Abstract
The rat has served as an excellent model for studies on animal physiology and as a model for human diseases such as diabetes and alcoholism; however, genetic studies have been limited because of the inability to knock out genes. Our goal was to produce heritable deficiencies in specific gene function in the rat using RNA interference to knock down gene expression in vivo. Lentiviral-mediated transgenesis was used to produce rats expressing a short hairpin RNA targeting Dazl, a gene expressed in germ cells and required for fertility in mice [Ruggiu, M., Speed, R., Taggart, M., McKay, S. J., Kilanowski, F., Saunders, P., Dorin, J. & Cooke, H. J. (1997) Nature 389, 73–77]. Germ-line transmission of the transgene occurred, and its expression correlated with significant reductions in DAZL protein levels and male sterility, and the knockdown was stable over multiple generations (F1–F3). This study demonstrates an efficient system by which directed reverse genetic analysis can now be performed in the rat.
Keywords: Dazl, fertility, RNA interference
The Norway rat was the first mammal to be domesticated for scientific purposes, and over the last 150 years it has proven an invaluable research model, in part because of its size, fecundity, and ability to learn new behaviors with relative ease (1). The generation of new rat genetic models for the study of human diseases has been possible using forward genetic methods such as random mutagenesis and conventional transgenesis or through the discovery of serendipitous spontaneous mutations (2). However, reverse genetic methods crucial for analyzing the function of specific genes of interest are severely limited in the rat. Our goal was to produce heritable and stable deficiencies in specific gene function in this animal model. Gene inactivation through homologous recombination in ES cells is still not possible in the rat because of the lack of pluripotent ES cell lines similar to those available from the mouse. Instead, we chose to test an RNA interference (RNAi) approach that relies on the generation of transgenic rats.
RNAi is a reverse genetic technology that allows one to down-regulate gene expression by introducing into cells a short (≈19- to 21-nt) double-stranded RNA that is complementary to a target gene (3). In combination with the recent expansion of available genome sequence information, RNAi has provided a powerful tool affecting many areas of biological research. Whereas methods are now well established for performing RNAi in vitro using short interfering RNA, the application of RNAi to knock down gene expression in vivo is far less common. In most cases RNAi is achieved in vivo only transiently and is targeted to a particular tissue. Stable RNAi is usually accomplished by genetic modification of cells such that they carry a piece of DNA that contains a ubiquitous promoter (e.g., the Pol III promoters U6 or H1) that drives expression of a short hairpin RNA (shRNA). The shRNA is then processed to short interfering RNA by cellular machinery. Recent studies have shown that genetic modification of mice to express shRNA can be effective in down-regulating gene expression (4–9). Here we demonstrate the utility of this method to deplete a specific gene product in the rat to generate a new genetic model with a heritable phenotype, thereby showing that the creation of rat models with depletions in specific gene function is now possible.
Results and Discussion
Development of a Vector That Efficiently Suppresses Dazl Expression in Vitro.
To test the principle of using RNAi technology to disrupt gene function in the rat, we chose to target Dazl. DAZL, an RNA binding protein, is highly conserved among diverse species. It is expressed specifically in germ cells from late embryogenesis to adults and is known to be required for fertility in the mouse (10–14). Also, Dazl has a haploinsufficient phenotype in the mouse: heterozygous knockout males contain an elevated percentage of abnormal sperm cells relative to wild-type mice (13), suggesting that a partial reduction in DAZL protein levels in the rat could cause a measurable phenotype such as infertility.
The vectors used in this study are derived from pLL3.7 and contain separate GFP and shRNA expression elements as well as elements required for lentiviral packaging (8). The CMV promoter driving GFP expression was replaced with the ubiquitin C (Ubc) promoter (pLLU2G), and double-stranded DNA oligonucleotides coding for two different shRNAs designed to target Dazl were each ligated downstream of a U6 promoter [pLLU2G-Dazl1 (in Fig. 1) and pLLU2G-Dazl2]. To test the efficacy of each of the shRNAs in knocking down Dazl expression, we transduced FR cells (a rat embryonic skin fibroblast cell line) with virus carrying shRNA or control vectors and then transiently transfected the cells with DNA encoding a myc-tagged DAZL. Cells transduced with either pLLU2G-Dazl1 or pLLU2G-Dazl2 exhibited almost complete suppression of DAZL-MYC expression based on Western blot analysis (Fig. 2A and data not shown). Transduced cells were viable, and tubulin levels were not altered, suggesting that there were no obvious off-target effects (Fig. 2A and data not shown). Methods for in vitro propagation of male germ stem cells that express Dazl have recently been established (15, 16), and pLLU2G-Daz1 was also effective at knocking down endogenous DAZL protein in germ cells propagated in vitro (>50% reduction) (data not shown). Therefore, we conclude that the U6 promoter is active in rat cells and that the shRNAs produced are effective at knocking down DAZL protein levels in vitro.
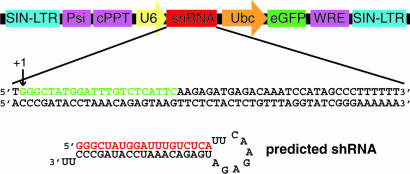
Fig. 1.
Dazl shRNA construct design. DNA oligonucleotides containing Dazl shRNA 1 (shown) and 2 (not shown) were inserted downstream of the U6 promoter in pLLU2G. The green sequence is equivalent to the Dazl mRNA target sequence. The presumed U6 transcription start site is designated as +1, and the predicted shRNA structure is shown. The red sequence corresponds to the antisense RNA probe generated for RNase protection assay. SIN-LTR, self-inactivating long terminal repeat; Psi, HIV packaging signal; cPPT, central polypurine track; U6, Pol III promoter; shRNA, small hairpin RNA; Ubc, Ubc promoter; eGFP, enhanced green fluorescent protein; WRE, woodchuck hepatitis virus response element.
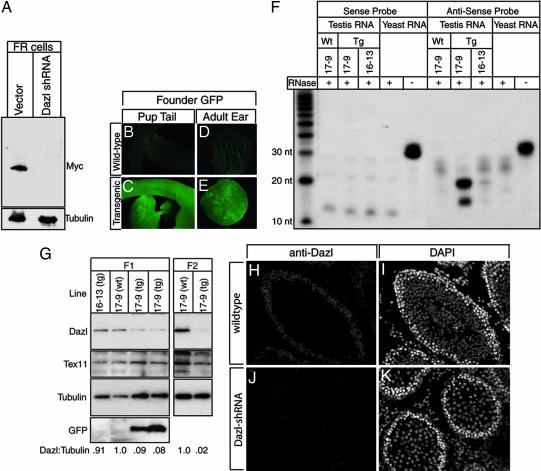
Fig. 2.
Transgene expression and DAZL knockdown. (A) Western blot analysis to detect DAZL-MYC production in cells transduced with virus carrying pLLU2G (Vector) or pLLU2G-Dazl1 (Dazl-shRNA) and transiently transfected with pCDNA-Dazl-myc. The blot was stripped and reprobed with anti-tubulin to verify equal loading of protein. (B–E) Fluorescence of wild-type (B and D) and transgenic (C and E) neonatal pups (B and C) or adult ear punch skin (D and E). (F) RNase protection assay using a probe to detect Dazl short interfering RNA (from Dazl shRNA 1) in testis RNA from transgenic rats (17-9 or 16-13 Tg), a wild-type sibling of the 17-9 transgenic rat (Wt), or yeast RNA (mock). The protected portion of the antisense probe sequence is shown in red in Fig. 1. The sense probe is reverse-complementary to the antisense probe. Addition of RNase is indicated by +. (G) Western blot analysis to detect Dazl, Tex11, tubulin, and GFP expression in the testis. Lanes were loaded as follows. (Left) First lane, transgenic 16-13; second lane, wild-type sibling of 17-9 F1; third and fourth lanes, two transgenic 17-9 F1s. (Right) First lane, wild-type sibling of 17-9 F2; second lane, transgenic 17-9 F2. (H–K) Immunofluorescence to detect DAZL protein in testis cryosections of a Dazl-shRNA rat (H) and a wild-type sibling (J). DAPI (DNA) staining of the same sections is also shown (I and K).
Production of Dazl-shRNA Rats.
To generate transgenic rats carrying pLLU2G-Dazl1, lentivirus was injected under the zona pellucida of fertilized pronuclear eggs, which were then transplanted into the oviducts of pseudopregnant surrogate females as described (17, 18). Transgenic founders were identified by PCR on DNA isolated from a tail biopsy, and Southern blotting demonstrated that the transgene likely integrated into a single genomic site in all founders (data not shown). Ten of the 75 (13%) rats produced were found to be transgenic (Table 1). Transgene expression was evaluated in each founder by observing GFP fluorescence in a biopsy of ear skin, and 6 of the 10 founders had detectable GFP expression (Table 1 and Fig. 2 D and E). All of the founders were bred to wild-type animals to assess fertility and to establish transgenic lines. Although no discernable effects on fertility were observed, it was apparent that several of the founders that expressed the transgene exhibited varying degrees of somatic mosaicism (data not shown). Therefore, the lack of germ-line transmission in several of the founders that had a mosaic pattern of expression is likely because of the mosaic contribution of the transgene in the germ line. These data are in agreement with other studies showing that injection of lentivirus is an effective method for generating transgenic rats, with mosaicism being a common occurrence (18, 19).
Table 1.
Fecundity and germ-line transmission of transgenic founders
| Founder |
GFP expression* |
Litter size |
Transmission† |
||||
|---|---|---|---|---|---|---|---|
| No. | Sex | Overall | Relative | Mean | No. | Tg/total F1 | % |
| 8-8 | M | − | 1.1 | 12 | 3 | 0/36 | 0 |
| 16-13 | M | − | 1.1 | 9 | 1 | 5/9 | 55 |
| 15-12 | M | − | 1.2 | 11 | 2 | 0/21 | 0 |
| 16-4 | F | − | 1.3 | 16 | 2 | 0/32 | 0 |
| 16-14 | M | + | 1.7 | 11 | 5 | 0/48 | 0 |
| 17-11 | M | + | 1.8 | 10 | 7 | 0/69 | 0 |
| 17-9 | F | + | 1.9 | 11 | 5 | 28/54 | 52 |
| 16-15 | F | + | 2.1 | 8 | 5 | 0/27 | 0 |
| 1-9 | M | + | 2.8 | 13 | 4 | 0/48 | 0 |
| 15-10 | F | + | 2.9 | 11 | 5 | 0/56 | 0 |
M, male; F, female; Tg, transgenic.
*Overall presence of detectable GFP expression was assessed by comparing fluorescence in transgenic rats with wild-type rats in 2-mm punches of ear skin using a stereoscope. Relative amounts of GFP expression were estimated by using imagej software to obtain a transgenic-to-wild-type ratio of fluorescence in an ear punch for each founder.
†F1 of total progeny genotyped by using PCR.
Two founders (16-13 and 17-9) transmitted the transgene to progeny in a Mendelian fashion, and lines were established. Line 16-13 exhibited almost no GFP expression (Fig. 2G); accordingly, there was no effect on fecundity, and no other phenotypes were observed. In striking contrast, males from line 17-9, which exhibited moderate levels of GFP expression (Fig. 2G), were completely sterile (n = 5 males), whereas females were fully fertile (n = 11 litters from three females with average litter size of 11 pups). These results were consistent with the possibility that Dazl expression was knocked down and germ cell development was perturbed in males. To determine whether the observed sterility was due to transgene-mediated RNAi, we first analyzed the testis for production of shRNA. Using a probe complementary to a portion of the shRNA (red sequence in Fig. 1) we were able to detect a small RNA (≈20 nt) in transgenic animals from line 17-9, but not 16-13, using an RNase protection assay (Fig. 2F). All subsequent experiments were focused on line 17-9, and males from this line we now refer to as “Dazl-shRNA” rats.
DAZL Protein Levels Are Significantly Reduced in Dazl-shRNA Rats.
DAZL protein was consistently (n = 8 animals) reduced in testes of Dazl-shRNA rats compared with wild-type siblings based on Western blot analysis (≈70% reduction) (see Materials and Methods). Importantly, knockdown was observed in F1, F2, and F3 progeny, indicating that the knockdown is stably inherited (Fig. 2G and data not shown). At the stage examined (6 weeks), the seminiferous tubules of transgenic rats comprised the normal distribution of germ cells (data not shown). Consistent with this observation, expression levels of another germ cell marker, Tex11 (see Materials and Methods), were unchanged, likely eliminating the possibility that the reduced levels of DAZL protein in Dazl-shRNA rats is merely a reflection of a change in germ cell numbers in total testis lysate. To confirm that the observed reduction in Dazl expression depended on robust transgene expression, we also examined DAZL protein levels in testes of males from line 16-13, which have minimal transgene expression. DAZL protein levels in testes of rats from this line were similar to wild-type animals (Fig. 2G), indicating that the knockdown was dosage-dependent.
A significant reduction in DAZL protein levels in Dazl-shRNA rats was also revealed by immunofluorescence analysis of cryosectioned testis (Fig. 2 H–K). DAZL was almost completely absent in spermatocytes in Dazl-shRNA rats, where it is normally expressed at very high levels. Low levels of DAZL protein remained in spermatogonia of Dazl-shRNA rats, which likely explains the incomplete knockdown observed by Western blot analysis of total testis lysates. The exact reasons for the different degrees of knockdown as a function of germ cell development are not known; however, a few possibilities are cell stage-dependent differences in the half-life of Dazl mRNA, or in the expression or processing of shRNA, or effectiveness of short interfering RNA in knocking down gene expression.
Male Dazl-shRNA Rats Are Sterile.
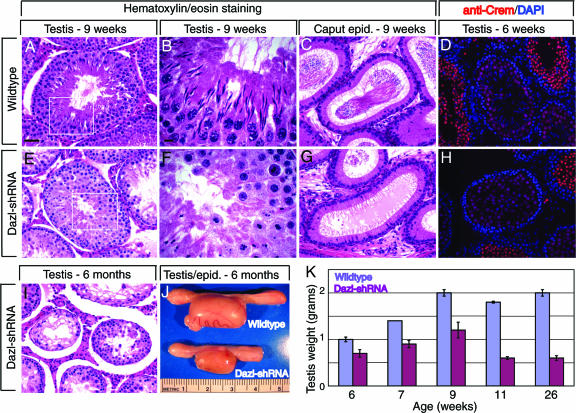
Over the course of the study Dazl-shRNA males never sired progeny, although they did produce copulatory plugs when paired with wild-type females (see Materials and Methods). Therefore, we further characterized the males to ascertain whether the cause of sterility was similar to that observed in Dazl knockout mice. The testes of transgenic males were noticeably smaller (67% and 30% at 6 weeks and 26 weeks, respectively) than those of wild-type siblings (Fig. 3J and K). To determine whether germ cells in Dazl-shRNA males could undergo meiosis, immunofluorescence analysis was used to detect SCP1 (synaptonemal complex 1, meiotic spermatocytes) and CREM-τ (haploid spermatids). At 6 weeks, transgenic and wild-type rats had a similar pattern of both markers, indicating that the transgenic germ cells could complete meiosis (Fig. 3 D and H and data not shown). However, histological staining of testis (Fig. 3 A, B, E, and F) or epididymis (Fig. 3 C and G) from 9-week-old Dazl-shRNA rats revealed that mature sperm were not produced. In the testes of wild-type siblings, the process of spermiogenesis can be visualized by dramatic changes in nuclear morphology (elongation). In contrast, Dazl-shRNA males had only abnormal cells with tightly rounded nuclear morphology in the lumen of the seminiferous tubules. Significant disruption of the stereotypical germ cell associations and cellular degeneration in the lumen were also observed (Fig. 3E). By 6 months the depletion of germ cells was much more pronounced in Dazl-shRNA rats, with many tubules appearing to contain only somatic cells (Fig. 3I). These observations suggest that during the first wave of spermatogenesis germ cells in Dazl-shRNA males can progress through meiosis but not spermiogenesis, and during subsequent spermatogenic waves the germ cells die at earlier points in development, leading to a nearly complete loss of germ cells. Thus, Dazl-shRNA rats have defects in germ cell development and reduced germ cell viability, consistent with phenotypes observed in Dazl knockout mice.
Fig. 3.
Analysis of Dazl-shRNA phenotype. (A–C, E–G, and I) Hematoxylin/eosin histological staining of testis (A, B, E, and F) and epididymis (C and G) from an adult (9-week-old) Dazl-shRNA rat (E–G) and a wild-type sibling (A–C). (D and H) CREM immunofluorescence (red) and overlaid DAPI staining (blue) in testis of a young (6-week-old) Dazl-shRNA rat (H) or a wild-type sibling (D). [Scale bars: 50 μm in A (for A, C–E, and G–I) and 10 μm in B (for B and F).] (I) Hematoxylin/eosin histological staining of testis from a 6-month-old Dazl-shRNA rat. (J) Testis and epididymis from 6-month-old wild-type (Upper) or Dazl-shRNA (Lower) rat. (K) Graphical representation of mean testis weight (in grams) at various ages. The following numbers of testes were examined: at 6 weeks old, 4 wild-type and 10 transgenic; at 7 weeks old, 2 wild-type and 6 transgenic; at 9 weeks old, 2 wild-type and 4 transgenic; at 11 weeks old, 4 wild-type and 10 transgenic; at 26 weeks old, 2 wild-type and 4 transgenic.
The reported phenotypes in Dazl knockout mice are highly variable (from embryogenesis through meiosis), dependent in large part on the genetic background (11, 13, 14). The germ cells in young Dazl-shRNA rats are capable of developing further than those in Dazl knockout mice, most likely because of the continued presence of low levels of DAZL protein. The defects we observed in nuclear morphology during spermiogenesis in Dazl-shRNA rats may reveal a previously unappreciated function for Dazl late in germ cell development and is in agreement with a report that DAZL protein is present in the acrosome region of elongating spermatids (12). It is conceivable that Dazl functions in several stages of germ cell development given the proposed function of Dazl as a translational regulator with multiple downstream targets (20). Although no effect on female fertility was observed in line 17-9, we observed that female descendents of founder 17-9 lacked detectable transgene (GFP) expression, possibly as the result of a sex-specific position effect. Therefore, it remains unclear whether Dazl is required for female fertility in rats, as it is in mice. The lack of transgene transmission from several other female and male founders with significant GFP expression (Table 1) is consistent with the idea that transgenic gamete production in these founders was disrupted by the expression of Dazl shRNA. However, we cannot rule out the possibility that the transgene never contributed to the germ line.
RNAi as a Tool for Studying Gene Function in Vivo.
Presently there are very few examples of endogenous gene knockdown in vivo using shRNA-producing transgenes. Therefore, this study represents one of a small but growing number of studies demonstrating that in vivo RNAi can be used to significantly and stably deplete an endogenous gene product, resulting in a heritable phenotype (4–7, 21). Furthermore, to our knowledge, this is the first report of endogenous gene knockdown in the rat, an excellent model system for human disease in need of reverse genetic methods. Because it is still not possible to produce gene knockouts through homologous recombination in the rat, the method we describe should allow for the efficient generation of new rat genetic models in which specific gene function is permanently disrupted. Based on the findings in this study, it is reasonable to estimate that ≈50% of transgenic founders express the transgene at detectable levels and that, even with a low transgene copy number, significant knockdown (>70%) can be achieved. With somewhat higher transgene copy number, it is likely that even greater levels of knockdown could be achieved, possibly approaching complete gene function ablation.
There are several advantages to using in vivo RNAi over traditional knockout methods. First, production of the transgene can be achieved quickly because of the simplicity of design. Second, shRNA expression vectors are small and therefore amenable to delivery by lentiviral packaging, an efficient method for generating transgenic animals (18). Finally, in vivo RNAi can generate animals with different degrees of gene deficiency. An allelic series of hypomorphs is often informative for modeling human disease phenotypes. For example, Hemann et al. (22) found that hematopoietic stem cell lines expressing shRNAs targeting the tumor suppressor gene Trp-53 produced a range of tumor phenotypes upon introduction into lethally irradiated mice depending on the degree of p53 knockdown. Our data also demonstrate the utility of RNAi in creating a hypomorph exhibiting a defect in spermiogenesis, a phenotype that may otherwise have been masked in the context of a null allele.
As is the case for gene knockouts, the method described here results in ubiquitous down-regulation of gene expression. However, for many gene function studies conditional gene knockdown would be preferable. One particularly promising new method involves using a chosen tissue-specific RNA polymerase II promoter to drive microRNA (miRNA) engineered to target a gene of interest (23, 24). Now that the foundation has been lain for reverse genetic analysis in rats through in vivo RNAi, regulated shRNA/miRNA expression will be an important next step for universally applying the method to gene function analysis.
Materials and Methods
Construction of Plasmids.
pLL3.7 (8) was modified by removing the CMV promoter by using NotI/NheI digestion, creating blunt ends by Klenow reaction, and ligating the human Ubc promoter (1.2 kb) derived from pUBC-6-V5-hisA and described by Nenoi et al. (25). The Ubc promoter is a polymerase II promoter thought to have a wider cellular activity than CMV (26), in particular in germ cells (Zhuoru Wu, personal communication, and D.L.G., unpublished observation), and is less likely to become silenced. The resulting vector, called pLLU2G, was further modified by introducing DNA oligos into the unique HpaI site to create pLU2G-Dazl1 and pLU2G-Dazl2. Oligos used were Dazl1 sense (5′-GGGCTATGGATTTGTCTCATTCAAGAGATGAGACAAATCCATAGCCCTTTTTT-3′) and Dazl2 sense (5′-TGTTGATCCAGGAGCTGACCTTCAAGAGAGGTCAGCTCCTGGATCAACTTTTTT-3′). Sense and antisense 5′ phosphorylated DNA oligos (Integrated DNA Technologies) were mixed at 1 μM into 70 mM Tris (pH 7.5), 10 mM MgCl2, and 100 mM KCl and annealed by using a thermal cycler (program: 95°C for 30 s, 60°C for 10 min, ramp from 60°C to 20°C at −1°C for 15 s). Annealed oligos were diluted 10-fold before ligating into linearized and dephosphorylated pLLU2G. Ligations (1 μl of a 20-μl reaction) were transformed into Fusion Blue cells (BD Biosciences). Bacterial colonies were screened for insert by PCR with the following primers: TD143, 5′-CAGTGCAGGGGAAAGAATAGTAGAC3′; TD144, 5′GCGGCCGCTTAAGCTTGGAACCC-3′. DNA subclones containing insert were sequenced to identify those with the correct sequence and insert orientation.
To construct pCDNA-Dazl-myc, Dazl was PCR-amplified from 25-day rat testis cDNA by using Expand High-Fidelity polymerase (Roche) and the following primers: TD197b, 5′-GGGAAGCTTATCATGCCAAACACTGTTTTTG-3′; TD198, 5′-CCCTCTAGAGATTTTTGCCTTTTGTGGGCC-3′. The PCR product and pCDNA4-myc/His (Invitrogen) vector were each digested with XbaI and HindIII and ligated by using standard procedures. The sequence of the insert was verified and matched the National Center for Biotechnology Information database entry XM_236905.1.
Lentiviral Production.
A total of 2 × 106 293 FT (Invitrogen) cells were plated on 10-cm gelatin-coated plates in 10 ml of DMEM plus 10% FBS. After 18 h, cells were transfected with a 1-ml (per plate) calcium phosphate precipitation mixture containing 3 μg of pMD2G, 5 μg of pMDLG/pRRE, 2.5 μg of pRS-REV, and 10 μg of pLLU2G (or pLLU2G-Dazl1/2). Ten milliliters of fresh DMEM plus 10% FBS was applied 24 and 48 h after transfection and collected at 48 h and 72 h after transfection. Collected medium was centrifuged at 2,500 × g for 15 min and filtered [Steriflip Duraflip 0.45 μM poly(vinylidene difluoride); Millipore]. Lentiviral particles were concentrated by subjecting filtered media (28 ml) to ultracentrifugation at 25,000 rpm for 2 h at 4°C in a Beckman SW28 swinging bucket rotor. Pellets were resuspended by incubating in 20–50 μl of PBS for 2–18 h at 4°C before storage at −80°C in 10-μl aliquots. Lentivirus was titered by transducing 4 × 105 FR (for experiments in FR cells) or 293 FT (for injections and other experiments) cells in 1 ml of medium containing 6 μg/ml polybrene in a 60-mm well and using FACS analysis to quantitate the percentage of GFP-positive cells 48–72 h after transduction.
In Vitro Knockdown in FR Cells.
FR cells (ATCC CRL-1213) are fibroblasts derived from rat embryonic skin. A total of 3 × 105 FR cells were plated in a 60-mm well. Cells were transduced (multiplicity of infection ≈ 6) with virus (pLLU2G or pLLU2G-Dazl1), transfected by using Lipofectamine 2000 (Invitrogen) with 2 μg of pCDNA-DazMyc plasmid DNA 48 h after transduction, and lysed for immunoblotting 72 h after transduction.
Production of Transgenic Rats and Genotyping.
Transgenic rats were produced in a Sprague–Dawley (Harlan) background by microinjection of lentiviral particles (≈3 × 105 transducing units per μl) under the zona pellucida of one-cell pronuclear eggs by using an intracytoplasmic sperm injection needle (Eppendorf). Embryos that survived injection were transferred into the oviduct of day-0.5 pseudopregnant females (17, 18). Founder rats were identified by PCR on tail genomic DNA by using the following primers: EGFP-S11, 5′-CTGACCCTGAAGTTCATCTGCACCAC-3′; EGFP-1-3, 5′-TCCAGCAGGACCATGTGATC-3′. Southern blot analysis was performed by using a probe corresponding to the GFP sequence to confirm the presence of the transgene and to estimate the number of transgene integration sites. Founders were bred to wild-type Sprague–Dawley rats to test for germ-line transmission and to establish lines. Animals were kept in conventional housing conditions with a 12-h/12-h light/dark cycle and fed a Teklad rodent diet (Harlan Teklad) ad libitum. All procedures involving animals were approved by the Institutional Animal Care and Use Committee of the University of Texas Southwestern Medical Center. In accordance with the guidelines of the Rat Genome and Nomenclature Committee, line 17-9 has been designated Tg(Ubc-eGFP/Dazl-shRNA)17-9Gar, and line 16-13 has been designated Tg(Ubc-eGFP/Dazl-shRNA)16-13Gar.
Fertility Tests and Testes Analysis.
Five Dazl-shRNA males (aged from 2 to 5 months) were housed with one or two wild-type females for 1 week to several months, and no progeny were produced. During parts of this time, females were examined for the presence of copulatory plugs. Each male produced at least one copulatory plug, confirming that their ability to mate was not affected. For Fig. 3K, testes were removed from transgenic and wild-type sibling rats and trimmed of fat before weighing.
RNase Protection Assay.
Total RNA enriched in small RNAs was isolated from 200 mg of testis by using the mirVana miRNA isolation kit (Ambion). The probes were constructed by using DNA oligonucleotides (TD358 for antisense, 5′-TGAGACAAATCCATAGCCCTTCCTGTCTC-3′, and TD341 for sense, 5′-GGGCTATGGATTTGTCTCACCTGTCTC-3′) and the mirVana miRNA probe construction kit (Ambion). The mirVana miRNA detection kit was used according to the standard recommendations with the following changes: 4.2 μg of RNA were present in each hybridization reaction, including 1.7 μg of small RNA from testis (or 0 μg for mock) and the balance being that of yeast RNA. Hybridization was performed at 42°C overnight. Decade markers (Ambion) were used as size standards.
Immunocytochemistry.
Tissues were fixed by immersion in 4% paraformaldehyde overnight at 4°C, equilibrated in 10% and then 30% sucrose/PBS, and embedded in OCT (optimal cutting temperature, Tissue Tek) in a HistoCool 2-methylbutane bath (−55°C). Cryosections (12 μm) were applied to SuperFrost/Plus slides and stored at −80°C. For immunostaining slides were rinsed in PBS, blocked in 0.1 M glycine/PBS for 15 min, rinsed in PBS, and permeabilized/blocked in PBT/block [PBT is 0.1% Triton X-100 in PBS, and block is 10 mg/ml IgG-free BSA (Jackson ImmunoResearch)] for 30 min. Primary antibody was applied overnight in PBT/block at 4°C followed by three washes in PBT. Secondary antibody was applied with DAPI (1 μg/ml) for 2 h in PBT/block followed by three washes in PBT. All incubations and washes were done at room temperature unless otherwise specified. Slides were mounted with Gel-mount (Biomeda). Antibodies used were rabbit anti-DAZL-3(1:200) (10), rabbit anti-SCP1 (1:400; 228; Novus Biologicals, Littleton, CO), rabbit anti-CREM (1:400; X-12; Santa Cruz Biotechnology), and Alexa Fluor 488- or Alexa Fluor 594-conjugated goat anti-rabbit IgG (1:1,000; Invitrogen). Images in Fig. 2 H and J were exposed and processed equivalently and are representative of several experiments.
Immunoblotting.
Protein lysates were obtained by homogenizing ≈200 mg of testis in 1 ml of cell disruption buffer (PARIS kit; Ambion) containing Complete protease inhibitors (Roche). After brief centrifugation to remove cellular debris, the protein concentration of each lysate was assessed by bicinchoninic acid assay (Pierce). Alternatively cells were lysed directly in protein-loading buffer (Fig. 2A). Equivalent amounts of protein (50–90 μg for testis lysates) were separated by SDS/PAGE on a 10–20% Tris-glycine gradient gel (Invitrogen) and transferred to poly(vinylidene difluoride). Membranes were incubated in PTW/block (PTW is 0.1% Tween 20 in PBS, and block is 5% nonfat dry milk) overnight at 4°C. Primary antibody was applied in PTW/block for 1 h followed by three washes in PTW, and secondary antibody was applied in PTW/block for 40 min followed by three washes in PTW. Antibodies used were mouse anti-Myc (1:2,000; 9E10), rabbit anti-DAZL-3 (1:1,000) (10), rabbit anti-TEX11 (1:1,000; see below), mouse anti-TUBULIN (1:5,000; DMIA, Bio-Genex), rabbit anti-GFP (1:7,500; ab290, Abcam), horseradish peroxidase-conjugated goat anti-rabbit IgG, and goat anti-mouse IgG. Horseradish peroxidase was detected by using SuperSignal West Pico (Pierce). Tex11 was first identified to be a germ cell-specific transcript in mouse by Wang et al. (27). A rabbit polyclonal antibody to rat TEX11 was generated by Kent Hamra in the D.L.G. laboratory, and it recognizes an antigen in rat germ cells (unpublished data). The Western blots in Fig. 2G were stripped and reprobed for DAZL, TEX11, and TUBULIN (in that order), and GFP was analyzed on a separate blot.
To estimate the relative change in DAZL protein levels in transgenic versus wild-type rat testis, two methods were used. In the first (Fig. 2G), imagej software (National Institutes of Health) was used to quantitate DAZL signal normalized to TUBULIN signal in the Western blots shown. This method suggested that there was a 90% reduction in DAZL protein levels. In a second quantitation method DAZL levels in 50 μg of transgenic testis lysate were compared to DAZL levels in a dilution series of wild-type testis lysate by Western blotting. We determined that 50 μg of transgenic testis lysate has less DAZL protein than 14 μg of wild-type testis lysate, suggesting an ≈70% reduction in transgenic animals.
Histology.
Testes and epididymis were fixed overnight in Bouin’s solution (room temperature) followed by extensive washing in 70% ethanol. Subsequent paraffin processing, embedding, and sectioning were performed by standard procedures (28, 29). All compound microscopy was performed on an AX70 Olympus microscope using magna-fire software for image acquisition.
Acknowledgments
We thank Dr. Kent Hamra for graciously providing the Tex11 antibody before publication, Laura Molyneaux for assistance with animal husbandry, and members of the University of Texas Southwestern Medical Center pathology core for assistance with tissue preparation and histological analysis. This work was supported by the Lalor Foundation, Inc., the Howard Hughes Medical Institute, and The Cecil H. and Ida Green Center for Reproductive Biology Sciences.
Abbreviations
- shRNA
short hairpin RNA
- miRNA
microRNA
- Ubc
ubiquitin C.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Hedrich H. History, Strains and Models. London: Academic; 2000. [Google Scholar]
- 2.Lazar J., Moreno C., Jacob H. J., Kwitek A. E. Genome Res. 2005;15:1717–1728. doi: 10.1101/gr.3744005. [DOI] [PubMed] [Google Scholar]
- 3.Dillon C. P., Sandy P., Nencioni A., Kissler S., Rubinson D. A., Van Parijs L. Annu. Rev. Physiol. 2005;67:147–173. doi: 10.1146/annurev.physiol.67.040403.130716. [DOI] [PubMed] [Google Scholar]
- 4.Carmell M. A., Zhang L., Conklin D. S., Hannon G. J., Rosenquist T. A. Nat. Struct. Biol. 2003;10:91–92. doi: 10.1038/nsb896. [DOI] [PubMed] [Google Scholar]
- 5.Fedoriw A. M., Stein P., Svoboda P., Schultz R. M., Bartolomei M. S. Science. 2004;303:238–240. doi: 10.1126/science.1090934. [DOI] [PubMed] [Google Scholar]
- 6.Kunath T., Gish G., Lickert H., Jones N., Pawson T., Rossant J. Nat. Biotechnol. 2003;21:559–561. doi: 10.1038/nbt813. [DOI] [PubMed] [Google Scholar]
- 7.Lu W., Yamamoto V., Ortega B., Baltimore D. Cell. 2004;119:97–108. doi: 10.1016/j.cell.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 8.Rubinson D. A., Dillon C. P., Kwiatkowski A. V., Sievers C., Yang L., Kopinja J., Rooney D. L., Ihrig M. M., McManus M. T., Gertler F. B., et al. Nat. Genet. 2003;33:401–406. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- 9.Tiscornia G., Singer O., Ikawa M., Verma I. M. Proc. Natl. Acad. Sci. USA. 2003;100:1844–1848. doi: 10.1073/pnas.0437912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamra F. K., Gatlin J., Chapman K. M., Grellhesl D. M., Garcia J. V., Hammer R. E., Garbers D. L. Proc. Natl. Acad. Sci. USA. 2002;99:14931–14936. doi: 10.1073/pnas.222561399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin Y., Page D. C. Dev. Biol. 2005;288:309–316. doi: 10.1016/j.ydbio.2005.06.032. [DOI] [PubMed] [Google Scholar]
- 12.Rocchietti-March M., Weinbauer G. F., Page D. C., Nieschlag E., Gromoll J. Int. J. Androl. 2000;23:51–56. doi: 10.1046/j.1365-2605.2000.00207.x. [DOI] [PubMed] [Google Scholar]
- 13.Ruggiu M., Speed R., Taggart M., McKay S. J., Kilanowski F., Saunders P., Dorin J., Cooke H. J. Nature. 1997;389:73–77. doi: 10.1038/37987. [DOI] [PubMed] [Google Scholar]
- 14.Saunders P. T., Turner J. M., Ruggiu M., Taggart M., Burgoyne P. S., Elliott D., Cooke H. J. Reproduction. 2003;126:589–597. doi: 10.1530/rep.0.1260589. [DOI] [PubMed] [Google Scholar]
- 15.Ryu B. Y., Kubota H., Avarbock M. R., Brinster R. L. Proc. Natl. Acad. Sci. USA. 2005;102:14302–14307. doi: 10.1073/pnas.0506970102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamra F. K., Chapman K. M., Nguyen D. M., Williams-Stephens A. A., Hammer R. E., Garbers D. L. Proc. Natl. Acad. Sci. USA. 2005;102:17430–17435. doi: 10.1073/pnas.0508780102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hammer R. E., Maika S. D., Richardson J. A., Tang J. P., Taurog J. D. Cell. 1990;63:1099–1112. doi: 10.1016/0092-8674(90)90512-d. [DOI] [PubMed] [Google Scholar]
- 18.Lois C., Hong E. J., Pease S., Brown E. J., Baltimore D. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- 19.van den Brandt J., Wang D., Kwon S. H., Heinkelein M., Reichardt H. M. Genesis. 2004;39:94–99. doi: 10.1002/gene.20037. [DOI] [PubMed] [Google Scholar]
- 20.Collier B., Gorgoni B., Loveridge C., Cooke H. J., Gray N. K. EMBO J. 2005;24:2656–2666. doi: 10.1038/sj.emboj.7600738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ventura A., Meissner A., Dillon C. P., McManus M., Sharp P. A., Van Parijs L., Jaenisch R., Jacks T. Proc. Natl. Acad. Sci. USA. 2004;101:10380–10385. doi: 10.1073/pnas.0403954101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hemann M. T., Fridman J. S., Zilfou J. T., Hernando E., Paddison P. J., Cordon-Cardo C., Hannon G. J., Lowe S. W. Nat. Genet. 2003;33:396–400. doi: 10.1038/ng1091. [DOI] [PubMed] [Google Scholar]
- 23.Rao M. K., Pham J., Imam J. S., MacLean J. A., Murali D., Furuta Y., Sinha-Hikim A. P., Wilkinson M. F. Genes Dev. 2006;20:147–152. doi: 10.1101/gad1367806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xia X. G., Zhou H., Samper E., Melov S., Xu Z. PLoS Genet. 2006;2:e10. doi: 10.1371/journal.pgen.0020010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nenoi M., Mita K., Ichimura S., Cartwright I. L., Takahashi E., Yamauchi M., Tsuji H. Gene. 1996;175:179–185. doi: 10.1016/0378-1119(96)00145-x. [DOI] [PubMed] [Google Scholar]
- 26.Schorpp M., Jager R., Schellander K., Schenkel J., Wagner E. F., Weiher H., Angel P. Nucleic Acids Res. 1996;24:1787–1788. doi: 10.1093/nar/24.9.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang P. J., McCarrey J. R., Yang F., Page D. C. Nat. Genet. 2001;27:422–426. doi: 10.1038/86927. [DOI] [PubMed] [Google Scholar]
- 28.Shehan D. C., Hrapchack B. B. Theory and Practice of Histotechnology. Columbia, WA: Battelle; 1980. [Google Scholar]
- 29.Woods A. E., Ellis R. C. Laboratory Histopathology: A Complete Reference. Oxford: Churchill–Livingston; 1996. [Google Scholar]