Abstract
To investigate the manner in which B cells with λ light (L) chains undergo receptor editing, we have studied hybridoma panels from 56R/κ-deleted (kdel) mice. 56R/kdel mice only produce four L chains (λ1, λ2, λ3, and λX). They also have a simplified heavy (H) chain repertoire: All B cells start out with a 56R anti-DNA H chain. A few frankly autoreactive 56R λ1 cells appear to escape into the periphery, but the majority of the peripheral B cell repertoire in 56R/kdel is made up of B cells expressing the 56R H chain with the λX L chain. Surprisingly, 56R λX B cells are multireactive, binding to a variety of self and nonself antigens, including dsDNA (albeit at reduced affinity compared with the other λ L chains). Another significant population in the 56R/kdel mouse consists of allelically included B cells that express λX along with another L chain. The multireactivity of both 56R λX and 56R λX/λ1 receptors could contribute to autoimmunity if these B cells were to become activated. Also found among 56R/kdel hybridomas are clones that have inactivated the H chain and secrete only L chains. These clones may represent products of exhaustive rearrangement. Multireactivity, allelic inclusion, and L chain secretion are three consequences of editing at the λ locus that may predispose toward the development of autoimmunity.
Keywords: autoimmunity, B cell, V(D)J recombination, antibody, anti-DNA
Tolerance to self-antigens can be achieved by replacing autoreactive heavy (H) chain and light (L) chain V genes with V genes coding for nonself receptors, a mechanism called receptor editing (1–4). Several recent studies support a pivotal role for receptor editing in the generation of a tolerant B cell repertoire (5–7). The κ L chain locus uses the nested organization of Vκ and Jκ gene segments for secondary rearrangements that can replace or inactivate a VκJκ that contributes to autoreactivity (Fig. 3 Top, which is published as supporting information on the PNAS web site). κ editing can also delete and/or inactivate an autoreactive κ chain by rearrangement to the κde (human) or to RS (mouse) (8–10). Secondary rearrangements to Jκ and deleting elements are presumed to be unidirectional; hence, editing enriches the repertoire in Jκ5 rearrangements. Editing also increases the frequency of B cells that are rearranged at both κ alleles (3, 11–13). Rearrangement at the second L chain isotype, λ, can rescue κ-inactivated and κ-deleted B cells (1, 14).
λ L chain genes also contribute to autoreactivity and require editing. The mouse λ locus has two separate clusters, each consisting of Vλ and JλCλ gene segments (Fig. 3 Middle) (15, 16). In one cluster, Vλ1 rearranges either to Jλ1 or Jλ3. In the other cluster, Vλ2 or VλX rearranges to Jλ2 (15). Rearrangements that leapfrog across clusters (for example, Vλ2 to Jλ1) are very infrequent (15). Thus a B cell with a λ rearrangement that yields a self-reactive receptor is in a dangerous position because of its inability to delete the primary λ rearrangement.
H chain variable region gene segment (VH) genes can also be edited because a VDJ gene can be replaced by secondary rearrangement of an upstream VH to a partial RSS embedded in VH genes (16–19). This mechanism of replacement is similar to heptamer-/nonamer-mediated rearrangement and can lead to both productive and nonproductive products. Hence, VH replacement can also inactivate an H chain and may play a role in tolerance induction (20). The 56R anti-DNA VH has been introduced into the H chain joining gene segment (JH) locus by homologous recombination and can serve as a substrate for VH replacement and D invasion (Fig. 3 Bottom) (21). This study differs from our previous studies (11, 12, 16, 22) in which the H chain transgene (tg) was not targeted to the H chain locus.
We have further simplified the 56R transgenic mouse by crossing it to mice without a κ locus (23). This model, 56R/κ-deleted (kdel), allows us to study the process of editing at the λ locus and the repertoire of edited anti-DNA B cells. We have analyzed hybridoma panels derived from these mice and found that the λ chain repertoire has a high frequency of the anti-DNA editor, VλX. In addition, a significant number of hybridomas express λX along with λ1 or λ2. Editing by λX yields 56 R-expressing B cells with multireactive receptors. We also provide genetic and biochemical evidence that some 56R/kdel B cell hybridomas only secrete L chains. In sum, λ editing produces by-products (allelic inclusion, λX expression, and free L chain secretion) that contribute to multireactivity and may promote autoimmune dysfunction.
Results
Most 56R/kdel Hybridomas Produce λX.
The mouse model used in these studies, 56R/kdel, has a simplified V gene repertoire consisting of a tg-encoded 56R H chain and only four different L chains (λ1, λ2, λ3, and λX). The 56R H chain was derived from the 3H9 H chain, which, in turn, was derived from a diseased MRL-lpr/lpr mouse and is typical of lupus anti-DNA antibodies (24, 25). 56R differs from 3H9 by a substitution of arginine for aspartate at position 56 of complementarity determining region 2 (CDR2), which confers a 10-fold higher affinity for dsDNA (26). Arginine residues in 3H9 and 56R provide most of the interaction with DNA, and these H chains bind DNA when paired with most L chains (11, 27). Of particular relevance in these experiments is that λ1 and λ2 sustain dsDNA binding when paired with 56R, whereas λX significantly reduces the affinity of 56R for dsDNA, i.e., it “edits” DNA binding (11). The CDRs of λX are highly acidic (pI = 4.62 compared with pI of 10.9 for λ1 and λ2). The low pI can be attributed to a high frequency of aspartate residues, a property shared with κ anti-DNA editors. These aspartate residues may interact with the arginine CDR residues to reduce or prevent DNA binding.
λX is barely detectable in normal mouse sera. Even sera from kdel mice (with wild-type H chain loci) have λX levels that are low compared with λ1 and λ2 (12, 28). Hybridomas from kdel mice also have a low frequency of λX rearrangements compared with λ1 and λ2 rearrangements; the observed frequency of λX rearrangements in hybridomas with a single λ rearrangement is 5% (Table 1 and refs. 12 and 29). Of note, half of the hybridomas with λX rearrangements also have rearrangements to other λ L chains (5/10), whereas less than one-fourth of hybridomas with λ1 rearrangements (20/86) have other λ rearrangements. The low frequency of λX in the peripheral repertoire could be because of late rearrangement of λX, a lower frequency of in-frame λX joins (30), or to selection events favoring λ1 over λX after rearrangement.
Table 1.
λ L chain rearrangements in LPS hybridomas from three different lines of kdel mice
| λ Genotype | Non-tg/kdel (%) | 3H9/kdel (%) | 56R/kdel (%) |
||||
|---|---|---|---|---|---|---|---|
| 56R− | 56R+ | ||||||
| Single rearrangements | |||||||
| λ1 | 66 (65) | 14 (23) | 11 (50) | 4 (11) | |||
| λ2 | 31 (30) | 7 (11) | 6 (27) | 1 (3) | |||
| λX | 5 (5) | 40 (66) | 5 (23) | 30 (86) | |||
| Total | 102 | 61 | 22 | 35 | |||
| Multiple rearrangements | |||||||
| λ1+λX | 0 (0) | 18 (41) | 3 (30) | 12 (52) | |||
| λ2+λX | 3 (13) | 13 (30) | 1 (10) | 3 (13) | |||
| λ1+λ2+λX | 2 (9) | 11 (25) | 2 (20) | 8 (35) | |||
| λ1+λ2 | 18 (78) | 2 (5) | 4 (40) | 0 (0) | |||
| Total | 23 | 44 | 10 | 23 | |||
The frequencies of different λ rearrangements were measured by PCR on genomic DNA in splenic hybridoma panels from 56R/kdel mice. Shown for comparison are previously published data from hybridoma panels in 3H9/kdel mice and non-tg kdel mice. [Data are reproduced with permission from ref. 12 (Copyright 1994, The Rockefeller University Press).] Numbers and percentages of clones that have rearranged only one type of λ L chain (single rearrangement) or have rearranged multiple L chains (multiple rearrangement) are shown. Single rearrangement assays are not sensitive to copy number (an unknown fraction of clones may have rearranged the same L chain on both alleles). The multiple rearrangement category includes clones with productive as well as nonproductive L chain rearrangements. The PCR assays do not distinguish λ2 from λ3 rearrangements. The 3H9 tg H chain is a conventional tg; hence, it cannot be deleted by editing. In contrast, the 56R tg has been knocked into the HC locus and can be inactivated and/or deleted by further rearrangement (16). Therefore, clones from 56R tg mice comprise two categories, 56R+ and 56R−.
Compared with kdel mice, 3H9/kdel and 56R/kdel mice have significantly increased λX usage (Table 1 and ref. 12). The higher affinity anti-DNA tg, 56R, has a repertoire that is more skewed toward λX. Furthermore, the frequency of λX usage among clones with one λ rearrangement is higher in hybridomas that retain the 56R tg DNA (“56R+” in 86%) than in hybridomas that lack 56R (“56R−” in 23%; Table 1). However, even among 56R− hybridomas, there is an increased frequency of λX usage compared with kdel (23% vs. 5%).
λX Usage Predominates Among 56R/kdel Hybrids with Multiple λ Rearrangements.
3H9/kdel and 56R/kdel mice also exhibit increased frequencies of clones with multiple λ rearrangements (Table 1). The increased frequency of clones with multiple λs is suggestive of an increased level of ongoing L chain rearrangement (editing). The theoretical basis for this increase is that most developing B cells in a 56R/kdel mouse will initially rearrange λ1, λ2, λ3, or a nonproductive λX. Because 56R+ cells with nearly all λs either express autoreactive or nonfunctional B cell receptors, they are subjected to further recombination. Among kdel hybridomas, the observed ratio of clones with multiple rearrangements that include a VλX rearrangement to clones with multiple rearrangements that do not is 1:3.6 (Table 1). Hybridomas with multiple rearrangements from the 3H9 and 56R transgenic mice have higher frequencies of λX rearrangements: 95% and 100%, respectively (Table 1). Therefore, most 56R B cells that do not eventually rearrange a productive λX L chain probably undergo negative selection. Another finding consistent with selection against non-λX clones is the ratio of 56R+ clones with one λX rearrangement to clones with multiple rearrangements. In 56R/kdel, this ratio is 1.3 to 1 (Table 1), which is very close to the expected ratio of 1.34 (see Materials and Methods).
H Chain Editing and Loss.
A B cell can also edit by replacing the 56R VH gene with an upstream VH. Alternatively, the 56R VH can be inactivated by D invasion or an out-of-frame VH replacement. A third pathway of 56R VH inactivation occurs via somatic mutation and/or deletion. In the hybridoma S43, the 56R sequence has multiple mutations and two deletions (20 and 33 nucleotides in length) when compared with the 56R tg sequence (Fig. 4, which is published as supporting information on the PNAS web site). These mutations in 56R are not a result of analyzing the germ-line version of the 3H9 H chain, because the sequenced H chain shares the in vitro site-directed mutations of the 56R tg. We refer to clones that have replaced or inactivated the 56R H chain as 56R−. Thirty-two of 90 LPS hybridomas are 56R− (36%; Table 1). Nineteen of 35 antibody-secreting spontaneous hybridomas are 56R− (54%; Table 2).
Table 2.
H chain and L chain rearrangements in 56R/kdel spontaneous hybridomas
| λ Genotype | 56R/kdel |
||
|---|---|---|---|
| 56R+ | 56R− | HC− | |
| λ1 | 0 | 9 (47) | 1 (4) |
| λ2 | 0 | 4 (21) | 0 |
| λX | 10 (63) | 1 (5) | 2 (8) |
| λ1+λX | 2 (13) | 2 (11) | 1 (4) |
| λ2+λX | 0 | 0 | 0 |
| λ1 or λ2 | 2 (13) | 1 (5) | 0 |
| λ− | 2 (13) | 2 (11) | 20 (83)* |
| Total | 16 | 19 | 24 |
λ Rearrangements were typed by PCR and confirmed in ambiguous cases by Western blotting or ELISA (see Materials and Methods). Clones marked λ− fail to amplify in any of the λ genotyping PCRs.
*HC− clones lack evidence of λ rearrangement and type negative in the 56R genotyping PCR assay.
The number and kinds of L chain rearrangements in 56R− hybrids suggest that H chain editing occurs at two stages of B cell development: early, at the pro-B stage, and late, during or possibly after L chain rearrangement. 56R− clones with multiple λ rearrangements are likely to have replaced or inactivated the 56R H chain during or after L chain rearrangement. The higher frequency of λX rearrangements among 56R− hybrids (34%) than among nontransgenic kdel hybrids (8%) favors the idea that at least some H chain editing is occurring after L chain rearrangement. On the other hand, 56R− clones that lack λX rearrangements or have a single λ1 or λ2 rearrangement may have edited the H chain before L chain rearrangement.
H chain replacement can also lead to H chain loss by inactivation of one or both alleles. Clones lacking serological evidence of H chains (denoted HC−) comprise 40% of 56R/kdel spontaneous hybridoma panel (Table 2). In ELISAs, these HC− hybridomas produce no detectable levels of IgM, IgG, or IgA (data not shown). Yet, clearly some HC− clones are B cells, because they harbor λ rearrangements (Table 2 and data not shown).
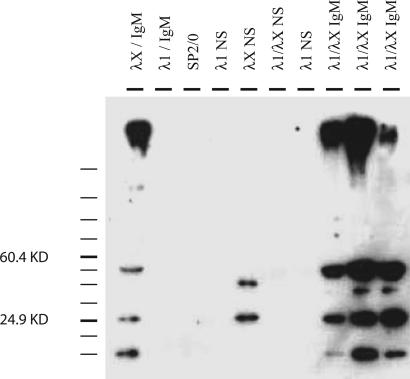
To confirm the absence of the H chain in HC− hybridomas, Western blotting was performed on supernatants, using a polyclonal antibody that recognized free λX L chains as well as λ L chains that are associated with the 56R H chain (Fig. 1 and ref. 31). The specificity of this antibody for λX is shown by its reactivity with λX/IgM and its lack of reactivity with λ1/IgM (Fig. 1). Of the four HC− clones shown, one, λX HC− (fifth lane), has one band at 25 kDa and another at ≈50 kDa, consistent with a λX monomer and dimer, respectively. There is no higher molecular weight band in λX HC− that corresponds to H2L2 complexes, confirming its status as an L chain-only secretor. Secretion of free L chain monomers and dimers is not unique to λX (data not shown and refs. 32–35). In contrast, the λ1/λX IgM antibody-secreting clones produce the predicted >150 kDa band, corresponding to H2L2 (Fig. 1, lanes 1 and 8–10). However, free L chain monomers and dimers are also seen in these clones. The ratios of free L chains to presumed dimers and to H+L chain pairs appear to vary from one clone to the next, even in clones with identical H and L chain genotypes. Also variable are the relative amounts of λ1 and λX L chains that are secreted in the various types of complexes (monomer, dimer, and H2L2).
Fig. 1.
Western blot analysis of 56R/kdel hybridoma supernatants. 56R/kdel hybridoma supernatants were run in nonreducing conditions and probed with anti-λX antibody (see Materials and Methods). Sample loading was normalized for L chain band intensity and overall protein content. Lanes 1–3, previously characterized clones that are known to express λ1/IgM, λX/IgM, and no antibody (Sp2/0). Lanes 4–7, the H chain deficient (HC−) clones. Lanes 8–10, allelically included clones (coexpressing λ1 and λX). Estimated sizes in kilodaltons (based on molecular mass standards; data not shown) are shown on the left of the gel. Predicted sizes of an L chain monomer, dimer, and fully assembled antibody (H2L2) are 25 kDa, 50 kDa, and ≈150 kDa, respectively.
The genetic basis for the loss of H chain expression in the HC− hybridomas is unresolved. None of the HC− clones types positive in the 56R genotyping PCR, suggesting that a rearrangement event has disrupted the H chain tg. The rearrangement may be some form of D-invasion, because PCRs with degenerate VH primers and 56R CDR3 or JH4 primers are repeatedly negative in these clones (see Materials and Methods and Fig. 3 for a description of the H chain PCR assays). HC− hybridomas are also found in B6.56R mice (36) and in spontaneous hybridomas from wild-type B6 mice (L. Yunk and E.T.L.P., unpublished data).
Editing at λ and Multireactivity.
The 56R/kdel mouse has a highly restricted repertoire that is heavily biased toward the production of anti-dsDNA antibodies. Despite these constraints, serum levels of anti-dsDNA antibodies are not elevated in BALB/c 56R/kdel compared with kdel littermate controls (data not shown). The majority of spontaneous 56R/kdel hybridomas have evidence of H chain editing (19 secretors are 56R−, 24 clones are HC−, and only the remaining 16 are 56R+) (Table 2).
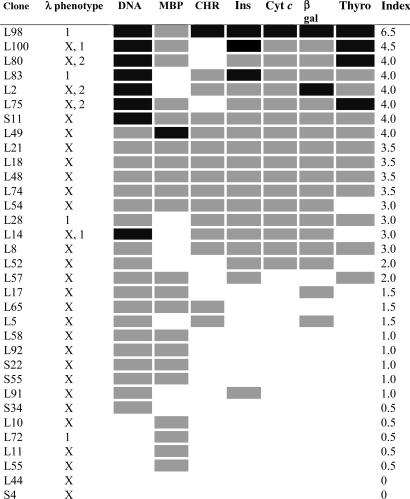
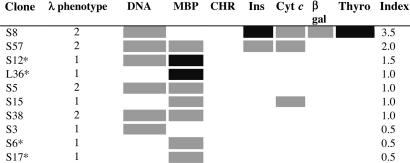
If 56R is left unmodified, L chain editing is insufficient to abrogate DNA binding. 56R in combination with any λ chain, including λX, produces antibodies that are multireactive. Tables 3 and 4 show the reactivity profiles of 56R+ hybridomas with different L chain phenotypes. The binding results from these assays are not normalized for Ig concentration and are not quantitative. Semiquantitative comparisons, based on three independent assays of individual clones and selected subclones, yield a spectrum of the most probable binding activities for each clone. The overall level of reactivity is estimated with an index shown in the rightmost column of the table.
Table 3.
Reactivity profile of 56R+ kdel hydridomas
The λ phenotype is based on the following assays: λ 1/2 ELISA, λ1 PCR, λ2 PCR, and λX PCR. PCR was performed on DNA and did not distinguish functional from nonfunctional rearrangements. Only clones with unambiguous phenotypes are shown. The functional status of λX was confirmed in a subset of clones by Western blot analysis (data not shown). Reactivity in solid phase ELISAs for dsDNA, myelin basic protein (MBP), chromatin, insulin (Ins), cytochrome c (Cyt c), and thyroglobulin (Thyro) are coded as follows: black (OD > 1.0), gray (OD 0.20–0.99), and white (OD < 0.2). OD values used represented the average of triplicate determinations. The index is an estimate of the overall strength of reactivity. OD values of ≥1.0 are given 1 point, OD values between 0.20 and 0.99 are given 0.5 points, and OD values <0.2 get no points. S, spontaneous 56R/kdel panel; L, LPS 56R/kdel panel.
Table 4.
Reactivity profile of 56R− kdel hybridomas
The λ phenotype is based on the following assays: λ 1/2 ELISA, λ1 PCR, λ2 PCR, and λX PCR. PCR was performed on DNA and did not distinguish functional from nonfunctional rearrangements. Only clones with unambiguous phenotypes are shown. The functional status of λX was confirmed in a subset of clones by Western blot analysis (data not shown). Reactivity in solid phase ELISAs for dsDNA, myelin basic protein (MBP), chromatin, insulin (Ins), cytochrome c (Cyt c), and thyroglobulin (Thyro) are coded as follows: black (OD > 1.0), gray (OD 0.20–0.99), and white (OD < 0.2). OD values used represented the average of triplicate determinations. The index is an estimate of the overall strength of reactivity. OD values of ≥1.0 are given 1 point, OD values between 0.20 and 0.99 are given 0.5 points, and OD values <0.2 get no points. S, spontaneous 56R/kdel panel; L, LPS 56R/kdel panel.
*Clones secreted IgG. All other clones secreted only IgM.
Tables 3 and 4 reveal multiple classes of cells, each with different reactivity profiles. In the most polyreactive group are cells that appear to have escaped tolerance altogether. They express 56R and λ1 and lack λX. Also highly multireactive are allelically included cells with multiple productively rearranged L chains. Cells with 56R and λX rearrangements exhibit moderate to low levels of multireactivity. Low reactivity is not due to low levels of antibody secretion, because these cells secrete comparable or higher levels of antibody than the more multireactive 56R λX clones (data not shown). Some minimally reactive clones may have productively rearranged the endogenous H chain allele or undergone somatic mutation of the 56R. Cells lacking 56R appear to have a lower level of multireactivity (Table 4). Alternatively, the heterogeneity observed in these reactivity patterns could be because of differences in the composition of the secreted L chain pool. Perhaps secreted L chains (L or L2) modify or compete with fully formed antibodies (H2L2) for antigen binding.
Discussion
Pathways of Editing in 56R/kdel Mice.
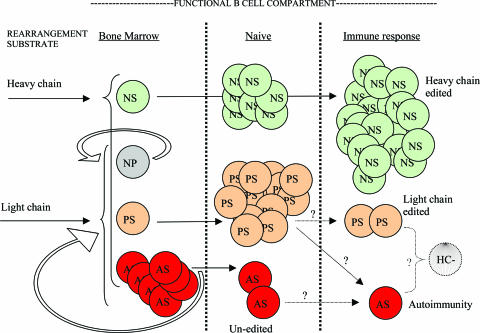
This study focused on receptor editing at the λ locus in 56R/kdel mice. The highly simplified immune repertoire of these mice was advantageous because it allowed us to characterize the full range of λ editing pathways and to correlate these with receptor structure and specificity. The repertoire in 56R/kdel includes nonself-reactive B cells (NS or “complete editing”) most likely consisting of H chain edited cells, partially edited B cells (PS, consisting of different classes of L chain edited 56R+ cells), and an occasional unedited cell (AS; Fig. 2). L chain editing in this model results either in 56R+ cells that express λX or 56R+ cells that express λX along with other λ L chains. The 56R λX antibody has anti-DNA activity; however, the relative affinity is low compared with Vλ1 and Vλ2 (11). In contrast to λX, certain κ editors (such as Vκ21D) completely veto DNA binding of 56R (21). On the other hand, other κ editors (Vκ38c and Vκ20) behave more like λX: They permit DNA binding but not to the degree that the cell is deleted from the peripheral repertoire in a self-tolerant mouse.
Fig. 2.
Hypothetical pathways and fates of edited B cells in 56R/kdel. 56R/kdel B cells start out with a 56R anti-DNA H chain and can rearrange λ1, λ2, λ3, or λX, resulting in one of three outcomes: AS (fully autoreactive in red, for example 56R λ1), PS (partially self-reactive in pink, for example allelically included cells such as 56R λX/λ1), or NP (nonproductive). AS cells can undergo additional L chain rearrangement (editing, depicted with the circular arrow). PS cells can emigrate from the bone marrow but may be sequestered in specialized compartments rather than participating fully in immune responses. 56R/kdel B cells can also undergo H chain rearrangement on the 56R allele or the endogenous allele. Successful H chain editing results in NS (nonself reactive) (green) B cells. Such NS cells likely predominate in immune responses in self-tolerant 56R mice (62). The route to fully activated AS cells (autoimmunity) is unclear. Possible sources for such cells included PS cells that have undergone somatic mutation or unedited AS cells. HC− cells (those lacking H chains) may represent terminal receptor editing products.
The extent to which different editor L chains contribute to the B cell repertoire may depend on the lupus-susceptibility of the mouse strain. For example, the lupus-susceptible strain, B6.56R, has a high frequency of incompletely edited B cells, whereas the lupus-resistant strain, BALB/c.56R, has mainly completely edited B cells (36). Incompletely edited B cells may serve as direct precursors of lupus anti-DNA antibodies, as in the case of the 3H9 anti-DNA antibody (26), and/or they may play a role in activating an autoimmune response. For example, the incompletely edited B cell could serve as a multipotential, antigen-presenting cell (37).
A second class of “incompletely” edited B cells expresses more than one λ. Allelically included B cells are ordinarily present at a relatively low frequency, ≈1–7% (5, 38), but are a significant part of the B cell repertoire of anti-DNA transgenics (4). The λ locus is a set-up for allelic inclusion, because of the low frequency of leapfrogging between the gene clusters (15). Isotypic inclusion can also be seen in 56R κ-sufficient mice that have B cells that express a κ editor and an autoreactive λ1 L chain (39). The included cells in 3H9/kdel and 56R/kdel produce λ1 and λX. These B cells, as in the case of 56R κ/λ double expressers, are partially autoreactive. Recent findings in the 3–83Igi model further support the idea that editing contributes to the development of autoreactive, allelically included B cells (13). However, 56R+ B cells that coexpress λX and λ1 appear to be tolerant because anti-DNA antibodies are not found in abundance in the 56R/kdel serum. Perhaps autoantigen fails to activate dual receptor B cells or targets the cell to a site in which they are unable to differentiate to a B cell that secretes a pathogenic autoantibody. One such site is the marginal zone, and most of the 56R κ/λ population resides in this site (39). Another site to which partially autoreactive B cells may home is the interface between B cell and T cell zones of the spleen. Autoreactive B cells in other systems have been shown to accumulate at the T cell–B cell interface (40–42), and the TB interface has been shown to include λ1 B cells in 3H9 transgenics (43). A recent repertoire analysis of peripheral blood B cells in healthy human subjects suggests that fewer IgM+CD27+ (“memory” cells) express autoantibodies than naive IgM+ B cells (44). Furthermore, a detailed study of rheumatoid factor (RF) B cells reveals that RF B cells readily form germinal centers in healthy mice but infrequently become antibody forming cells (45). In contrast, on the MRL/lpr background, RF B cells not only produce antibody-forming cells but undergo more extensive intraclonal somatic mutation (45). These studies and findings in the 56R mouse model (refs. 36 and 46 and this paper), indicate that autoreactive B cells can survive in the periphery but are subjected to additional tolerance checkpoints to prevent them from participating in (auto)immune responses.
A Consequence of Incomplete Editing Is Multireactivity.
Incomplete editing can account for the acquisition of new autospecificities. Edited antibodies with a high ratio of aspartate to arginine residues may have unsequestered aspartate residues that can access basic proteins, for example. λX antibodies might bind myelin basic protein (MBP) if the VH arginine content is low. To determine whether the interaction between λX and MBP is simply because of an excess of asparate residues (without regard for conformation), we tested λX antibodies for binding to other proteins. As summarized in Tables 3 and 4, 56R λX antibodies bind a variety of proteins.
In the same sense that anti-DNA reactivity of 56R is edited by pairing with a negatively charged L chain, λX, the anti-MBP, or other self-protein binding of λX may be edited by the positive charges in 56R. But just as incomplete H chain editing leads to antibodies with residual anti-DNA activity, editing of the MBP activity may also be incomplete. These antibodies may also be subject to negative regulation. Indeed selection against B cells with editor L chains might explain the low serum levels of λX and other multireactive editor L chains (J.H. and M.G.W., unpublished data).
The special properties of partially edited B cells, including multireactivity, may provide the structural basis for what are referred to as natural autoantibodies (47, 48). Natural autoantibodies, by virtue of their ability to recognize conserved bacterial and, in some cases also self-antigens, play an important role in innate immune defense. Cells expressing natural autoantibodies are characteristically found in regions of the body where immune cells play a sentinel role (body cavities, the gut, and the marginal zone of the spleen, for example). Natural IgM autoantibodies also appear to play a protective role in autoimmunity (49), promoting the clearance of apoptotic debris and potentially decreasing inflammation and atherosclerosis (50–52). The production of natural autoantibodies and expansion of marginal zone or B1 B cells may be favored in the setting of a highly restricted B cell repertoire and may also occur in the 56R/kdel mouse (53).
It will be of interest to determine which cells within the 56R/kdel repertoire correspond to those that cause autoimmunity on an autoimmune-prone genetic background. By their sheer frequency, one might expect extensively L chain edited B cells [the PS cells in Fig. 2] to pose the most significant risk: In humans, an accumulation of autoreactive B cells confers an increased risk of autoimmune disease (54). On the other hand, among the most multireactive were 56R λ1 cells that had failed to edit AS cells in Fig. 2. A failure of editing could result in autoimmunity and a repertoire enriched in AS cells with primary rearrangements (55). On the other hand, if editing is operative and driven by autoreactivity, then edited PS cells should dominate the autoreactive population. It is unresolved how or if partially edited cells serve as substrates for progression to full-blown autoimmunity.
Terminal Editing Products?
The analysis of 56R/kdel also revealed a significant population of HC− cells. HC− hybridomas did not secrete detectable quantities of H chains and lacked the 56R H chain DNA. On the other hand, λ L chain expression persists in some of these HC− cells because the organization of the λ locus does not allow deletion or replacement. HC− hybridomas are not unique to the 56R/kdel model and may represent a population that has undergone prolonged division and rearrangement. HC− cells may be prevalent in autoimmune individuals, and mice and could account for the serum free L chains that are found in autoimmune diseases (refs. 56–60 and C.M.D. and M.G.W., unpublished observations).
Materials and Methods
Mice.
Transgenic mice were crossed to homozygous kdel mice to produce mice with the 56R/kdel genotype and genotyped as described (21). To type mice for the κ locus deletion, two separate PCR assays were performed as described (23). All mice used in these experiments have been backcrossed at least 10 generations onto the BALB/c background and are 6–12 weeks old.
Hybridomas.
B cell hybridomas were prepared from spleens of 56R/kdel mice. Spontaneous hybridomas were produced by fusing splenocytes with Sp2/0 myeloma cells (61) immediately on harvesting from the mouse. For the LPS panel, splenocytes were harvested and cultured for 3 days in the presence of 20 μg/ml Escherichia coli LPS (Sigma). Hybridomas were plated and selected, and DNA was prepared as described (8, 12).
PCR Assays.
λ Genotyping was performed as described (12). 56R PCR amplifications were carried out by using 3H9 LD, a VH leader primer, and a tg-specific CDR3 reverse primer as described (22). To detect the presence of the 56R tg allele in 56R− hybridomas, amplification by using a tg-specific CDR3 forward primer and a JH4 or H constant (JHCH) region reverse primer were used as described (16).
Sequencing.
56R, VλX, and Vλ1 were amplified from genomic DNA, and products were separated by agarose gel electrophoresis and purified by using a QIAEX II gel extraction kit (Qiagen). The primers used for amplification and sequencing of the 56R HC were VH5.3, a degenerate H chain (FW1) primer, and JH4 or H constant (JHCH) (16). PCR products amplified by VH5.3 and JHCH were also sequenced by using the tg-specific CDR3 forward and reverse primers. λX was amplified by using VLXup (31) and Jλ2 (using the same PCR conditions as VλX and Jλ2) and sequenced by using VλX and Jλ2. λ1 was amplified and sequenced by using Vλ1/2 + Jλ1.
ELISA.
For H chain isotyping of hybridoma supernatants, Immulon 2 plates (Thermo Electron, Waltham, MA) were coated with 3.3 μg/ml unlabeled goat anti-mouse Ig H and L chain antibody (Southern Biotech, Birmingham, AL) in coating buffer (15 mM Na2CO3, 35 mM NaHCO3, and 0.004% NaN3, all at pH 9.6). Plates were blocked with 1% BSA in PBS for 1.5 h at 37°C. Undiluted hybridoma supernatants were added and incubated for 1–2 h at 37°C. After washing, alkaline phosphatase-conjugated goat anti-mouse IgM, IgG, or IgA (Southern Biotech) was added and incubated for 1–2 h at 37°C. Plates were developed with alkaline phosphatase substrate (Sigma) at 1.0 mg/ml in 0.1 M carbonate buffer (pH 9.6) and 10 mM MgC12. Alkaline phosphatase-conjugated anti-mouse λ (Southern Biotech) was used as the secondary antibody in the λ ELISAs. The same method was used to detect binding to MBP, chromatin, cytochrome c, thyroglobulin, and insulin. Plates were coated overnight with 5 μg/ml of each antigen (except for MBP, which was at 10 μg/ml) and blocked with 8% casein in PBS for 1–2 h at 37°C. Supernatant was added, and binding was detected by using alkaline phosphatase-conjugated anti-mouse IgM or anti-mouse IgG secondary. dsDNA binding was detected by a solution phase ELISA, as described (26). Samples were tested in all assays on the same day. Negative controls include Sp2/0 supernatant, binding to the blocked plate (without antigen), and the use of an irrelevant anti-H chain antibody (anti-IgG for an IgM antibody). Assays were performed in duplicate or triplicate in three independent experiments. Readings (not normalized for antibody concentration) are presented by using a semiquantitative scale in Tables 3 and 4. Strength of binding was analyzed in a subset of clones by titrating the supernatant in each of the antigen-binding assays and standardizing the results between clones based on H chain concentration. In some supernatants, a high level of background binding was observed when the supernatant was applied directly to the blocked plate.
Western Blot.
Hybridoma supernatants, diluted 1/10 and 1/25, were resolved in nonreducing conditions on a 4–20% polyacrylamide gradient gel (Cambrex, Rockland, ME) and transferred to a poly(vinylidene difluoride) membrane. λX proteins were detected by using a polyclonal anti-VλX antibody (31) followed by horseradish peroxidase (HRP)-conjugated goat anti-rabbit antibodies (Bio-Rad) and developed by using SuperSignal West Pico chemiluminescent substrate (Pierce). λ1, λ2, and λ3 proteins were detected by using an alkaline phosphatase-conjugated goat anti-mouse Vλ1/2 (Southern Biotech) or an HRP-conjugated anti-mouse Vλ1/2 (Southern Biotech). Hybridoma supernatants were monitored for overall protein concentration using Ponceau S staining, and loading volumes were adjusted based on serial analysis of the same samples on a reducing gel, such that each lane had comparable levels of the 25-kDa λX band.
λ Rearrangement Probability Calculations.
To calculate the predicted ratio of λX-only clones to clones with λX plus other L chains, we assume that a B cell has an equal chance of rearranging each of the four λ L chains, that the chance of a productive (in-frame) rearrangement is 33%, that there is no allele preference, that there is prompt termination of rearrangement if a productive λX rearrangement is obtained or that rearrangement is continued for one more cycle if λX is not expressed, that cells that fail the first round automatically go on to the second round, that allelic and isotypic inclusion are permitted, and that only cells with a productive λX rearrangement survive to be counted among our hybridomas. With these assumptions, the following “survival” genotypes and corresponding probabilities for two cycles of λ rearrangement are as follows: X (0.33); X, X (0.0367); 1, X (0.11); 2, X (0.055); 3, X (0.11). Clones with (1, X), (2, X), or (3, X) genotypes can have either functional or nonfunctional rearrangements to the non-λX L chain. Based on these probabilities, the ratio of “λX only” (including clones with two λX rearrangements) to “λX + other” is 1.34.
Supplementary Material
Acknowledgments
We thank Yoram Louzoun, Mark Shlomchik, David Nemazee, and Michel Nussenzweig for helpful comments and discussions. We also thank Dongyao Ni for technical assistance and Yijin Li for advice. The funding for this project was provided by National Institutes of Health Grants GM02964-29 (to M.G.W.) and 5T32 AI07090 (to C.M.D.).
Abbreviations
- AS
autoreactive
- HC−
H chain deficient
- κ deleted
kdel
- MBP
myelin basic protein
- VH
H chain variable region gene segment
- JH
H chain joining gene segment
- tg
transgene
- NP
nonproductive
- PS
partially self-reactive.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Tiegs S. L., Russell D. M., Nemazee D. J. Exp. Med. 1993;177:1009–1020. doi: 10.1084/jem.177.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nemazee D., Weigert M. J. Exp. Med. 2000;191:1813–1817. doi: 10.1084/jem.191.11.1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Radic M. Z., Erikson J., Litwin S., Weigert M. J. Exp. Med. 1993;177:1165–1173. doi: 10.1084/jem.177.4.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gay D., Saunders T., Camper S., Weigert M. J. Exp. Med. 1993;177:999–1008. doi: 10.1084/jem.177.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casellas R., Shih T. A., Kleinewietfeld M., Rakonjac J., Nemazee D., Rajewsky K., Nussenzweig M. C. Science. 2001;291:1541–1544. doi: 10.1126/science.1056600. [DOI] [PubMed] [Google Scholar]
- 6.Halverson R., Torres R. M., Pelanda R. Nat. Immunol. 2004;5:645–650. doi: 10.1038/ni1076. [DOI] [PubMed] [Google Scholar]
- 7.Ait-Azzouzene D., Verkoczy L., Peters J., Gavin A., Skog P., Vela J. L., Nemazee D. J. Exp. Med. 2005;201:817–828. doi: 10.1084/jem.20041854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siminovitch K. A., Bakhshi A., Goldman P., Korsmeyer S. J. Nature. 1985;316:260–262. doi: 10.1038/316260a0. [DOI] [PubMed] [Google Scholar]
- 9.Durdik J., Moore M. W., Selsing E. Nature. 1984;307:749–752. doi: 10.1038/307749a0. [DOI] [PubMed] [Google Scholar]
- 10.Moore M. W., Durdik J., Persiani D. M., Selsing E. Proc. Natl. Acad. Sci. USA. 1985;82:6211–6215. doi: 10.1073/pnas.82.18.6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen C., Radic M. Z., Erikson J., Camper S. A., Litwin S., Hardy R. R., Weigert M. J. Immunol. 1994;152:1970–1982. [PubMed] [Google Scholar]
- 12.Prak E. L., Trounstine M., Huszar D., Weigert M. J. Exp. Med. 1994;180:1805–1815. doi: 10.1084/jem.180.5.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu S., Velez M. G., Humann J., Rowland S., Conrad F. J., Halverson R., Torres R. M., Pelanda R. J. Immunol. 2005;175:5067–5076. doi: 10.4049/jimmunol.175.8.5067. [DOI] [PubMed] [Google Scholar]
- 14.Retter M. W., Nemazee D. J. Exp. Med. 1998;188:1231–1238. doi: 10.1084/jem.188.7.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanchez P., Nadel B., Cazenave P. A. Eur. J. Immunol. 1991;21:907–911. doi: 10.1002/eji.1830210408. [DOI] [PubMed] [Google Scholar]
- 16.Chen C., Nagy Z., Prak E. L., Weigert M. Immunity. 1995;3:747–755. doi: 10.1016/1074-7613(95)90064-0. [DOI] [PubMed] [Google Scholar]
- 17.Reth M. G., Jackson S., Alt F. W. EMBO J. 1986;5:2131–2138. doi: 10.1002/j.1460-2075.1986.tb04476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reth M., Gehrmann P., Petrac E., Wiese P. Nature. 1986;322:840–842. doi: 10.1038/322840a0. [DOI] [PubMed] [Google Scholar]
- 19.Taki S., Schwenk F., Rajewsky K. Eur. J. Immunol. 1995;25:1888–1896. doi: 10.1002/eji.1830250715. [DOI] [PubMed] [Google Scholar]
- 20.Monestier M., Zouali M. Scand. J. Immunol. 2002;55:425–431. doi: 10.1046/j.1365-3083.2002.01070.x. [DOI] [PubMed] [Google Scholar]
- 21.Li H., Jiang Y., Luning Prak E. T., Radic M., Weigert M. Immunity. 2001;15:947–957. doi: 10.1016/s1074-7613(01)00251-5. [DOI] [PubMed] [Google Scholar]
- 22.Erikson J., Radic M. Z., Camper S. A., Hardy R. R., Carmack C., Weigert M. Nature. 1991;349:331–334. doi: 10.1038/349331a0. [DOI] [PubMed] [Google Scholar]
- 23.Chen J., Trounstine M., Kurahara C., Young F., Kuo C. C., Xu Y., Loring J. F., Alt F. W., Huszar D. EMBO J. 1993;12:821–830. doi: 10.1002/j.1460-2075.1993.tb05722.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Radic M. Z., Mascelli M. A., Erikson J., Shan H., Weigert M. J. Immunol. 1991;146:176–182. [PubMed] [Google Scholar]
- 25.Shlomchik M. J., Aucoin A. H., Pisetsky D. S., Weigert M. G. Proc. Natl. Acad. Sci. USA. 1987;84:9150–9154. doi: 10.1073/pnas.84.24.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Radic M. Z., Mackle J., Erikson J., Mol C., Anderson W. F., Weigert M. J. Immunol. 1993;150:4966–4977. [PubMed] [Google Scholar]
- 27.Ibrahim S. M., Weigert M., Basu C., Erikson J., Radic M. Z. J. Immunol. 1995;155:3223–3233. [PubMed] [Google Scholar]
- 28.Sanchez P., Drapier A. M., Cohen-Tannoudji M., Colucci E., Babinet C., Cazenave P. A. Int. Immunol. 1994;6:711–719. doi: 10.1093/intimm/6.5.711. [DOI] [PubMed] [Google Scholar]
- 29.Sanchez P., Marche P. N., Le Guern C., Cazenave P. A. Proc. Natl. Acad. Sci. USA. 1987;84:9185–9188. doi: 10.1073/pnas.84.24.9185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanchez P., Marche P. N., Rueff-Juy D., Cazenave P. A. J. Immunol. 1990;144:2816–2820. [PubMed] [Google Scholar]
- 31.Li Y., Louzoun Y., Weigert M. J. Exp. Med. 2004;199:337–346. doi: 10.1084/jem.20031712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cowan N. J., Secher D. S., Milstein C. J. Mol. Biol. 1974;90:691–701. doi: 10.1016/0022-2836(74)90533-6. [DOI] [PubMed] [Google Scholar]
- 33.Ma J., Kearney J. F., Hendershot L. M. Mol. Immunol. 1990;27:623–630. doi: 10.1016/0161-5890(90)90004-j. [DOI] [PubMed] [Google Scholar]
- 34.Schiffer M., Girling R. L., Ely K. R., Edmundson A. B. Biochemistry. 1973;12:4620–4631. doi: 10.1021/bi00747a013. [DOI] [PubMed] [Google Scholar]
- 35.Argon Y., Burrone O. R., Milstein C. Eur. J. Immunol. 1983;13:301–305. doi: 10.1002/eji.1830130406. [DOI] [PubMed] [Google Scholar]
- 36.Sekiguchi D. R., Yunk L., Gary D., Charan D., Srivastava B., Allman D., Weigert M. G., Luning Prak E. T. J. Immunol. 2006;176:6879–6887. doi: 10.4049/jimmunol.176.11.6879. [DOI] [PubMed] [Google Scholar]
- 37.Chan O. T., Hannum L. G., Haberman A. M., Madaio M. P., Shlomchik M. J. J. Exp. Med. 1999;189:1639–1648. doi: 10.1084/jem.189.10.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rezanka L. J., Kenny J. J., Longo D. L. Cell Immunol. 2005;238:38–48. doi: 10.1016/j.cellimm.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 39.Li Y., Li H., Weigert M. J. Exp. Med. 2002;195:181–188. doi: 10.1084/jem.20011453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cyster J. G., Goodnow C. C. Immunity. 1995;3:691–701. doi: 10.1016/1074-7613(95)90059-4. [DOI] [PubMed] [Google Scholar]
- 41.Cyster J. G., Hartley S. B., Goodnow C. C. Nature. 1994;371:389–395. doi: 10.1038/371389a0. [DOI] [PubMed] [Google Scholar]
- 42.Huang H., Kearney J. F., Grusby M. J., Benoist C., Mathis D. Proc. Natl. Acad. Sci. USA. 2006;103:3734–3739. doi: 10.1073/pnas.0600214103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mandik-Nayak L., Bui A., Noorchashm H., Eaton A., Erikson J. J. Exp. Med. 1997;186:1257–1267. doi: 10.1084/jem.186.8.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsuiji M., Yurasov S., Velinzon K., Thomas S., Nussenzweig M. C., Wardemann H. J. Exp. Med. 2006;203:393–400. doi: 10.1084/jem.20052033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.William J., Euler C., Primarolo N., Shlomchik M. J. J. Immunol. 2006;176:2142–2151. doi: 10.4049/jimmunol.176.4.2142. [DOI] [PubMed] [Google Scholar]
- 46.Fukuyama H., Nimmerjahn F., Ravetch J. V. Nat. Immunol. 2005;6:99–106. doi: 10.1038/ni1151. [DOI] [PubMed] [Google Scholar]
- 47.Kearney J. F. Springer Semin. Immunopathol. 2005;26:377–383. doi: 10.1007/s00281-004-0184-0. [DOI] [PubMed] [Google Scholar]
- 48.Baumgarth N., Tung J. W., Herzenberg L. A. Springer Semin. Immunopathol. 2005;26:347–362. doi: 10.1007/s00281-004-0182-2. [DOI] [PubMed] [Google Scholar]
- 49.Notkins A. L. Trends Immunol. 2004;25:174–179. doi: 10.1016/j.it.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 50.Pisetsky D. S. Rheum. Dis. Clin. N. Am. 2004;30:575–587. doi: 10.1016/j.rdc.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 51.Binder C. J., Silverman G. J. Springer Semin. Immunopathol. 2005;26:385–404. doi: 10.1007/s00281-004-0185-z. [DOI] [PubMed] [Google Scholar]
- 52.Manson J. J., Mauri C., Ehrenstein M. R. Springer Semin. Immunopathol. 2005;26:425–432. doi: 10.1007/s00281-004-0187-x. [DOI] [PubMed] [Google Scholar]
- 53.Herzenberg L. A., Baumgarth N., Wilshire J. A. Curr. Top Microbiol. Immunol. 2000;252:3–13. doi: 10.1007/978-3-642-57284-5_1. [DOI] [PubMed] [Google Scholar]
- 54.Yurasov S., Wardemann H., Hammersen J., Tsuiji M., Meffre E., Pascual V., Nussenzweig M. C. J. Exp. Med. 2005;201:703–711. doi: 10.1084/jem.20042251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bensimon C., Chastagner P., Zouali M. EMBO J. 1994;13:2951–2962. doi: 10.1002/j.1460-2075.1994.tb06593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cohen P. L., Eisenberg R. A., Dermer G. B. Clin. Exp. Immunol. 1984;55:509–515. [PMC free article] [PubMed] [Google Scholar]
- 57.Solling K., Solling J., Romer F. K. Acta Med. Scand. 1981;209:473–477. doi: 10.1111/j.0954-6820.1981.tb11632.x. [DOI] [PubMed] [Google Scholar]
- 58.Hopper J. E., Golbus J., Meyer C., Ferrer G. A. J. Clin. Immunol. 2000;20:123–137. doi: 10.1023/a:1006686514743. [DOI] [PubMed] [Google Scholar]
- 59.Abraham R. S., Geyer S. M., Price-Troska T. L., Allmer C., Kyle R. A., Gertz M. A., Fonseca R. Blood. 2003;101:3801–3808. doi: 10.1182/blood-2002-09-2707. [DOI] [PubMed] [Google Scholar]
- 60.Berggard I., Peterson P. A. J. Biol. Chem. 1969;244:4299–4307. [PubMed] [Google Scholar]
- 61.Kohler G. Proc. Natl. Acad. Sci. USA. 1980;77:2197–2199. doi: 10.1073/pnas.77.4.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sekiguchi D. R., Jainandunsing S. M., Fields M. L., Maldonado M. A., Madaio M. P., Erikson J., Weigert M., Eisenberg R. A. J. Immunol. 2002;168:4142–4153. doi: 10.4049/jimmunol.168.8.4142. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.