Abstract
In this report, we show that yccV, a gene of unknown function, encodes a protein having an affinity for a hemimethylated oriC DNA and that the protein negatively controls dnaA gene expression in vivo.
The minimum replication origin of Escherichia coli (oriC) has an elevated number of DNA adenine methylation (Dam) sites (18) that become hemimethylated immediately after initiation of replication and stay in this state for one-quarter of the generation time; these sites differ from most Dam sites located elsewhere on the chromosome (6). The finding that the hemimethylated oriC interacts with an outer membrane preparation of E. coli (20) prompted a search for hemimethylated DNA binding proteins in the membrane fraction. The existence of such a category of proteins in the membrane has been reported previously; in fact, Southwestern blot analysis of the membrane preparation demonstrated the presence of 25- and 16-kDa peptides reacting with a hemimethylated oriC probe (9). These proteins might participate in the hemimethylated oriC membrane sequestration that prevents initiation of chromosome replication in E. coli (16).
Lu et al. (17) isolated the seqA gene that codes for a 21-kDa peptide and has an affinity for the hemimethylated oriC DNA (24). In a seqA mutant, the duration of the hemimethylation period of oriC is shortened, and the reinitiation of replication occurs at oriC repeatedly in a single replication cycle (17). These phenotypes may be explained by a lack of hemimethylated oriC sequestration by either SeqA or a SeqA membrane complex; the membrane obtained from the seqA mutant failed to bind the hemimethylated oriC DNA in vitro (2, 24). Although these results show the predominant role of the SeqA protein in oriC sequestration, the auxiliary role played by the membrane in the process should not be disregarded. Shakibai et al. (23) observed that the hemimethylated oriC binding activity of SeqA is stimulated by addition of a membrane protein preparation designated SeqB. It has also been found that the membrane obtained from a seqA mutant stimulates the protective activity of a His-tagged SeqA protein against DNase I attack of hemimethylated oriC DNA (7).
In order to isolate auxiliary factors of SeqA, we adopted a strategy of searching for factors among the dnaA46 suppressor gene products. The rationale behind this approach was based on the observation that the seqA mutation partially suppresses the temperature sensitivity of dnaA46 (17). Likewise, mutation of the auxiliary factor gene should also result in suppression of dnaA46. Previously, this type of work has been undertaken by Katayama's group, who used a system involving random insertion of Tn10-Tetr into the chromosome. In this way, they found one new suppressor mutation for dnaA46, hslU (12).
Isolation of dnaA46 suppressors by mini-Tn10 insertion.
Briefly, random insertion of mini-Tn10 into the chromosome containing dnaA46 (KA413ΔH) (Table 1) was accomplished by infection of the mutant with a lambda phage (λ1098) carrying mini-Tn10 (27), followed by plating of the culture on Luria-Bertani (LB) agar plates containing tetracycline and incubation at 40°C. Twenty-three colonies were isolated. To identify the mini-Tn10 insertion site on the chromosome, PstI chromosome fragments containing mini-Tn10 were cloned into the Bluescript plasmid (pSK+) or pUC18 (Table 1), and DNA sequences surrounding mini-Tn10 were determined and compared to the E. coli whole genome sequence (3) by using the BLAST program (Genetics Computer Group [GCG], University of Wisconsin, Madison, Wis.). To determine these sequences, DNA fragments surrounding mini-Tn10 were amplified by PCR with one primer, P15 (5′ GATCATATGACAAGATGTGTATCCACC), homologous to IS10R and a second primer, P10 (5′ACGCAAACCGCCTCTCCCCG) or P11 (5′ GCGAAAGGGGGATGTGCTGC), homologous to the vectors on either side of the polylinker. These fragments were sequenced with the same primers.
TABLE 1.
Bacterial strains, phage, and plasmids
| Strain, phage, or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| KH5402-1 | thyA thr trpE9829(Am) ilv tyrA(Am) metE deo supF6(Ts) | T. Katayama 19 |
| KA413 | KH5402-1 dnaA46 ilv+ | 11 |
| KA413ΔH | KA413 aphA::Km | This study |
| M2-24 | Temperature-resistant revertant of KA413ΔH | This study |
| M2-5 | Temperature-resistant revertant of KA413ΔH | This study |
| M2-11 | Temperature-resistant revertant of KA413ΔH | This study |
| JM109 | 28 | |
| C600 | 1 | |
| C600 dam | C600 dam-13 | This study |
| C600 yccV | C600 yccV::mini-Tn10 | This study |
| C600 dam yccV | C600 dam-13 yccV::mini-Tn10 | This study |
| WM1032 | dnaA508 ilvB or ilvG lac supD thi trp | W. Messer |
| WM1032 yccV | WM1032 yccV::mini-Tn10 | This study |
| WM2007 | TC3145 lac his RB1 proB902 rpsL supF81(Ts) thi trp(Am) tsX | W. Messer (RB220 derivativec) |
| WM2007 yccV | WM2007 yccV::mini-Tn10 | This study |
| Phage | ||
| λ1098 | Pam80 cI857(Ts) nin5 mini-Tn10 | 27 |
| Plasmids | ||
| pSK+ | Bluescript, pBR322 derivative vector | Stratagene |
| pUC18 | pBR322 derivative vector | 28 |
| pSK24 | 6.7-kbp Pstl chromosomal fragment carrying yccV::mini-Tn10 cloned on pSK+a | This study |
| pSK11 | Deletion part of a Pstl chromosomal fragment carrying mutS::mini-Tn10 cloned on pSK+ | This study |
| pUC5 | Deletion part of a Pstl chromosomal fragment carrying rpoN::mini-Tn10 cloned on pUC18 | This study |
| pDK5 | pBR322 derivative expression vector | 13 |
| pES6ΔHE | Derived from pDK5 by inserting between EcoRI and HindIII two complementary oligonucleotides, generating an XbaI site flanked by a hexahistidine coding sequence | This study |
| pES6ΔHEE1 | RBS from PT7 and YccV ORF cloned between the EcoRI and XbaI sites of pES6ΔHEb | This study |
The fragment was from PstI position 8776 of GenBank sequence ae000198.gb_ba2 to PstI position 1008 of GenBank sequence ae000199.gb_ba2.
RBS, ribosome binding site; ORF, open reading frame.
See reference 5.
The locations were confirmed by Southern hybridization of chromosomal PstI digests with the corresponding probes.
At present, we have identified three sites of mini-Tn10 insertion that suppress the dnaA46 thermosensitivity; they are yccV (in strain M2-24, with the corresponding PstI fragment cloned into pSK+, resulting in plasmid pSK24), rpoN (in strain M2-5, with the corresponding PstI fragment partially deleted after cloning into pUC18, resulting in plasmid pUC5), and mutS (in strain M2-11,with the corresponding PstI fragment also partially deleted after cloning into pSK+, resulting in pSK11), and they are located at 22, 74, and 69 min, respectively, on the genetic map.
The insertion mutants were further analyzed for genetic linkage between the tetracycline-resistant element (mini-Tn10) and the suppressor mutations. P1 phages prepared from M2-24, M2-5, or M2-11 were used for transduction of the tetracycline resistance marker in KA413ΔH at 30°C and then tested for growth at 40°C. The cotransduction frequencies between tetracycline resistance and growth capacity at 40°C were 100% for deletions of yccV and rpoN and 50% for the mutS deletant. Therefore, the mini-Tn10 insertion in either yccV or rpoN is responsible for suppression of dnaA46 thermosensitivity, whereas the insertion in mutS is not sufficient for suppression, indicating that an unidentified mutation closely linked to mutS is necessary.
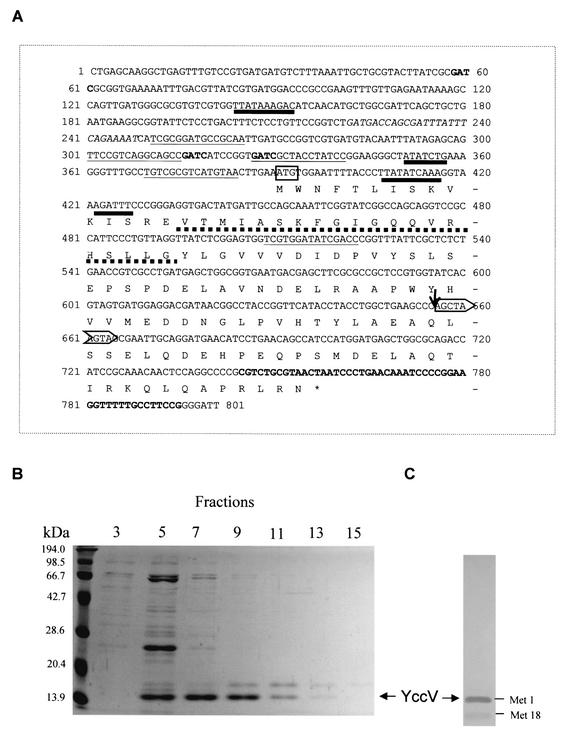
Precise insertion site of mini-Tn10 and reading frame of yccV.
As shown in Fig. 1, mini-Tn10 was found at almost the middle of the yccV gene in the M2-24 suppressed strain according to our DNA sequencing data. The yccV gene, including its upstream region, has some interesting features; first, it has two possible translational starts, one at Met1-391nt (Fig. 1A) and the other at Met18-442nt, which produce 122- and 104-amino-acid peptides, respectively. Second, the upstream region of yccV contains two DnaA boxes, two ATP-DnaA boxes (22, 25), three Dam sites, and surprisingly, five FIS binding sites (10). A putative helix-turn-helix motif is located near the N-terminal region of yccV, which is partially deleted when the second translation start is considered. Except with yccV paralogs, no significant homology could be found in the GenBank-EMBL database with yccV coding sequence by a BLAST search when the GCG program was used.
FIG. 1.
yccV sequence and purification of the His-tagged YccV protein. (A) Coding region (122 amino acids) and flanking sequences. The start codon is enclosed in a box. A putative helix-turn-helix motif (according to the hthscan program in GCG sequence analysis software) is underlined with a dotted line. The vertical arrow indicates the location of the mini-Tn10 insertion in the nucleotide sequence. The horizontal arrow indicates the 9 bp duplicated upon transposon insertion. A putative transcription promoter sequence described by Blattner et al. (3) is indicated by italics. A putative transcription terminator (according to the GCG software) is indicated by boldface type. GATC sites are indicated by boldface type. Sequences that exhibit a reasonable match with suggested consensus binding sequences for DnaA protein (22, 25) and for FIS (10) are underlined with thick and thin lines, respectively. (B) Purification. Strain JM109 harboring plasmid pES6ΔHEE1 (Table 1) was grown in LB medium containing ampicillin (100 μg/ml). At an optical density at 600 nm of 0.8, 0.5 mM isopropyl-β-d-thiogalactopyranoside was added to induce synthesis of the fusion protein for 3 h at 37°C. The YccV protein was purified according to the manufacturer's instructions by using 5 ml of chelating Sepharose FF (Pharmacia) coupled with nickel ion. The results of polyacrylamide gel electrophoresis of the eluted fractions followed by Coomassie blue staining are shown. Fractions 7 to 9 containing His-tagged YccV (14.9 kDa) were pooled for further experiments. The protein was dialyzed against buffer containing 50 mM Tris-HCl, 5 mM EDTA, 1 mM dithiothreitol, and 1 M NaCl (pH 7.5) supplemented with 10% glycerol before freezing at −80°C. (C) Western blot analysis of the purified fractions with monoclonal anti-His tag antibodies.
We constructed a vector (pES6ΔHEE1) that produced YccV (from Met1-391nt) fused to hexahistidine at the C-terminal end. Western analysis of the purified protein with anti-His tag antibodies revealed that the two translation starts may have been active in our conditions, producing two peptides of the expected length, the more active of which was Met1-391nt (Fig. 1B and C). Construction of plasmid pES6ΔHEE1 was performed as follows. The polylinker of expression vector pDK5 (13) was modified in order to add a polyhistidine tail at the carboxy-terminal end of the cloned gene (at an XbaI site [underlined nucleotides]) by insertion of two annealed complementary oligonucleotides, P28 [5′ AATTCCTCTAGA(CAT)6TGA] and P29 [5′ AGCTTCA(ATG)6TCTAGAGG], downstream of the Ptac promoter between the EcoRI and HindIII restriction sites, resulting in plasmid pES6ΔHE. The yccV gene was then PCR amplified from the E. coli chromosome with primers P36 (5′GGAATTCGAAGGAGATATACATATGTGGAATTTTACCTTATA) and P31 (5′ GCTCTAGAGTTACGCAGACGCGGGGC). The PCR fragment, containing a ribosome binding site (indicated by boldface type in the P36 sequence), was cut by EcoRI and XbaI (underlined nucleotides) and cloned into pES6ΔHE cut by EcoRI and XbaI to obtain pES6ΔHEE1.
Overexpression of dnaA in yccV mutant.
dnaA46 aphA yccV::mini-Tn10 (KA413ΔH yccV) can form colonies on LB agar plates at 40°C but not at 42°C. Suppression does not depend on the aphA mutation (21) since yccV::mini-Tn10 can suppress the thermosensitivity of KA413. yccV::mini-Tn10 also suppresses another dnaA thermosensitive mutation, dnaA508 (strain WM1032) at 40°C. The main mechanism of suppression of temperature sensitivity of dnaA(Ts) is either induction of stable DNA replication (14) or overexpression of dnaA(Ts) (8).
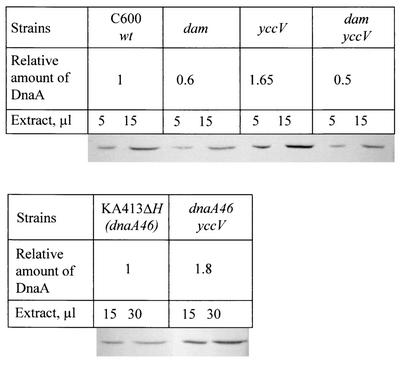
We examined the possibility of overexpression by measuring cellular concentrations of the DnaA46 protein in both the mutant and revertant cultures by immunoblotting analysis followed by densitometric quantification. We found 1.8 times more DnaA46 protein in the revertant culture than in the parental strain (Fig. 2). This overexpression can explain the reversion of temperature sensitivity at 40°C.
FIG. 2.
Quantitation of DnaA protein. Overnight cultures were diluted 100-fold in LB medium and grown to an optical density at 650 nm of 0.8 at 37°C (C600 derivatives) or at 30°C (KA413 derivatives). Cells were collected by centrifugation and lysed in buffer containing 25 mM Tris-HCl (pH 6.8), 4% sodium dodecyl sulfate, 20% glycerol, and 50 mM β-mercaptoethanol. Total protein contents were estimated by the Bradford assay, and cell extracts were normalized on the basis of their protein concentrations. Extracts were serially diluted into sodium dodecyl sulfate-polyacrylamide gel electrophoresis loading buffer and boiled. After sodium dodecyl sulfate-polyacrylamide gel electrophoresis, the samples were transferred to a polyvinylidene difluoride membrane for immunoblot analysis. DnaA protein was detected on the membrane by using an anti-DnaA antiserum with the aide of a peroxidase-conjugated secondary antibody in the presence of 1,4-dichloronaphthol. The signal on the membrane was scanned and quantified with the Imagequant program (Molecular Dynamics).
Since the dnaA promoter region contains many Dam sites like oriC and its activity is affected by Dam methylation (5, 15), we tested the effect of introducing the dam-13 mutation into the yccV single mutant. Again, quantitative immunoblot analysis (Fig. 2) demonstrated that introduction of the dam-13 mutation resulted in reductions in the amount of the DnaA protein in both the wild-type and yccV strains to the same level, indicating that Dam methylation is required for overexpression of dnaA even in the presence of the yccV deletion.
In order to confirm that the overexpression of dnaA46 in the yccV deletion mutant operates at the transcriptional level, we compared expression of lacZ under the control of dnaAP1 and -P2 (4) in strain WM2007 with and without yccV deletion and found that the beta-galactosidase activity was consistently 1.5 times higher in WM2007 yccV than in WM2007, indicating that there was a higher level of transcription of dnaA in the yccV deletion mutant (data not shown).
The effect of the yccV mutation is not specific for dnaA since it also increases malE expression.
YccV as a hemimethylated DNA binding protein.
The observation that the dam-13 mutation cancels dnaA overexpression provoked by the yccV deletion suggests that YccV can negatively regulate dnaA gene expression when its promoter region is either methylated or hemimethylated. This could occur through binding of YccV itself to fully or hemimethylated DNA.
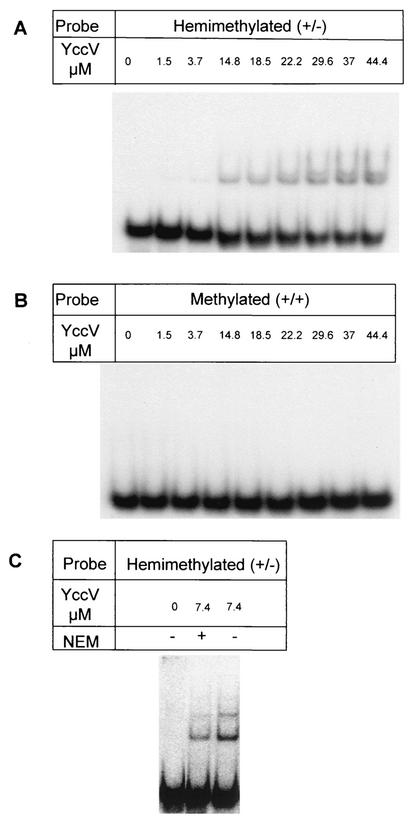
We examined this possibility by performing gel retardation experiments using as a probe either fully methylated or hemimethylated DNA corresponding to a part of oriC, in the presence of His-tagged YccV. An oriC probe was used because it contains multiple Dam sites.
The results of this experiment demonstrated that in fact YccV bound preferentially to the hemimethylated DNA probe, forming two complexes, whereas it failed to bind to the fully methylated DNA (Fig. 3A and B). A 50% shift of the hemimethylated probe (50 pM) was obtained at a high concentration of YccV (40 μM), indicating that the affinity of YccV for the hemimethylated DNA was relatively low compared to the affinity of SeqA. However, this shift was not due to contamination of SeqA during preparation of the His-tagged YccV since pretreatment of the preparation with N-ethylmaleimide (10 mM) for 10 min at 37°C, which inactivates SeqA (2), did not affect the binding activity (Fig. 3C).
FIG. 3.
Gel shift experiments. A 32P-labeled double-stranded oligonucleotide (75 bp) corresponding to oriC (nucleotide positions 74 to 148) was used as the probe for gel retardation experiments in the presence of different amounts of YccV protein. Hemimethylated (A and C) or fully methylated (B) probe was generated by annealing equal amounts of the complementary oligonucleotides, including (if necessary) N6-methyl deoxyadenosine, at the four GATC sites. After hybridization, the double-stranded oligonucleotide was purified by 12% polyacrylamide gel electrophoresis before labeling with [γ-32P]ATP (3,000 Ci/mmol; Amersham) for 1 h at 37°C in the presence of T4 polynucleotide kinase (Promega). The labeled oligonucleotide was then purified through a Micro Bio-Spin 30 chromatography column (Bio-Rad). Gel retardation assays were performed as described by Taghbalout et al. (26) by using 20-μl reaction mixtures containing 10 mM Tris-HCl (pH 7.6), 50 mM KCl, 1 mM EDTA, 1 mM dithiothreitol, 10% glycerol, 5 mM MgCl2, and 0.1% bovine serum albumin, the radioactive probe (5,000 cpm/0.5 fmol), and poly(dI-dC) as a competitor added at a 1,000-fold excess. After 10 min of incubation at room temperature, the reaction mixtures were loaded onto native 5% polyacrylamide gel electrophoresis gels, and electrophoresis was carried out at 4°C with a constant voltage in 0.5× Tris-borate-EDTA buffer. (A and B) YccV prepared as described in the legend to Fig. 1. (C) YccV preparation dialyzed against the final buffer without dithiothreitol was preincubated with 10 mM N-ethylmaleimide (NEM) (+) for 15 min at 37°C. −, no NEM treatment.
We examined the sequence specificity of YccV by performing DNase I footprinting with an oriC probe and did not find any protection even at a high concentration of YccV. Therefore, we cannot tell the sequence of YccV binding sites.
ADDENDUM
After our oral presentation concerning the yccV mutant at an EMBO workshop in 2000, T. Katayama communicated to us that he and coworkers isolated a yccV deletion as a suppressor of dnaA46 a few years ago.
Acknowledgments
This work was supported by grants from Association pour la Recherche sur le Cancer and by grant CR52 1090 from the DGA. A.T. thanks La Fondation pour la Recherche Médicale for support. E.D. is on the Institut National de la Recherche Agronomique staff and is on leave.
REFERENCES
- 1.Bachmann, B. J. 1987. Linkage map of Escherichia coli K-12, p. 1190-1219. In F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology. American Society for Microbiology, Washington, D.C.
- 2.Bahloul, A., J. Meury, R. Kern, J. Garwood, S. Guha, and M. Kohiyama. 1996. Co-ordination between membrane oriC sequestration factors and a chromosome partitioning protein, TolC (MukA). Mol. Microbiol. 22:275-282. [DOI] [PubMed] [Google Scholar]
- 3.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 4.Braun, R. E., K. O'Day, and A. Wright. 1985. Autoregulation of the DNA replication gene dnaA in E. coli K-12. Cell 40:159-169. [DOI] [PubMed] [Google Scholar]
- 5.Braun, R. E., and A. Wright. 1986. DNA methylation differentially enhances the expression of one of the two E. coli dnaA promoters in vivo and in vitro. Mol. Gen. Genet. 202:246-250. [DOI] [PubMed] [Google Scholar]
- 6.Campbell, J., and N. Kleckner. 1990. E. coli oriC and the dnaA promoter are sequestered from Dam methyltransferase following the passage of the chromosomal replication fork. Cell 62:967-979. [DOI] [PubMed] [Google Scholar]
- 7.d'Alençon, E., A. Taghbalout, R. Kern, and M. Kohiyama. 1999. Replication cycle dependent association of SeqA to the outer membrane fraction of E. coli. Biochimie 81:841-846. [DOI] [PubMed] [Google Scholar]
- 8.Hansen, F. G., S. Koefoed, and T. Atlung. 1992. Cloning and nucleotide sequence determination of twelve mutant dnaA genes of Escherichia coli. Mol. Gen. Genet. 234:14-21. [DOI] [PubMed] [Google Scholar]
- 9.Herrick, J., R. Kern, S. Guha, A. Landoulsi, O. Fayet, A. Malki, and M. Kohiyama. 1994. Parental strand recognition of the DNA replication origin by the outer membrane in Escherichia coli. EMBO J. 13:4695-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hübner, P., and W. Arber. 1989. Mutational analysis of a prokaryotic recombinational enhancer element with two functions. EMBO J. 8:577-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katayama, T., and T. Nagata. 1991. Initiation of chromosomal DNA replication which is stimulated without oversupply of DnaA in Escherichia coli. J. Biol. Chem. 269:12698-12703. [DOI] [PubMed] [Google Scholar]
- 12.Katayama, T., T. Kubota, M. Takata, N. Akimitsu, and K. Sekimizu. 1996. Disruption of the hslU gene, which encodes an ATPase subunit of the eukaryotic 26S proteasome homolog in Escherichia coli, suppresses the temperature-sensitive dnaA46 mutation. Biochem. Biophys. Res. Commun. 229:219-224. [DOI] [PubMed] [Google Scholar]
- 13.Kleiner, D., P. Wyatt, and M. J. Merrick. 1988. Construction of multicopy expression vectors for regulated overproduction of proteins in Klebsiella pneumoniae and other enteric bacteria. J. Gen. Microbiol. 134:1779-1784. [DOI] [PubMed]
- 14.Kogoma, T., and K. G. Lark. 1975. Characterization of the Escherichia coli DNA replication in the absence of protein synthesis: stable DNA replication. J. Mol. Biol. 94:243-256. [DOI] [PubMed] [Google Scholar]
- 15.Landoulsi, A., P. Hughes, and M. Kohiyama. 1989. dam methylation and the initiation of DNA replication on oriC plasmids. Mol. Gen. Genet. 216:217-223. [DOI] [PubMed] [Google Scholar]
- 16.Landoulsi, A., A. Malki, R. Kern, M. Kohiyama, and P. Hughes. 1990. The E. coli cell surface specifically prevents the initiation of DNA replication at oriC on hemimethylated DNA templates. Cell 63:1053-1060. [DOI] [PubMed] [Google Scholar]
- 17.Lu, M., J. L. Campbell, E. Boye, and N. Kleckner. 1994. SeqA: a negative modulator of replication initiation in E. coli. Cell 77:413-426. [DOI] [PubMed] [Google Scholar]
- 18.Messer, W., M. Meijer, H. E. Bergmans, F. G. Hansen, K. von Meyenburg, E. Beck, and H. Schaller. 1979. Origin of replication, oriC, of the Escherichia coli K-12 chromosome: nucleotide sequence. Cold Spring Harbor Symp. Quant. Biol. 43:139-145. [DOI] [PubMed] [Google Scholar]
- 19.Murakami, Y., H. Ohmori, T. Yura, and T. Nagata. 1987. Requirement of the Escherichia coli dnaA gene function for ori-2-dependent mini-F plasmid replication. J. Bacteriol. 169:1724-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ogden, G. B., M. Pratt, and M. Schaechter. 1988. The replicative origin of the E. coli chromosome binds to cell membranes only when hemimethylated. Cell 54:127-135. [DOI] [PubMed] [Google Scholar]
- 21.Reshetnyak, E., E. d'Alençon, R. Kern, A. Taghbalout, P. Guillaud, and M. Kohiyama. 1999. Hemi-methylated oriC DNA binding activity found in nonspecific acid phosphatase. Mol. Microbiol. 31:167-175. [DOI] [PubMed] [Google Scholar]
- 22.Schaefer, C., and W. Messer. 1991. DnaA protein/DNA interaction. Modulation of the recognition sequence. Mol. Gen. Genet. 226:34-40. [DOI] [PubMed] [Google Scholar]
- 23.Shakibai, N., K. Ishidate, E. Reshetnyak, S. Gunji, M. Kohiyama, and L. J. Rothfield. 1998. High-affinity binding of hemimethylated oriC by Escherichia coli membranes is mediated by a multiprotein system that includes SeqA and a newly identified factor, SeqB. Proc. Natl. Acad. Sci. USA 95:11117-11121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slater, S., S. Wold, M. Lu, E. Boye, K. Skarstad, and N. Kleckner. 1995. E. coli SeqA protein binds oriC in two different methyl-modulated reactions appropriate to its role in DNA replication initiation and origin sequestration. Cell 82:927-936. [DOI] [PubMed] [Google Scholar]
- 25.Speck, C., C. Weigel, and W. Messer. 1999. ATP- and ADP-DnaA protein, a molecular switch in gene regulation. EMBO J. 18:6169-6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taghbalout, A., A. Landoulsi, R. Kern, M. Yamazoe, S. Hiraga, B. Holland, M. Kohiyama, and M. Malki. 2000. Competition between the replication initiator DnaA and the sequestration factor SeqA for binding to the hemimethylated chromosomal origin of E. coli in vitro. Genes Cells 5:873-884. [DOI] [PubMed] [Google Scholar]
- 27.Way, J. C., M. A. Davis, D. E. Roberts, and N. Kleckner. 1984. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene 32:369-379. [DOI] [PubMed] [Google Scholar]
- 28.Yanisch-Perron, C., J. Viera, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequence of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]