Abstract
Flagellin is a major surface antigen for many bacterial species. The pathogen Salmonella enterica switches between two alternative, antigenic forms of its flagellin filament protein, either type B or C. This switching (flagellar phase variation) is achieved by stochastic inversion of a promoter that produces both type B flagellin (FljB) and an inhibitor (FljA) of type C flagellin formation. When the fljB-fljA operon is expressed, only type B flagella are produced; when the operon is not transcribed, the gene for type C flagellin (fliC) is released from inhibition and forms type C flagella. Long thought to be a transcription repressor, the FljA inhibitor is shown here to block both translation and use of the FliC protein by binding to an mRNA region upstream from the translation start codon. Bypass mutants resistant to this inhibition alter this mRNA region, and some prevent FljA–RNA binding. Other bypass mutations are duplications within the leader mRNA that make FljA essential for FliC assembly. Certain bypass mutations allow FljA to block FliC-dependent motility without blocking production of the FliC protein, per se. Other mutations in the FliC mRNA leader block expression of the unlinked fljB gene. Results suggest that mRNAs for types B and C flagellin compete for occupancy of a site that directs the product toward assembly and that FljA influences this competition. This mechanism may serve to prevent assembly of flagella with a mixture of subunit types, especially during periods of switching from one type to the other.
Keywords: FljA, translation
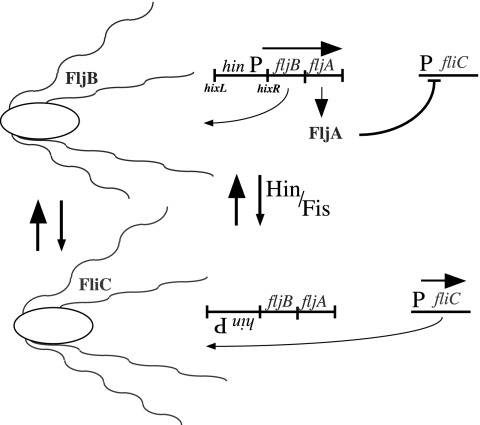
Many bacteria stochastically vary the nature of the cell surface by switching on and off the genes for alternative structural features (antigenic variation) (1). One of the first examples of this switching is flagellar phase variation of Salmonella typhimurium, which switches between two flagellar types (2). The alternative expression of the two flagellin antigens (FliB and FliC) (3) is achieved by inverting a chromosomal region that includes the promoter for a small operon that encodes one type of flagellin (FljB) and a protein (FljA) that inhibits synthesis of the other type (FliC), encoded by an unlinked gene (4, 5). This system is the premier example of gene control by site-specific DNA recombination (Fig. 1). Historically, the inhibitor FljA has been assumed to repress transcription of the fliC gene (6, 7). However, recent results suggest that FljA acts posttranscriptionally (8). In this work, we show that FljA interacts with fliC mRNA but not with DNA for the corresponding region. We characterize mutations that circumvent the inhibition by FljA and allow expression of FliC. Results suggest that FljA inhibits translation of fliC mRNA and also controls later use of the FliC protein. This mechanism may serve to minimize production of flagella with a mixture of flagellin types immediately after phase switching.
Fig. 1.
Flagellar phase variation in Salmonella. The reversible, Hin-mediated inversion of a 996-bp segment of the S. typhimurium chromosome results in the inversion of a promoter driving the expression of the fljB flagellin gene and fljA, which encodes an inhibitor of the alternative flagellin gene fliC. The Hin recombinase in conjunction with the Fis protein catalyzes a site-specific recombination reaction between the hixL and hixR recombination sites. (Upper) fljBA is transcribed to produce FljB flagellin, whereas fliC expression is inhibited by the action of FljA. (Lower) The promoter for the fljBA transcript is removed, and only fliC is expressed.
Results
FljA-Bypass Mutants in the fliC 5′ UTR.
A mutation that allows synthesis or use of FliC flagellin in the presence of the inhibitory protein FljA is an FljA-bypass mutation. Nine independently arising FljA-bypass mutations were isolated based on production of LacZ (β-gal) from the fliC::Mud(lac) insertion, and they proved to be closely linked to the fliC locus. The mutations affected a region upstream from the fliC coding sequence within the 5′ UTR of the fliC message (Fig. 2).
Fig. 2.
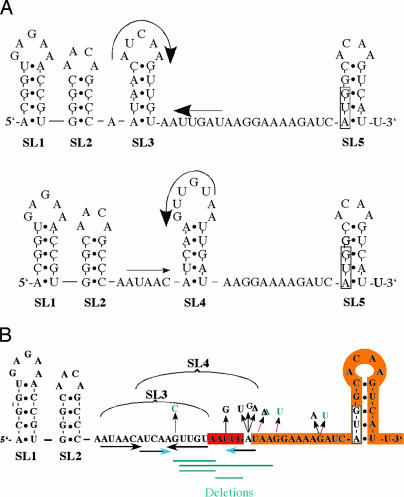
The 5′ UTR region of the fliC gene. (A) The RNA sequence of the 5′ UTR region of fliC from the +1 site of transcription through 12 bases after the translation-initiating AUG codon was subjected to the mfold RNA-folding program of Zucker (www.bioinfo.rpi.edu/applications/mfold). Five potential SL structures (SL1–SL5) were identified, corresponding to the two lowest-energy folds. SL3 and SL4 are competing SL structures and are depicted in Upper and Lower, respectively. The translation-initiating AUG codon is boxed. (B) Mutational changes in the fliC 5′ UTR that allow fliC-lacZ expression (Lac+) in cells expressing the FljA protein. Mutations in green were isolated in this study, and mutations in the fliC 5′ UTR that allow fliC+ expression (motility) in cells expressing the FljA protein (10) are depicted in black. The regions of the two duplications are highlighted in red [DUP-(−15 to −19)] and orange [DUP-(−13 to +15)].
This region of the mRNA includes five potential stem–loop regions (SL1–SL5; Fig. 2A), which were identified by using mfold software (Version 3.1; ref. 9). Two of the predicted SLs, SL3 and SL4, overlap and possess equal energies. They are mutually exclusive and would compete in their formation. All nine of the new FljA-bypass mutations affect the SL3 and SL4 regions (Fig. 2B, green). Seven additional FljA-bypass mutations described previously were found to affect bases between the SL3 loop and the AUG start codon of the fliC gene (Fig. 2B, black). These previously isolated mutants were selected based on their ability to restore FliC-dependent motility in the presence of FljA, and therefore they had to restore both the synthesis and the use of FliC (10). The mutants described here were isolated as lactose-utilizing (Lac+) revertants in a fliC-lac fljB+ FljAON background, and thus they were required to restore only translation of the fliC-lac fusion gene. The map positions of the FljA-bypass mutations isolated in both selections are consistent with FljA acting at the level of DNA (to block transcription) or at the level of mRNA (to block translation).
The FljA Inhibitor Binds to the RNA but Not the DNA Version of the fliC+ mRNA.
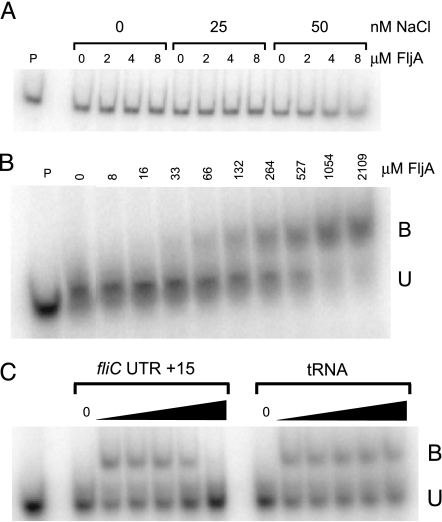
Purified FljA was tested for the ability to bind to the RNA and DNA versions of the sequence found at the 5′ end of the fliC mRNA (5′ UTR sequence). We detected no binding of FljA to a DNA fragment, including the fliC promoter, 5′ UTR, and N-terminal coding region (−160 through +100, relative to the AUG translation start codon; Fig. 3A). In contrast, His6-FljA did bind (with an estimated Kd of 370 nM) to a synthetic radiolabeled RNA whose sequence included the 5′ UTR through −62 to +15, relative to the AUG (Fig. 3B). This binding was sequence-specific in that it was prevented by providing an excess of unlabeled RNA of the same sequence, but not by a similar excess of tRNA (Fig. 3C). For the competition experiments, the RNA gel-shift reactions were performed as above, but they included a 20-, 200-, 2,000-, and 20,000-fold molar excess of either fliC RNA or tRNA.
Fig. 3.
Gel-shift assays of purified FljA binding to DNA and RNA. (A) Assay of purified His6-FljA binding to a DNA fragment containing the fliC promoter, 5′ UTR, and N-terminal coding sequences from −160 to +100 relative to the ATG start codon. (B) Assay of purified His6-FljA binding to an RNA sequence including the 5′ UTR of fliC and the first 15 bases of the coding sequence. B, bound; U, unbound. (C) Competition assays of purified His6-FljA binding to the RNA sequence in B with either unlabeled RNA of the same sequence (Left) or tRNA (Right) as the chase. All reactions contained 264 nM His6-FljA; excess unlabeled RNA was added at 20-, 200-, 2,000-, and 20,000-fold molar excess with respect to the concentration of the added fliC transcript (see Materials and Methods).
Binding of FljA to the fliC 5′ UTR of Bypass Mutants.
Several sorts of mutation allow FliC-LacZ expression in strains expressing the FljA inhibitor. Deletion, duplication, and single-base substitution mutations within the fliC 5′ UTR allowed FliC-dependent motility, regardless of whether or not FljA was present. One remarkable mutation, a large duplication including both 5′ UTR sequences and the first 15 bases of coding sequence, allowed FliC-dependent motility, but only in the presence of FljA. Thus, in this mutant strain, FljA behaved as an activator (rather than an inhibitor) of fliC gene expression.
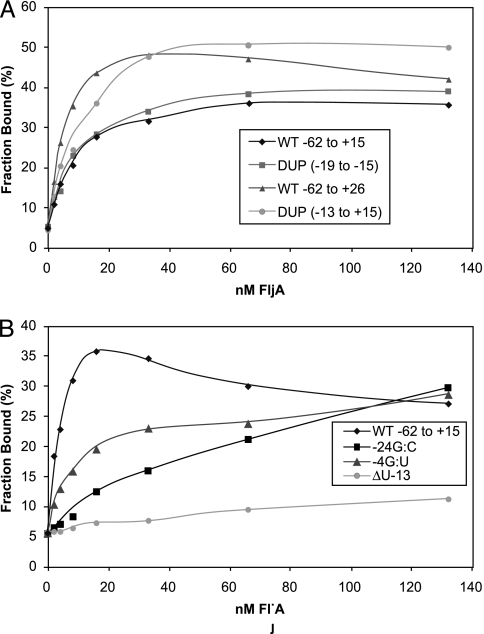
Purified RNAs of fliC 5′ UTR regions for 5 FljA-bypass mutants, −24G → C, −10G → U, and Δ(−13U), DUP-(−15 to −19) and DUP-(−13 to +15) (mutation positions are relative to the AUG start codon) were tested for binding His6-FljA in vitro by filter-binding assays (Fig. 4). All three single-base mutations were defective in binding His6-FljA relative to the wild-type (WT) fliC 5′ UTR sequence (Fig. 4B).
Fig. 4.
Filter-binding assays of purified FljA binding to 5′ UTR duplication mutants and single-base mutants. (A) Assay of purified His6-FljA binding to in vitro-transcribed RNA from the following fliC 5′ UTR mutants: −62 to +15 bp from AUG of fliC for WT and the fliC duplication mutants DUP-(−15 to −19) and DUP-(−13 to +15). Because DUP-(−13 to +15) is a longer transcript, a second WT transcript, WT −62 to +26, was used as a control. (B) Assays of purified His6-FljA binding to in vitro-transcribed RNA from WT, −24G → C, −4G → U, and Δ(−13U).
Bypass Mutants That Require FljA for Expression of FliC-Dependent Motility.
In contrast to the point mutations and deletions (above), two bypass mutations (both duplications) allowed motility in the presence of FljA. One duplication, DUP-(−15 to −19), was a repeat of the 5 bases in the 3′ stem of SL4 (Fig. 2B, red); this mutant was among those whose isolation is described here. Another mutant, DUP-(−13 to +15), was isolated in a previous hunt (Fig. 2B, orange) (10). Because one of these duplications showed FljA-dependent motility, it seemed likely that FljA would bind to the mutant RNA.
Consistent with our prediction, mRNA from mutant DUP-(−13 to +15) interacted normally with FljA protein (histidine-tagged) (Fig. 4A). Because the duplication extended into the fliC coding region, we used a WT region of equal length as a control in the binding test. Interestingly, this longer transcript bound FljA with higher affinity than the shorter WT RNA, suggesting that the RNA sequences in the very N-terminal coding region are important for FljA binding to the 5′ UTR, perhaps by affecting the folding of the UTR sequences.
Surprisingly, the second duplication, Dup-2 [DUP-(−19 to −15)], showed a phenotype that was distinct from that of all of the others. This duplication allowed fliC expression in the presence or absence of FljA, but it did not prevent binding of FljA protein to mRNA (Fig. 4A). This result suggests that FljA interacts with other sequences downstream from Dup-2 and that the spacing of the FljA interaction sites is essential for the negative regulation of fliC translation but not binding.
FljA Toeprint to fliC mRNA Reveals Binding to the Ribosome-Binding Site Region.
The 5′ end of the RNA sequence in the fliC 5′ UTR, to which FljA binds, was determined, and it is known as the FljA toeprint in the fliC 5′ UTR sequence. By allowing FljA to bind RNA containing the fliC 5′ UTR and then allowing primer extension by reverse transcriptase to proceed from a primer that hybridizes to the fliC coding sequence, the primer extension will stop when reverse transcriptase is blocked by FljA. In the absence of FljA, the primer-extension reaction will proceed to the 5′ end of the RNA sequence. A toeprint assay was performed to identify the 5′ position(s) of bound FljA on the fliC mRNA transcript (Fig. 5). Comparison of toeprint reactions in which FljA was present (+FljA lanes) or absent (−FljA and R lanes) revealed an FljA-specific signal ≈4 bases upstream of the AUG start codon (blue circles, Fig. 5) on all transcripts except that containing the Δ(−13U) mutation. These toeprint data suggest that FljA binds to the translation-initiation region of the fliC 5′ UTR to control fliC mRNA translation. The two duplications DUP-(−15 to −19) and DUP-(−13 to +15), which bind FljA as efficiently as WT, retain the WT toeprint. Indeed, DUP-(−13 to +15) has a duplicated toeprint corresponding to this duplicated site (Fig. 5, red circle).
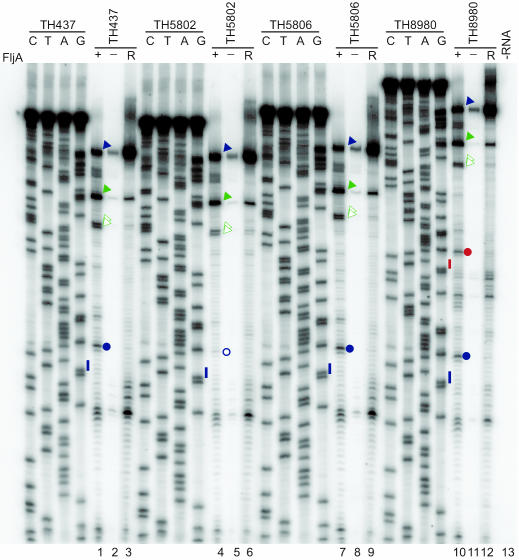
Fig. 5.
Toeprint analysis of FljA binding to the fliC mRNA 5′ UTR. Equal amounts of synthetic RNA transcripts and 32P-labeled oligonucleotide primer were incubated with FljA bound to Ni-NTA–agarose (+FljA; lanes 1, 4, and 7) or Ni-NTA–agarose alone (−FljA; lanes 2, 5, and 8), and the associated RNA was toeprinted. The RNAs analyzed contained WT fliC 5′ UTR (TH437; lanes 1–3), fliC5751[FljA-BP Δ(−13U) from AUG] (TH5802; lanes 4–6), fliC5755[FljA-BP DUP-(−15 to −19) from AUG] (TH5806; lanes 7–9), and fliC6323[DUP-(−13 to +15) relative to AUG] (TH8980; lanes 10–12). A reaction lacking RNA but containing radiolabeled oligonucleotide and FljA bound to Ni-NTA–agarose (−RNA, lane 13) showed that all signals in lanes 1–9 were RNA-specific. Primer-extension analysis of RNA in the presence of Ni-NTA–agarose (R; lanes 3, 6, 9, and 12) was used as a control to identify protein-specific signals. The PCR templates used to produce RNA were sequenced with the oligonucleotide used for primer extension so that toeprint sites could be precisely mapped. The nucleotide complementary to the dideoxynucleotide added to each sequencing reaction is indicated above the corresponding lane (C, T, A, and G) so that the sequence of the template can be directly deduced; the 5′–3′ sequence reads from top to bottom. The positions of an identically placed FljA-dependent signal observed with TH437, TH5806, and TH8930 are indicated with filled blue circles; the corresponding position in the TH5802 experiments is indicated with an open blue circle; FliC start codon positions are indicated with blue lines. The duplicated region in TH8980 includes the start codon (red line) and the FljA toeprint site (red filled circle). Blue arrowheads indicate the primer-extension products corresponding to mRNA 5′ ends; filled and open green arrowheads indicate toeprint sites at SL1 and SL2, respectively.
Effect of the FljA-Bypass Mutants in fliC on FliC-Dependent Motility.
All of the FljA-bypass mutations described previously were isolated by using motility as a criterion for mutants that can express FliC in the presence of inhibitory FljA (10). Motility requires expression of FliC as well as the ability to assemble a functional flagellum by using the produced protein. The FljA-bypass mutations described here were isolated by using expression of fliC-lac as a criterion for mutants that can express FliC in the presence of inhibitory FljA. To determine whether FliC expression is sufficient to provide motility, we constructed the newly isolated bypass mutants based on FliC protein production upstream from a fliC+ gene, and we tested the resulting mutants for their ability to provide motility.
Assuming that FliC protein production is sufficient for motility, it was expected that these mutants, like those isolated on the basis of motility, would affect the upstream mRNA region and that the resulting mutations would provide motility (when given an intact FliC-coding sequence). Surprisingly, we found that two of the nine FljA-bypass alleles that were isolated based on FliC-LacZ protein production did not provide motility when the fusion allele was replaced by an intact FliC-coding sequence, which suggests that FljA not only inhibits FliC production but also impairs a step later in the flagellum assembly. Some control mutants restored the ability to synthesize the protein without restoring the ability to use the resulting protein. The mutations that restored FliC production without motility were the single-base substitutions −10 G → U and the deletion Δ-(11–23) (Fig. 6A).
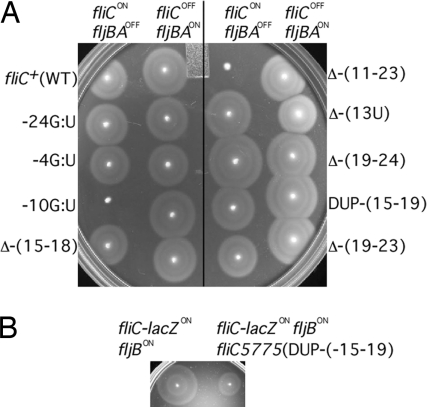
Fig. 6.
Motility of FljA-bypass mutants in the fliC operator. (A) Nine FljA-bypass mutants were placed in strains containing either Δhin-5717::FRT (fljBAOFF) or Δhin-5718::FRT (fljBAON) and assayed for the ability to swim in motility agar. (B) Effect of the 5-bp duplication DUP-(−15 to −19) in a fliC-lacZ gene fusion on expression of the unlinked fljB flagellin gene.
Effect of the Dup-2 FljA-Bypass Mutant in fliC on FljB-Dependent Motility.
The experiments above support the idea that FljA inhibits the translation and some aspect of the posttranslational use of the FliC flagellin. This inhibition is thought to occur when the stochastic switch that controls phase variation is in the position that expresses both FljB flagellin and FljA, the inhibitor of FliC expression. In the opposite phase, neither FljB nor FljA is expressed, and FliC is produced without impediment. One does not expect the FljA protein, or the mutations that affect its action on FliC, to play any role in expression of FljB.
Contrary to this expectation, Dup-2 [DUP-(−15 to −19)] inhibited FljB-dependent motility. This behavior was seen in strains that carried Dup-2 in combination with the insertion mutation fliC::MudK, which forms the fliC-lacZ gene fusion described above (Fig. 6B). WT cells in the FljB-(FljA) phase express FljB flagellin (and inhibit FliC production) and are motile. In this phase, cells with the Dup-2 mutation at the fliC::MudK insertion are expected to be motile (because of FljB) and express the FliC-LacZ fusion protein but not functional FliC protein. Surprisingly, the Dup-2 mutation impaired the expected fljB-dependent motility. Thus, a mutation near fliC appeared to act in trans on the expression of fljB.
This effect could reflect a competition between the FljB and FliC transcripts for translation and assembly of the product into a growing flagellum. The FljA regulator may support translation and assembly of FljB and inhibit translation and assembly of FliC (as described here). When both messages are produced because of the bypass mutations, the FljB and FliC transcripts may compete with each other for translation coupled to assembly; the secondary structure of the transcripts is crucial to the outcome of this competition.
Discussion
Initial work on the mechanism by which FljA prevents FliC expression revealed mutations like those reported here that affected the region between the promoter and the fliC coding sequence (10). This finding was interpreted in terms of transcriptional repression, and the mutations were assumed to alter an operator site. Recent work showed that FljA acts posttranscriptionally (8). Results presented here demonstrate that FljA binds particular sites in the FliC mRNA and impairs not only translation of the adjacent coding sequence but also some posttranslational aspect of assembly of the produced polypeptide into a functional flagellum.
The fliC 5′ UTR sequence includes several potential SL structures, two of which are formed alternatively. All of the FljA-bypass mutations identified affect the two competing SL structures, which are located immediately upstream from the translation-initiation site where the ribosome binds. This result suggests that FljA may prevent fliC mRNA translation by stabilizing one of the two alternative SL mRNA conformations.
The ability of FljA to bind the critical regions of the mRNA was demonstrated for RNA but not for the corresponding DNA sequences. The mRNA-binding specificity was demonstrated because binding of FljA to labeled RNA sequences was eliminated only by cognate competing RNA and because this binding was not seen for mutant versions of the sequence that eliminated in vivo inhibition by FljA.
Two FljA-bypass mutants had duplications of 5 and 27 nucleotides, respectively, within the control region. In these mutants, FljA still bound to the mRNA; however, binding did not prevent synthesis of the FliC protein. These results suggest that the position of FljA binding dictates the consequence for expression of FliC. Consistent with this possibility, toeprint analyses showed that with both duplications, FljA was bound at its original site; in the larger duplication, for which FljA was required for translation, the binding site was duplicated upstream.
Two aspects of the data suggest that this control mechanism includes more subtleties. Some FljA-bypass mutants altered the upstream region of the fliC mRNA so as to allow translation and production of the FliC protein without allowing that protein to contribute to a functional flagellum. A second finding is that several of the bypass mutations impair expression of the FljB protein, which is encoded by an unlinked gene. We suggest that these two observations reflect some kind of competition in the synthesis of the two flagellar proteins and that flagellar messages, fljB and fliC, are translated at specialized sites that direct the product toward excretion from the cell or assembly into the growing flagellum. If the two types of message competed for such sites, then the inhibitor FljA could help direct traffic by inhibiting the translation and direction of FliC protein and perhaps supporting the production and processing of FljB protein. We suggest that only one of the two flagellin types, FljB or FliC, will be expressed at a given flagellum so that flagella of mixed FljB and FliC are never produced. In this way, the cell might ensure that only one type of flagellum is produced, even during the time when switching is occurring, and both mRNA types are present.
We suggest that the mechanism described here and the possibilities mentioned above provide the cell with an exquisitely sensitive way of assembling a flagellum consisting entirely of one flagellin type or the other and avoiding the possibility of assembling a hybrid flagellum with both protein types. The biological value of flagellar phase variation is thought to be to allow cells to escape environments that are hostile to a particular flagellar type, when hosts are immune to one antigen or local surfaces bind one type tightly. By switching flagellar types, the cells can escape the environment. Such escape would be impossible if both flagellar types were made simultaneously during the changeover period. Alternatively, the two flagellar types may be functionally distinct such that either pure type functions well under some conditions, but a hybrid organelle is less valuable.
Materials and Methods
Bacterial Strains and Plasmids.
Bacterial strains used in this study and their origins are listed in Table 1.
Table 1.
Bacterial strains
| S. typhimurium genotype | Source* |
|---|---|
| LT2 (WT) | John Roth† |
| TH714 fljB5001∷MudJ | Ref. 11 |
| TH1077 fliC5050∷MudJ | Ref. 11 |
| TH2151 fla-2039(Δtar-flhD) fljB5001∷MudJ | |
| TH2592 fljB5001∷MudJ fljBe,n,xvh2 | |
| TH2788 fliY5221∷Tn10dTc | |
| TH3770 (ΔflgM5301) fljB5001∷MudJ | |
| TH4753 fliC5532∷tetRA fljB5001∷MudJ | |
| TH4754 fliC5533∷tetRA fljB5001∷MudJ | |
| TH4773 Δhin-132∷FRT (fljBAON) | Ref. 8 |
| TH5237 Δhin-132∷FRT fliC5469∷MudK | |
| TH5802 Δhin-132∷FRT fliC5469∷MudK | |
| fliC5751[FljA-BP Δ(−13U) from AUG] | |
| TH5806 Δhin-132∷FRT fliC5469∷MudK | |
| fliC5755[FljA-BP DUP(−15 to −19) from AUG] | |
| TH6297 Δhin-5717∷FRT (fljBAOFF)ΔfliC5533∷tetRA | |
| TH6298 Δhin-5718∷FRT (fljBAON) ΔfliC5533∷tetRA | |
| TH6303 Δhin-5717∷FRT (fljBAOFF)ΔfliC5532∷tetRA | |
| TH6305 Δhin-5717∷FRT fliC5050∷MudJ ΔfliC5532∷tetRA | |
| TH6307 Δhin-5717∷FRT fliC5469∷MudK ΔfliC5532∷tetRA | |
| TH6726 fliC5569∷tetRA | |
| TH6699 fliC5747∷Tn10dTc fliC5050∷MudJ | |
| TH6700 fliC5747∷Tn10d Tc fliC5469∷MudK | |
| TH8980 fliC6323 [DUP-(−13 to +15) relative to ATG] |
*Unless indicated otherwise, all strains were constructed during the course of this work.
†University of California, Davis.
Media and Standard Genetic Manipulations.
Media, growth conditions, transductional methods, and motility assays are described in refs. 11 and 12. The generalized transducing phage of S. typhimurium P22 HT105/1 int-201 was used in all transductional crosses (13). Tetracycline-sensitive (TcS) transductants were selected by mixing 0.1 ml of an appropriately diluted transducing phage stock with 0.1 ml of an overnight culture of recipient cells to achieve a final multiplicity between 1 and 10 phage per cell. As a control for spontaneous TcS mutants, 0.1 ml of 0.85% saline solution was mixed with 0.1 ml of the same overnight culture. After 30 min at room temperature for phage adsorption, the mixture was diluted 10- and 100-fold, plated on TcS selection plates, and incubated at 42°C for up to 48 h (14). The expression of the lac operon by strains carrying a fliC::MudK insertion was assessed by using 0.1% X-Gal and MacConkey/lactose (Difco) or tetrazolium/lactose indicator plates (15).
β-Gal Assays.
β-Gal assays were performed in triplicate on mid-logarithmic-phase cells as described in ref. 14. β-Gal activities are expressed as nmol·min−1·(OD650·ml)−1.
Isolation of fliC 5′ UTR Mutants Defective in Inhibition by FljA.
The transposon MudK was used to make lacZ translational (gene) fusions (16). Strain TH5237 carries a fliC::MudK translational fusion that adds the N-terminal 228 amino acids of FliC to a complete LacZ reading frame (8). This strain also carries a deletion of the hixL recombination site and part of the hin gene such that the fljBA promoter is oriented so as to express the fljBA structural genes, and it cannot invert to the fljBAOFF orientation. The FljA protein produced from this operon prevents production of β-gal from the unlinked fliC-lacZ fusion, resulting in a Lac− phenotype. Lac+ revertants, expected to allow FliC production to escape inhibition, were isolated by plating 0.1 ml from 70 independent overnight cultures of strain TH5237 on minimal (NCE) medium with 0.2% lactose as the sole carbon source and incubating at 37°C for 48 h. Two expected classes of Lac+ revertants were common: loss-of-function mutations in fljA and rare FljA-bypass mutations (gain-of-function mutations allowing expression of fliC despite the presence of the FljA inhibitor). To identify the rare fliC-linked mutations, the Lac+ revertants on each plate were pooled and used as transductional donors in a cross with an FljAON recipient strain (TH4773); selection plates included kanamycin to demand inheritance of the fliC::Mud(lac) fusion and bypass mutations in the immediate neighborhood. Transductants were screened to identify those that were Lac+ and able to express the fusion despite FljA produced from the unlinked fljBA operon.
Separation of FljA-Bypass Mutations from the fliC::MudK Insertion.
To separate the bypass mutations from the adjacent fliC5469::MudK insertion, P22 transducing phage was grown on the Lac+ revertants and used to transduce either TH6297 [Δhin-5717::FRT (fljBAOFF) ΔfliC5533::tetRA] or TH6298 [Δhin-5718::FRT (fljBAON) ΔfliC5533::tetRA] to tetracycline sensitivity on TcS selection plates containing X-Gal. Because the ΔfliC5533::tetRA allele is a deletion of the region from base −2 (relative to the fliC AUG initiation codon) through the −10 region of the fliC promoter, this transduction resulted in a replacement of the ΔfliC5533::tetRA allele with the entire mutated fliC 5′ UTR of the donor strain. The X-Gal-white, TcS transductants were isolated and screened for kanamycin sensitivity. Inheritance of the fliC::MudK would result in an X-Gal-blue (Lac+) phenotype, so the X-Gal-white phenotype screens against this (Lac−). X-Gal-white, kanamycin-sensitive, TcS transductants were further screened by PCR to verify that the ΔfliC5533::tetRA allele was replaced by the 5′ UTR region of fliC from the donor strain.
Construction of His6-FljA.
Plasmid construction of pJK604 for the purification of His6-FljA was as follows. The coding region of fljA from S. typhimurium LT2 was PCR-amplified by using primers HTV5′fljA (5′-ACA CGC TAG CGA AAA CCT GTA TTT TCA GGG CGC CAT GGA ATG TAT GGC TGT AAA TG-3′) and EcoRI3′fljA (5′-ACA GGA TTC TTA TTC AGC GTA GTC CGA AG-3′). The 582-bp PCR-amplified DNA fragment was digested with NheI and EcoRI and ligated into PT7-His6 expression vector pET28c (Novagen, EMD Biosciences, San Diego) digested with NheI and EcoRI.
Purification of His6-FljA.
Plasmid pJK604 in strain DE3 (Novagen, EMD Biosciences) was grown in 1-liter batches in LB plus 50 μg/ml kanamycin/12.5 μg/ml chloramphenicol (Sigma) at 37°C until the OD600 reached 0.8. Cells were induced with 1 mM isopropyl β-d-thiogalactoside and incubated at 30°C for 2 h. The cells were centrifuged at 10,000 × g for 5 min at 4°C, and the cell pellet was used immediately or stored at −80°C. After induction, His6-FljA was found in the soluble fraction at ≈50%. His6-FljA was purified under native conditions by using nickel–nitrilotriacetic acid (Ni-NTA)–agarose (Qiagen, Valencia, CA) according to the manufacturer’s instructions and buffer recipes, with the exception that all buffers included 5 mM 2-mercaptoethanol. The His6-FljA was step-eluted in 4 × 2.5 ml of elution buffer (50 mM NaH2PO4, pH 8.0/300 mM NaCl/250 mM imidazole/5 mM 2-mercaptoethanol), collecting 0.25-ml fractions. Fractions were assayed on 10% N-tris(hydroxymethyl)methylglycine (Tricine) gels (17), and the His6-FljA-containing fractions were pooled, dialyzed in storage buffer (100 mM Tris·HCl, pH 8.98 at 4°C/5 mM MgCl2/0.1 mM EDTA/5 mM DTT/150 mM NaCl/50% glycerol), and stored at −80°C.
RNA Gel-Shift Assays.
RNA probes for RNA gel-shift assays and filter-binding assays were made as follows. Primer sets T7UTRFLIC1 (5′-TAA TAC GAC TCA CTA TAG GGA CGG TGA GAA ACC GTG GG-3′), FliC+15R (5′-AAT GAC TTG TGC CAT GAT CT-3′), and T7UTRFLIC1 and FliC+26R (5′-CTG TTT GTA TTA ATG ACT TG-3′) were used to PCR-amplify the fliC region of S. typhimurium LT2 with the promoter of phage T7 tagged to the 5′ UTR end of fliC. The T7-tagged UTR-fliC PCR products were subsequently used as a template for a T7 RNAP-dependent in vitro transcription reaction (Promega) using [α-32P]UTP (NEN, PerkinElmer) to generate 32P-labeled RNA.
The RNA–protein-binding reaction was performed in a final volume of 10 μl containing 0.5–1.25 fmol of 32P-labeled RNA/His6-FljA in 10 mM Tris·HCl, pH 7.2/1 mM MgCl2/2.5 mM DTT/2 units of RNasin (Promega)/200-fold molar excess of Escherichia coli tRNA. The reaction was incubated at room temperature for 15 min, and 2.5 μl of 25% glycerol/0.1% xylene cyanol/0.1% bromophenol blue was added. Twelve microliters was loaded onto an 8% polyacrylamide/0.5× TBE gel (90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3) and electrophoresed at a constant 125 V at 4°C until the bromophenol blue ran to the bottom of the gel. The gels were dried and analyzed on a Storm 840 PhosphorImager (Molecular Dynamics) with ImageQuant software (GE Healthcare).
RNA Filter-Binding Assays.
RNA probes were prepared as described in RNA Gel-Shift Assays. The RNA–protein-binding reactions were performed in triplicate in a final volume of 10 μl containing 0.5 fmol of 32P-labeled RNA/His6-FljA in 10 mM Tris·HCl, pH 7.2/1 mM MgCl2/2.5 mM DTT/2 units of RNasin/200-fold molar excess of E. coli tRNA. The reactions were incubated at room temperature for 15 min and filtered onto a prewetted 0.2-μm Protran BA83 nitrocellulose filter (Schleicher & Schuell). The filters were washed with 1 ml of 10 mM Tris·HCl, pH 7.2/1 mM MgCl2/2.5 mM DTT, dried, and added to 5 ml of scintillation fluid, and their radioactivities were measured in a liquid-scintillation counter.
Determination of FljA-Binding Site in the fliC 5′ UTR.
Reverse transcriptase is often used to map transcription start sites by primer extension that stops at the 5′ end nucleotide of the mRNA sequence. If a protein binds with high affinity to the mRNA sequence, it will block the primer extension by reverse transcriptase. Reverse transcriptase will read the mRNA sequence up to the point where it is blocked by the bound protein. This point is called the toeprint of that protein for the 5′ end of the RNA sequence to which it binds (18). The FljA toeprint for the fliC 5′ UTR sequence was determined by using an RNA template that was prepared by first cloning the fliC 5′ UTR and part of the FliC-coding region (nucleotides −62 to +289) under the control of a T7 RNA polymerase promoter to generate RNA substrate for the binding to FljA and used to determine the FljA toeprint on this sequence. The primers used to amplify the fliC 5′ UTR and part of the FliC-coding region (nucleotides −62 to +289) were T7UTRFLIC1 (5′-TAA TAC GAC TCA CTA TAG GGA CGG TGA GAA ACC GTG GG-3′) and FliC+289R (5′-CCG CCA GTT CAC GCA C-3′). Synthetic RNA was synthesized in a T7 RNA polymerase-dependent in vitro transcription reaction (Promega) with [α-32P]UTP (MP Biomedicals). General procedures for primer-extension inhibition are described in ref. 18. Purified His-tagged FljA (70 μg) was incubated with 20 μl of Ni-NTA–agarose (50% slurry; Qiagen) at 4°C for 60 min in 200 μl of 50 mM Tris·HCl, pH 7.5/150 mM NaCl/1% Triton X-100. The resin was washed once with 200 μl of 50 mM sodium phosphate, pH 7.8/300 mM NaCl (wash buffer) and resuspended in 20 μl of binding buffer (10 mM Tris·HCl, pH 7.2/1 mM MgCl2/2.5 mM DTT/0.1 unit/μl RNasin/1.6 μg/μl yeast tRNA). For controls, FljA was omitted from the procedure. RNA (300 fmol) was preannealed for 5 min to oligonucleotide primer FliC+69R (5′-GGA CTG GAT TTG TTC AGG-3′) (9 × 106 cpm) in 5 μl of 50 mM Tris·HCl, pH 8.3/75 mM KCl/10 mM MgCl2/10 mM DTT/0.25 mM dNTP/1 unit/μl RNasin (RT buffer) and added to Ni-NTA containing or lacking bound FljA. After a 20-min incubation at room temperature, unbound material was removed, and the Ni-NTA was washed once with 200 μl of wash buffer. As controls to evaluate primer extension in the presence of Ni-NTA, RNA preannealed with primer was added directly to washed Ni-NTA at this point. Toeprint reactions were performed directly on the Ni-NTA samples by the addition of 10 μl of RT buffer containing 100 units of SuperScript II reverse transcriptase (Invitrogen) after a 30-min incubation at 37°C. Primer-extension products were obtained by phenol/chloroform extraction of the entire Ni-NTA reaction mixture and analyzed on denaturing 6% polyacrylamide gels.
Acknowledgments
We thank John Roth for constructive criticism during manuscript preparation. This work was supported by Public Health Service National Institutes of Health Grants GM62206 (to K.T.H.) and GM47498 (to M.S.S.).
Abbreviations
- Lac+
lactose-utilizing
- Ni-NTA
nickel–nitrilotriacetic acid
- SL
stem–loop
- TcS
tetracycline-sensitive.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.van der Woude M. W., Baumler A. J. Clin. Microbiol. Rev. 2004;17:581–611. doi: 10.1128/CMR.17.3.581-611.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrewes F. W. J. Pathol. Bacteriol. 1922;25:1509–1514. [Google Scholar]
- 3.Stocker B. A. D. J. Hyg. 1949;47:398–413. doi: 10.1017/s002217240001473x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zieg J., Hilmen M., Simon M. Cell. 1978;15:237–244. doi: 10.1016/0092-8674(78)90098-3. [DOI] [PubMed] [Google Scholar]
- 5.Zieg J., Silverman M., Hilmen M., Simon M. Science. 1977;196:170–172. doi: 10.1126/science.322276. [DOI] [PubMed] [Google Scholar]
- 6.Simon M., Silverman M. In: Gene Function in Prokaryotes. Beckwith J., Davies J., Gallant J. A., editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1983. pp. 211–227. [Google Scholar]
- 7.Macnab R. M. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhart F. C., Curtis R. I., Ingraham J. L., Lin E. C. C., Low K. B., Magasnik B., Reznikoff W. S., Riley M., Schaechter M., Umbarger H. E., editors. Washington, DC: Am. Soc. Microbiol; 1996. pp. 123–145. [Google Scholar]
- 8.Bonifield H. R., Hughes K. T. J. Bacteriol. 2003;185:3567–3574. doi: 10.1128/JB.185.12.3567-3574.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zuker M. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inoue Y. H., Kutsukake K., Iino T., Yamaguchi S. Gene. 1989;85:221–226. doi: 10.1016/0378-1119(89)90485-x. [DOI] [PubMed] [Google Scholar]
- 11.Gillen K. L., Hughes K. T. J. Bacteriol. 1991;173:2301–2310. doi: 10.1128/jb.173.7.2301-2310.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gillen K. L., Hughes K. T. J. Bacteriol. 1991;173:6453–6459. doi: 10.1128/jb.173.20.6453-6459.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis R. W., Botstein D., Roth J. R. Advanced Bacterial Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1980. [Google Scholar]
- 14.Maloy S. R., Stewart V. J., Taylor R. K. Genetic Analysis of Pathogenic Bacteria. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1996. p. 59. [Google Scholar]
- 15.Karlinsey J. E., Pease A. J., Winkler M. E., Bailey J. L., Hughes K. T. J. Bacteriol. 1997;179:2389–2400. doi: 10.1128/jb.179.7.2389-2400.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Groisman E. A. Methods Enzymol. 1991;204:180–212. doi: 10.1016/0076-6879(91)04010-l. [DOI] [PubMed] [Google Scholar]
- 17.Schägger H., Jagow G. Anal. Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 18.Sachs M. S., Wang Z., Gaba A., Fang P., Belk J., Ganesan R., Amrani N., Jacobson A. Methods. 2002;26:105–114. doi: 10.1016/S1046-2023(02)00013-0. [DOI] [PubMed] [Google Scholar]