scores in generating a multialignment of sequences. By default, the bases in the pseudoreads are given a very low default quality score of three (a quality score of 20 is considered average). This allows the assembly algorithm to bias toward Sanger data over 454 data, because the overall error rate is believed to be lower in Sanger sequencing reads. Read incorporation percentages were studied to see whether the pseudoreads were incorporated at the same rate as Sanger reads. In a hybrid assembly, it was found that, for larger, more poorly assembled organisms, 454 data were incorporated at a higher percentage than Sanger data alone. For organisms with few gaps, it was found that 454 reads incorporated at a rate of 1–2% lower than Sanger data alone.
Assembly.
Each microbial genome was sequenced to 8× sequence coverage with Sanger reads, 7.5× from a 3–4 kb plasmid library and 0.5× from a 36–40 kb fosmid library. The plasmid reads were divided into five evenly distributed input packages for the Celera Assembler representing 0%, 20%, 40%, 60%, 80%, and 100% of the total reads sequenced. All fosmid reads were used in each assembly to help with scaffolding of contigs. One hundred and eight assemblies were carried out on the six microbial genomes using the six different levels of plasmid coverage, fosmid coverage, and either zero, one, or two 454 runs, corresponding to 0 bp, ≈20 Mbp, or ≈40 Mbp, respectively (6–18× coverage per 454 run depending upon the size of the genome).
Assembly analysis.
For each of the 108 assemblies, we collected information about the quality of the assembly such as gap number, gap size, N50 scaffold, contig numbers, etc. Each gap was further examined and categorized as either a sequencing gap (having clone coverage but no sequence coverage) or a physical gap (having neither clone nor sequence coverage). Each sequencing gap was then evaluated to determine whether it exhibited hard stop characteristics. The amount of missing sequence lost to gaps, based on the estimated genome size, as well as the number of Q20 (high-quality assembled bases) found in scaffold contigs, was then calculated. General repeat structures were characterized by manual review using visualization tools that are distributed with the Celera Assembler and The Institute for Genome Research repeat detection tools.
Acknowledgments
We express our appreciation to the following people for their donation of sample material: Dr. Rick Cavicchioli (University of New South Wales), Dr. Penny Chisholm (Massachusetts Institute of Technology, Cambridge, MA), Dr. Jang-Cheon Cho (Oregon State University, Corvallis, OR), Dr. Stephen J. Giovannoni (Oregon State University), Dr. Staffan Kjelleberg (University of New South Wales), Dr. Mary Ann Moran (University of Georgia, Athens, GA), and Dr. Art Yayanos (Scripps Institution of Oceanography, La Jolla, CA). In addition, we thank the Gordon and Betty Moore Foundation for the support of primary 3730xl genomic sequencing for the Marine Microbial Sequencing Project. This effort was also supported by the J. Craig Venter Science Foundation.
Footnotes
Conflict of interest statement: No conflicts declared.
**Although we labeled these as 5.3× coverage, there may have been more if slight contamination of the sample skewed our initial estimates of the genome size. This coverage estimate is for the main organism only; no contaminant is included.
References
- 1.Sanger F., Nicklen S., Coulson A. R. Proc. Natl. Acad. Sci. USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Margulies M., Egholm M., Altman W. E., Attiya S., Bader J. S., Bemben L. A., Berka J., Braverman M. S., Chen Y. J., Chen Z., et al. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ronaghi M., Karamohamed S., Pettersson B., Uhlen M., Nyren P. Anal. Biochem. 1996;242:84–89. doi: 10.1006/abio.1996.0432. [DOI] [PubMed] [Google Scholar]
- 4.Ronaghi M., Uhlen M., Nyren P. Science. 1998;281:363–365. doi: 10.1126/science.281.5375.363. [DOI] [PubMed] [Google Scholar]
- 5.Sambrook J., Fritsch E. F., Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 6.Huson D. H., Reinert K., Kravitz S. A., Remington K. A., Delcher A. L., Dew I. M., Flanigan M., Halpern A. L., Lai Z., Mobarry C. M., et al. Bioinformatics. 2001;17:S132–S139. doi: 10.1093/bioinformatics/17.suppl_1.s132. [DOI] [PubMed] [Google Scholar]