Abstract
The sudden increase in information derived from the completed Mycobacterium tuberculosis (Mtb) genome sequences has revealed the need for approaches capable of converting raw genome sequence data into functional information. To date, an experimental system for studying protein–protein association in mycobacteria is not available. We have developed a simple system, termed mycobacterial protein fragment complementation (M-PFC), that is based upon the functional reconstitution of two small murine dihydrofolate reductase domains independently fused to two interacting proteins. Using M-PFC, we have successfully demonstrated dimerization of yeast GCN4, interaction between Mtb KdpD and KdpE, and association between Esat-6 and Cfp-10. We established the association between the sensor kinase, DevS, and response regulator, DevR, thereby demonstrating the potential of M-PFC to study protein associations in the mycobacterial membrane. To validate our system, we screened an Mtb library for proteins that associate with the secreted antigen Cfp-10 and consistently identified Esat-6 in our screens. Additional proteins that specifically associate with Cfp-10 include Rv0686 and Rv2151c (FtsQ), a component and substrate, respectively, of the evolutionary conserved signal recognition pathway; and Rv3596c (ClpC1), an AAA-ATPase chaperone involved in protein translocation and quality control. Our results provide empirical evidence that directly links the Mtb specialized secretion pathway with the evolutionary conserved signal recognition and SecA/SecYEG pathways, suggesting they share secretory components. We anticipate that M-PFC will be a major contributor to the systematic assembly of mycobacterial protein interaction maps that will lead to the development of better strategies for the control of tuberculosis.
Keywords: Cfp-10, DevR, interaction, secretion, virulence
Mycobacterium tuberculosis (Mtb) is responsible for 2–3 million deaths annually (World Health Organization, Progress Report, 2004), yet little is known about the molecular basis of its pathogenicity. Despite major technical advancements in extracting information from genome sequences, gene function can be attributed to only ≈58% of the ORFs of Mtb (1). The latter emphasizes the need to develop novel approaches to better understand gene function and thus virulence and is a key challenge in the postgenomic era. It has become increasingly clear that virulence pathways are mediated by networks of interacting proteins. Furthermore, the physical association between a protein of unknown function and a known protein indicates that the former often has a function related to that of the latter. Therefore, we believe that the development of an in vivo technology to study the protein–protein association of genetically intractable pathogens such as Mtb will enhance the dissection of virulence pathways and significantly advance our understanding of the mechanisms of disease.
Importantly, an abundance of studies have shown that some protein associations do not occur in vitro or in unrelated surrogate hosts and require a more “natural” intracellular environment (2–4). Until now, an effective mycobacterial equivalent of the yeast two-hybrid (Y2H) method system has not existed. It is clear that bacterial protein interaction mapping has not attained the level of complexity as has the yeast counterpart. An exception is the protein network of Helicobacter pylori (5) that yielded >1,000 Y2H interactions, connecting close to half of the proteome. Previously, we successfully exploited the Y2H system to study Mtb virulence (6) and signal transduction (7). Nevertheless, yeast does have certain limitations: (i) interactions occur in the nucleus, (ii) membrane proteins represent a problem, (iii) bacterial proteins do not undergo appropriate posttranslational modification, (iv) self activation can be a significant problem, and finally (v) high G+C DNA is sometimes not well tolerated.
In this study, we have developed a simple and rapid method termed mycobacterial protein fragment complementation (M-PFC) to study Mtb protein–protein association in mycobacteria. We have shown that when two mycobacterial interacting proteins are independently fused with domains of murine dihydrofolate reductase (mDHFR), functional reconstitution of the two mDHFR domains can occur in mycobacteria, thereby allowing us to select for mycobacterial resistance against trimethoprim (TRIM). To establish M-PFC as an effective method to functionally dissect and connect virulence pathways, we screened an Mtb H37Rv library for proteins that associate with the virulence determinant Cfp-10 and performed a series of validation experiments using the Y2H system and in vivo pull-down assays. Our data demonstrate that we have successfully developed a simple and robust system that enables the study of protein–protein association in mycobacteria. We anticipate that M-PFC will significantly contribute toward the dissection and linking of Mtb virulence pathways, impact high-throughput screening approaches, and contribute to emerging disciplines such as systems biology.
Results
M-PFC.
mDHFR is a small 21-kDa monomeric protein that contains three structural fragments (F[1], F[2], and F[3]) comprising two domains, an adenine-binding domain (F[2]) and a discontinuous domain (F[1] and F[3]; ref. 8). Reconstitution of mDHFR activity has been used previously to study protein–protein association in vivo (9). We exploited the biochemical properties of mDHFR to develop a system for dissecting protein–protein association in vivo using the model organism Mycobacterium smegmatis (Msm) as host. The strategy is based on the principle that if two mycobacterial interacting proteins A and B are independently fused to the mDHFR complementary fragments F[1,2] and F[3], respectively, and cotransformed into mycobacteria, in vivo association of A and B will facilitate the functional reassembly of the F[1,2] and F[3] into active mDHFR enzyme, conferring resistance to TRIM. We anticipated that the addition of small tags (F[1,2], 105 amino acids; and F[3], 80 amino acid) to interacting proteins will minimally affect protein–protein association. Despite the fact that both prokaryotic and eukaryotic DHFRs are targets of the antifolate drug, TRIM, it has been shown that mammalian DHFR has a ≈12,000-fold lower affinity for TRIM than does bacterial DHFR (10). The selective targeting of mycobacterial DHFR by TRIM is the basis of the in vivo assay and allows for the identification of recombinant mycobacteria expressing mDHFR by screening for growth in the presence of TRIM concentrations that inhibit endogenous Msm DHFR (Fig. 1). Mycobacterial cells expressing complementary fragments of mDHFR fused to interacting proteins will survive in medium containing TRIM. Msm is an ideal host for M-PFC experiments, because this strain is sensitive to TRIM (≈50 μg/ml on 7H11) and is a nonpathogenic rapid grower that is an excellent surrogate for studying Mtb gene regulation and secretion (11, 12).
Fig. 1.
Diagram illustrating the principle of M-PFC. Interacting proteins A and B are fused to complementary mDHFR fragments F[1,2] and F[3]. Cotransformation of AF[1,2] and BF[3] fusions in Msm results in the functional reconstitution of mDHFR activity and subsequent growth on TRIM plates, whereas proteins that do not interact (A and C) will not reconstitute F[1,2] and F[3] and will not allow transformants to survive on TRIM plates.
Protein–Protein Association in Mycobacteria.
To study protein association in mycobacteria, we constructed a series of Escherichia coli–mycobacterial M-PFC vectors, pUAB100 and pUAB200 (Fig. 2). Previously, the mDHFR fragments were engineered such that the interacting protein pairs and F[1,2] and F[3] were separated by a flexible glycine linker peptide of 10 amino acids (9). In proof-of-concept experiments designed to demonstrate the feasibility of M-PFC to detect a diverse range of protein–protein associations in Msm, we selected several well characterized interacting partners, namely Saccharomyces cerevisiae GCN4 (9), Mtb two-component proteins KdpD (Rv1028c)/KdpE (Rv1027c) (7) and Mtb secreted antigens Esat-6 (Rv3875)/Cfp-10 (Rv3874) (13). We generated these bait and prey plasmids as C-terminal fusions with the complementary fragments of mDHFR to generate the interacting protein pairs GCN4[F1,2]/GCN4[F3], KdpD[F1,2]/KdpE[F3], and Esat-6[F1,2]/Cfp-10[F3]. The corresponding plasmids were cotransformed into Msm, and transformants were selected on 7H11 medium supplemented with kanamycin (KAN) and hygromycin (HYG; 7H11/KAN/HYG). Transformants were then subcultured onto 7H11KAN/HYG plates containing 50 μg/ml TRIM (7H11/TRIM; Fig. 3). Growth was observed in all cases, strongly suggesting that GCN4[F1,2]/GCN4[F3], KdpD[F1,2]/KdpE[F3], and Esat-6[F1,2]/Cfp-10[F3] associate in Msm. Importantly, the control clones expressing only one protein fused to F[1,2] (GCN4[F1,2], KdpD[F1,2], or Esat-6[F1,2]) and carrying an empty plasmid producing only F[3] showed no growth on TRIM plates. Furthermore, pairing clones with unrelated proteins, (e.g., Esat-6 and KdpD) or swapping F[1,2] or F[3] indicates that association of [F1,2] and [F3] does not occur spontaneously, and that only interacting domains independently fused to both mDHFR fragments reconstitute F[1,2] and F[3]. Thus, M-PFC is capable of specifically detecting protein–protein associations in Msm.
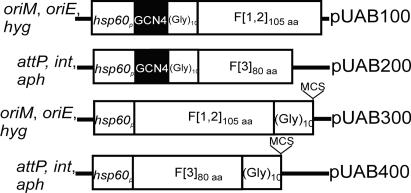
Fig. 2.
Schematic diagram of the plasmids used in M-PFC. pUAB100 and pUAB300 are episomal mycobacterial–E. coli shuttle plasmids, whereas pUAB200 and pUAB400 are integrating mycobacterial–E. coli shuttle plasmids. The preferred plasmid choice for making DNA libraries is pUAB300. F[1,2] and F[3] are the complementary fragments of mDHFR; GCN4 is the S. cerevisiae leucine-zipper sequence; (GLY)10 is the flexible 10-aa Gly linker; aph confers resistance to KAN; hyg confers resistance to HYG; hsp60p is the hsp60 promoter; oriM is the origin of replication for propagation in mycobacteria; oriE is the origin of replication for propagation in E. coli; and int and attP are the integrase and phage attachment sites, respectively, from mycobacteriophage L5.
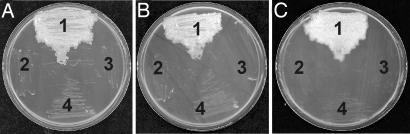
Fig. 3.
Exploiting M-PFC to demonstrate protein–protein association of yeast and Mtb proteins in Msm. Msm cells were independently transformed with M-PFC plasmids producing GCN4[F1,2]/GCN4[F3], KdpD[F1,2]/KdpE[F3], and Esat-6[F1,2]/Cfp-10[F3]. Transformants were subcultured on 7H11 medium containing TRIM and incubated at 37°C for 3–6 days. Growth is indicative of protein–protein association. (A) (1) GCN4[F1,2]/GCN4[F3], (2) GCN4[F1,2]/hsp60[F3], (3) GCN4[F3]/hsp60[F1,2], and (4) hsp60[F1,2]/hsp60[F3]. (B) (1) KdpD[F1,2]/KdpE[F3], (2) KdpD[F1,2]/hsp60[F3], (3) KdpE[F3]/hsp60[F1,2], and (4) hsp60[F1,2]/hsp60[F3]. (C) (1) Esat-6[F1,2]/Cfp-10[F3], (2) Esat-6[F1,2]/hsp60[F3], (3) Cfp-10[F3]/hsp60[F1,2], and (4) hsp60[F1,2]/hsp60[F3].
M-PFC Can Detect Association Between Membrane and Cytoplasmic Components of a Mycobacterial Signal Transduction Pathway.
To assess whether M-PFC is capable of detecting interactions between Mtb cytosolic and membrane signaling proteins, we studied the interaction between the Mtb membrane-spanning sensor histidine kinase, DevS (Rv3132c), and its corresponding response regulator DevR (Rv3133c; Fig. 4A). The Mtb DevR/DevS proteins comprise a two-component signaling system that was shown to be required for triggering the Mtb genetic response to hypoxia and nitric oxide, two environmental cues that may contribute to latency in vivo (14). Cotransformation of plasmids encoding DevRF[3] and DevSF[1,2] into Msm and subsequent growth of transformants on 7H11/TRIM demonstrated that association between DevRF[3] and DevSF[1,2] functionally reconstitutes mDHFR, thereby resulting in Msm resistance to TRIM (Fig. 4A). In contrast, clones containing empty control plasmids or unrelated proteins (e.g., KdpE; data not shown) showed no growth on 7H11/TRIM plates, strongly suggesting that the interaction is specific. Therefore, M-PFC is effective in detecting association between a membrane-spanning sensor protein and the corresponding cytoplasmic response regulator. Interestingly, clones producing DevRF[3] and DevSF[1,2] grew slightly slower on 7H11/KAN/HYG/TRIM compared to clones producing KdpDF[3]/KdpEF[1,2] and Esat-6F[3]/Cfp-10F[1,2], suggesting that the DevRF[3]/DevSF[1,2] interaction is weaker compared to the KdpDF[3]/KdpEF[1,2] and Esat-6F[3]/Cfp-10F[1,2] interactions (as shown later).
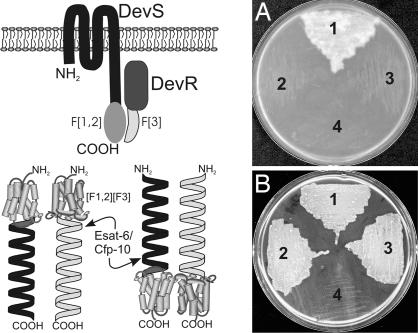
Fig. 4.
Effect of localization and orientation on interaction. (A) M-PFC is capable of detecting protein–protein association between two-component proteins in the mycobacterial membrane. The membrane-localized sensor histidine kinase, DevS, and its cognate response regulator, DevR, were fused to F[1,2] and F[3] to produce DevRF[3] and DevSF[1,2]. Cotransformation of plasmids producing these fusions into Msm and subsequent growth of transformants on 7H11/HYG/KAN/TRIM confirm the association between DevR and DevS. (1) DevRF[3] and DevSF[1,2], (2) DevRF[3] and hsp60F[1,2], (3) hsp60F[3] and DevSF[1,2], and (4) hsp60[F1,2]/hsp60[F3]. (B) Orientation of N- or C-terminal fusions does not influence Esat-6/Cfp-10 association. Esat-6 and Cfp-10 were fused to either the N or C terminus of F[1,2] or F[3] to generate Esat-6F[1,2], F[1,2]Esat-6, Cfp-10F[3], and F[3]Cfp-10. Cotransformation of interacting pairs Esat-6F[1,2]/Cfp-10F[3] and F[1,2]Esat-6/F[3]Cfp-10 and subsequent growth of transformants on 7H11/HYG/KAN/TRIM are indicative of protein–protein association (1). Esat-6F[1,2]/Cfp-10F[3], (2) F[1,2]Esat-6/F[3]Cfp-10, (3) GCN4[F1,2]/GCN4[F3], and (4) hsp60[F1,2]/hsp60[F3].
Fusion of F[1,2] and F[3] to the N or C Terminus of Esat-6 and Cfp-10 Does Not Significantly Influence Association.
To determine whether M-PFC is influenced by the N- or C-terminal orientation of the small F[1,2] or F[3] fusions, Esat-6 and Cfp-10 were cloned into pUAB100, pUAB300, and pUAB200, pUAB400 to generate Esat-6F[1,2], F[1,2]Esat-6 and Cfp-10F[3], F[3]Cfp-10 fusions, respectively. As shown in Fig. 4B, clones cotransformed with plasmids generating Esat-6F[1,2]/Cfp-10F[3] and F[1,2]Esat-6/F[3]Cfp-10 showed robust growth, whereas the controls showed no growth on 7H11/TRIM. Thus, M-PFC is a flexible system that, at least so far as the Esat-6 and Cfp-10 interaction is concerned, appears to be unaffected by the orientation of the DHFR domains.
Developing a Quantitative Assay for M-PFC.
The data showed that reconstitution of mDHFR due to protein–protein association in Msm could be easily monitored by a survival-based assay on 7H11/TRIM plates. However, the study of the effect of single amino acid substitutions or protein modifications on protein–protein association or domain-swapping experiments necessitates a more sensitive and quantitative assay. As a result, a colorimetric and fluorescent plate assay based on the widely used oxidation/reduction indicator Alamar blue (AB) was developed. AB is a compound that has been widely used in quantitative and qualitative assays to assess the sensitivity of mycobacteria to antimycobacterial compounds in a 96-well format (15). Msm harboring the interacting clones GCN4[F1,2]/GCN4[F3], KdpD[F1,2]/KdpE[F3], Esat-6[F1,2]/Cfp-10[F3], and DevRF[3]/DevSF[1,2] were cultured in 7H9/HYG/KAN and freshly inoculated into 96-well microtiter plates containing 7H9/KAN/HYG/TRIM. A change from nonfluorescent blue to fluorescent pink color indicates reduction of AB. The intensity of the pink color directly correlates with the extent of bacterial growth, which in turn depends upon the degree of reconstitution of mDHFR (F[1,2] and F[3]) driven by the interacting mycobacterial proteins (Fig. 5). Msm clones containing interacting partners grew in 7H9/KAN/HYG/TRIM media, as was evident by the development of a pink color due to the reduction of AB (Fig. 5). More importantly, there was virtually no color change observed in the vector control confirming the specificity of the assay. Furthermore, to accurately measure the strength of interaction, fluorescence was quantified at an excitation wavelength of 530 nm and an emission wavelength of 590 nm. Therefore, the reduction of AB can be used to detect a broad range of protein–protein associations of varying strengths in mycobacteria.
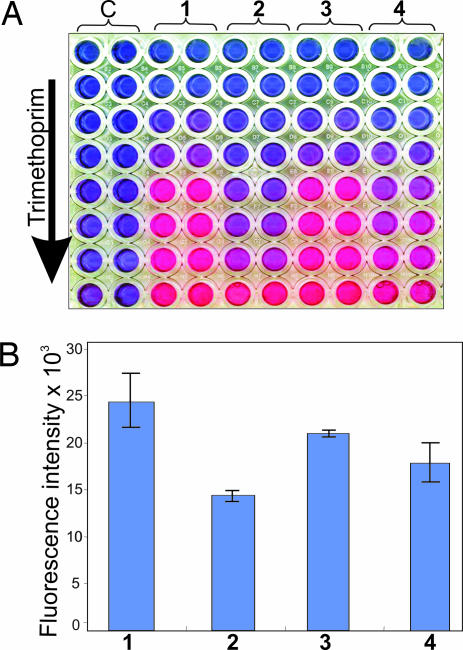
Fig. 5.
Adapting the AB fluorescent assay to quantify the strength of protein–protein association in mycobacteria. (A) Samples were processed in 96-well plates according to Methods. A change from nonfluorescent blue to fluorescent pink indicates reduction of AB and is indicative of protein–protein interaction. Msm cells were cotransformed with plasmids producing the corresponding protein partners. (1) GCN4[F1,2]/GCN4[F3], (2) DevS[F1,2]/DevR[F3], (3) Esat-6[F1,2]/Cfp-10[F3], and (4) KdpD[F1,2]/KdpE[F3]; C indicates the empty control constructs hsp60[F1,2]/hsp60[F3]. (B) The change in color intensity as shown in the microtiter plate was measured by using a microplate reader at an excitation wavelength of 530 and emission wavelength of 590 nm. TRIM concentration used ranged from 600 μg/ml (row 1) to 4.68 μg/ml (row 8). Samples were analyzed in duplicate, and all wells contained equal numbers of Msm cells (106 colony-forming units).
Exploiting M-PFC to Dissect an Mtb Virulence Pathway.
An advantage of protein interaction technologies is that genes involved in particular virulence pathways or signaling cascades can be identified, studied, and linked even if they are essential. We evaluated the applicability of M-PFC to discover virulence pathway components not previously accessible to standard approaches (i.e., gene deletion techniques). To do so, we tested the hypothesis that M-PFC will identify additional essential gene products participating in the secretion of Esat-6 and Cfp-10, two effector components of a yet-to-be-defined Mtb specialized secretion system (16, 17). Esat-6 (Rv3875) is the known partner of Cfp-10 (13, 16, 18), whereas Cfp-10 (Rv3874) is a major T cell antigen encoded by esxB present in the RD1 region and is required for full virulence of Mtb (17). An Mtb H37Rv genomic DNA prey library consisting of 5 × 105 independent clones was made in pUAB300, transformed into Msm cells containing the stable integrative M-PFC bait vector pUAB400-Cfp10, and screened for interacting proteins (see Methods). Proteins that were found to associate with Cfp-10 are listed in Table 1. Importantly, multiple overlapping clones of the same gene were identified (see legend of Table 1), thereby providing support that the association is indeed true. We note that Esat-6 was identified multiple times in the library screen, providing additional evidence that M-PFC can effectively be exploited to find interacting clones from an Mtb library. The specificity of these interactions was verified by determining the ability of individual Cfp-10F[3] interacting proteins to associate with unrelated proteins (e.g., KdpE) and cotransformation of Cfp-10F[3] interacting clones with the corresponding empty vector. In all cases, we did not observe any growth on 7H11/TRIM plates, thereby confirming the specificity of the interactions. Despite the fact that multiple overlapping clones were identified in the Cfp-10 screen, a finding that strongly suggests that the interactions are biologically significant, we used the Y2H system and in vivo pull-down assays to: (i) independently verify the Cfp-10/ClpC1, Cfp-10/Pks13, Cfp-10/FtsQ Cfp-10/Rv2240c, and Cfp-10/Rv0686 interactions (see supporting information, which is published on the PNAS web site) and (ii) identify interacting proteins that absolutely require the specific intracellular environment of mycobacteria. The Y2H data confirmed that Cfp-10/ClpC1, Cfp-10/FtsQ, Cfp-10/Rv2240c, and Cfp-10/Rv0686 do indeed associate. However, we were unable to confirm the Cfp-10/Pks13 interaction in yeast. Because Esat-6 and Cfp-10 form a tight complex, we hypothesized that the Cfp-10-interacting proteins might also associate with Esat-6. Indeed, of the six Cfp-10-interacting clones, Pks13, ClpC1, and FtsQ (and Cfp-10) also specifically associate with Esat-6. In addition, as suggested (19), our data show that Esat-6 oligomerizes (Fig. 6). To further investigate the role of ClpC1, we demonstrated that ClpC1 associates with the proteolytic component ClpP2 but not ClpP1 (Fig. 6). We did not identify Snm2 (Rv3871), a known Cfp-10-interacting protein (16), in our library screen. Because an exhaustive screen was not performed, this was not an unexpected finding. Therefore, we cloned snm2 as a F[1,2] fusion and confirmed that Snm2 strongly associates with Cfp-10 (Fig. 6). Finally, as anticipated, we identified TRIM-resistant clones that contained Mtb dfrA (Rv2763c), encoding DHFR. Conveniently, these TRIM-resistant clones act as internal controls for library complexity, and we used colony PCR to easily eliminate these clones from further analysis. In sum, 56% of the putative interacting clones contained Mtb dfrA, and 24% of the clones represent antisense and out-of-frame clones, whereas ≈20% of the clones contained in-frame clones.
Table 1.
Cfp-10-interacting Mtb proteins identified in an M-PFC screen
| Rv no. | Gene | Annotation and putative function |
|---|---|---|
| Rv3875* | esat-6 | Secreted antigen, known Cfp-10-interacting clone |
| Rv0686† | Belongs to COG0541, which contains members of the SRP-GTPase family. The family includes SRP and its receptor, which targets membrane proteins to bacterial or eukaryotic membranes for insertion: contains required IMP/GMP reductase domain and hydrophobic N terminus | |
| Rv2151c‡ | ftsQ | Cytoplasmic membrane protein; plays a role in cell division, delivered to membrane by the SRP pathway, and interacts with SecYEG translocase during insertion into the membrane |
| Rv3596c§ | clpC1 | AAA-ATPase chaperone with dual function: (i) ortholog of a component of protein translocation complex present in the outer and inner chloroplast and can facilitate translocation of precursor proteins across membranes by ClpC-dependent ATP hydrolysis, or (ii) is involved in protein quality control by retrotranslocating misfolded proteins into the degradation chamber of the proteolytic subunit, ClpP |
| Rv3800c¶ | pks13 | Polyketide synthase that catalyzes the last step of mycolic acid biosynthesis. Contains acyltransferase domain |
| Rv2240c‖ | Membrane protein of unknown function |
*Two classes of overlapping clones were identified: fusion junction at Esat-6 amino acid 9 and amino acid 13.
†Two classes of overlapping clones were identified: fusion junction at Rv0686 amino acid 201 and amino acid 334.
‡One class of interacting clone was identified: fusion junction at FtsQ amino acid 220.
§Four classes of overlapping clones were identified: fusion junction at 17, 38, 55, and 117 bp upstream of ATG generated as in-frame fusion with ClpCl.
¶One class of interacting clone was identified: fusion junction at Pks 13 amino acid 1340.
‖One class of interacting clone was identified: fusion junction at Rv2240c amino acid 127.
Fig. 6.
Using yed graph editor software to generate a miniinteraction network of Mtb proteins that associate with Cfp-10. The dashed arrows indicate self association. The red nodes refer to a previous Y2H study (16), whereas the yellow nodes refer to interactions found in this study. We confirmed the previously reported Cfp-10/Rv3871 association (16).
Discussion
Until now, an experimental system for studying protein–protein association in mycobacteria has not existed. This study shows that we have developed such a system, M-PFC, which allows us to study Mtb virulence mechanisms and other functional pathways through the analysis of protein–protein interaction. In M-PFC, the independent genetic coupling of mDHFR complementary fragments ([F1,2] and [F3]) with two mycobacterial-interacting proteins leads to the reconstitution of the mDHFR activity in vivo, at a concentration where endogenous mycobacterial DHFR activity is inhibited. We thoroughly tested M-PFC using well documented protein–protein interactions such as eukaryotic GCN4 (9), as well as the protein interactions of Mtb KdpD/KdpE (7), and DevR/DevS (14) and the secretory antigens Esat-6 and Cfp-10 (13). Using a number of controls, including unrelated proteins and empty vectors, we were able to confirm that these proteins specifically associate in mycobacteria. The system proved to be sensitive and robust, because we can study the association between membrane-located sensor kinases (KdpD and DevS) and their corresponding response regulators (KdpE and DevR), respectively. Moreover, it seems that the orientation of fusions has little to no effect on the refolding of F[1,2] and F[3] and is in agreement with a previously reported eukaryotic study (4). Nonetheless, we acknowledge that some proteins may require a free N or C terminus for interactions, a situation that would be apparent with testing. A major advantage of studying protein association in mycobacteria rather than in surrogate hosts such as yeast and E. coli is that appropriate modifications, cofactors, and the exclusive intracellular environments such as the mycobacterial cytoplasm and membrane may dictate the outcome of interactions. Nonetheless, using the Y2H system and pull-down assays, we were able to confirm all of the tested interactions with the exception of the Cfp-10/Pks13-interacting pair that may represent an example of an interaction that requires the mycobacterial cytoplasmic environment. A second advantage is the simplicity and robustness of the system; a typical screen is completed within 2 weeks and requires minimum manipulation.
Virulence pathways are mediated by complex networks of molecular interactions, which upon disruption alter protein–protein associations. Thus, protein–protein interactions typically suggest a direct link or role in a pathway. As a result, one of the central aims of this work was to thoroughly test the capacity of M-PFC to reveal undefined Mtb virulence mechanisms. Several studies (16, 17, 20–22) have shown that proteins in and outside of the Mtb RD1 region are involved in the secretion of the immunogenic antigens, Esat-6 and Cfp-10. An important feature of this yet-undefined specialized secretion system is that these small effector proteins contain no signal peptide. We characterized the Mtb specialized secretion system by performing a genome-wide screen for Cfp-10-interacting clones. We repeatedly identified Esat-6 as one of its interacting partners, which is consistent with previous studies (13, 16, 18) and validates M-PFC as an effective tool for screening the Mtb genome for interacting proteins. More importantly, we identified several previously undescribed components of the Mtb secretory pathway. For example, Rv0686, a member of the signal-recognition pathway (SRP)-GTPase family, was found to specifically interact with Cfp-10. Members of this family include SRP and its receptor, SR, and are involved in cotranslational targeting of proteins in the bacterial plasma membrane and eukaryotic endoplasmic reticulum membrane for secretion or membrane insertion (23). In Bacillus subtilis, both SRP and SR are involved in targeting the majority of the secreted proteins to the Sec translocase (24). Our data suggest that the SRP-GTPase ortholog (Rv0686) may facilitate targeting of Cfp-10 to the membrane by the SRP. In support of the above results, we identified a second substrate of the SRP, the cell division protein FtsQ, which has been widely used as a model protein to dissect SRP-dependent translocation of integral membrane proteins (25). Because FtsQ has been shown to interact with components of SecYEG translocon (25), it is possible that FtsQ participates in the delivery of Cfp-10 to the SecYEG translocon. Furthermore, we detected a positive interaction between Cfp-10 and the AAA-ATPase chaperone ClpC1 (a member of the Clp/Hsp100 family of proteins), which are involved in diverse functions, including secretion, gene regulation, protein refolding, and degradation (26, 27) and have been shown to associate with the translocation machinery in the chloroplast membrane (28). It may well be that Mtb ClpC1 facilitates Cfp-10 secretion by associating with the translocation complex in the membrane. Because ClpC can also target misfolded proteins into the chamber of proteolytic subunit, ClpP, for degradation (29), it can be argued that association with Cfp-10 is spurious. However, that ClpC1 selectively associated with the ClpP2 protease subunit (Fig. 5) but not with other proteolytic subunits (e.g., ClpP1) or unrelated control proteins strongly suggests the association is specific.
The interaction of Cfp-10 with Pks13 is unexpected. However, the association of Esat-6 also with Pks13 lends support to the idea that the interaction is physiologically relevant, especially because it was previously demonstrated that acetylation affects the interaction of Cfp-10 with Esat-6 (18). We hypothesize that the acyltransferase domain in the Pks13 may modify Cfp-10 through acylation. However, this assertion will require detailed experimental verification. Interestingly, N-terminal acylation of eukaryotic proteins occurs frequently and is required for proper translocation of proteins that lack a recognizable secretory signal sequence. Cfp-10 contains an Ala after the Met at the N terminus, whereas Esat-6, the known partner of Cfp-10, is acetylated at the N-terminal Thr residue (18). In eukaryotes, these amino acids account for ≈95% of the N-terminal acetylated residues (30).
In sum, M-PFC enabled us to shed light on the mechanism of protein secretion in Mtb and identified several components of the Cfp-10 secretory pathway. Our results directly link the Mtb specialized secretion pathway with the evolutionary conserved SRP and SecA/SecYEG pathways and provide strong evidence for an overlap between these pathways, suggesting they may share secretory components. Also, that M-PFC could implicate essential gene products such as Pks13, FtsQ, ClpC1, and the SecYEG complex (30) in this pathway demonstrates the potential of M-PFC to dissect virulence mechanisms that are not amenable to gene disruption strategies.
We have demonstrated the utility of a simple experimental system, M-PFC, for studying protein–protein association in mycobacteria. In so doing, we have identified previously uncharacterized players in a unique virulence and secretion pathway, thus opening avenues for future investigation. We anticipate that M-PFC will play an important role in the targeted disruption of protein interactions essential to Mtb virulence and latency and represents a potentially fruitful and as-yet-unexplored area of antimycobacterial drug development. We conclude that we have developed an experimental system not yet described in the mycobacterial field, one that has strong potential to identify and characterize novel virulence pathways involved in the persistence and pathogenesis of Mtb.
Methods
Strains and Media.
Cultivation and transformation of Mtb H37Rv and Msm mc2155 were performed as described (7). When necessary, Middlebrook medium was supplemented with KAN (25 μg/ml), HYG (50 μg/ml), or TRIM (40–50 μg/ml). E. coli DH10B was grown in LB supplemented with KAN (25 μg/ml) or HYG (150 μg/ml).
Plasmid Constructs for M-PFC.
The plasmids pKSFR (1, 2) and pKSFR (3) (kindly provided by Stephen W. Michnick, Département de Biochimie, Université de Montréal, Québec, QC, Canada) contain mDHFRF(1, 2) (F[1,2]) and mDHFRF(3) (F[3]), respectively, fused in-frame with a 10-aa flexible Gly linker (Gly-10) and GCN4 leucine-zipper coding sequence; these plasmids were used as templates for generating M-PFC plasmids. Complementary oligonucleotides containing the restriction enzyme sites BamHI and AccI were used to PCR amplify F[1,2] along with a flexible Gly linker and leucine-zipper (GCN4) sequences (GCN4-[Gly]10-F[1,2]). The PCR fragment was digested and ligated to BamHI/ClaI-digested pMV261 (7), generating the episomal vector pUAB100. Similarly, a PCR amplicon containing F[3] along with the flexible glycine linker and GCN4 sequences (GCN4-[Gly]10-F[3]) was digested and ligated with MfeI/HpaI-digested pMV361 to generate the integrating vector pUAB200. These constructs contain the GCN4 homodimerization domains fused to the N terminus of mDHFR fragments. When necessary, the GCN4 domains from pUAB100 and pUAB200 were replaced with bait or prey DNA sequences. For library screens, the M-PFC vectors were modified by cloning the F[3]-[Gly]10 sequence into the E. coli–mycobacterial shuttle vector pMV761 (7) to generate the integrating vector pUAB400. Similarly, the complementary fragment of mDHFR, i.e., F[1,2] along with the flexible glycine linker (F[1,2]-[Gly]10), was PCR-amplified and cloned in pMV762 (7) to create the episomal plasmid pUAB300.
M-PFC Library Screen.
The Mtb genomic DNA library containing 5 × 105 independent clones was prepared as described (6) and cloned into the unique ClaI site of pUAB300. The bait plasmid was constructed by PCR-amplifying cfp-10 and subsequent ligation to MunI/ClaI linearized pUAB400 to create pUAB400-Cfp10. The bait plasmid was transformed into Msm. The subsequent transformed strain was then electroporated with the Mtb library DNA. Interacting clones were selected by plating transformants on 7H11 media containing KAN, HYG, and TRIM (50 μg/ml). Colonies that showed growth on 7H11/TRIM plates were lysed, and plasmid DNA was isolated, amplified in E. coli, and again transformed into Msm containing pUAB400-Cfp10. Clones were again streaked on 7H11/TRIM plates and assessed for growth. Library plasmids from Msm clones that showed growth on TRIM plates were sequenced.
AB Assay.
Msm clones containing interacting plasmids were cultured in Middlebrook 7H9 medium containing HYG and KAN to an OD600 nm of 0.8. Cells were diluted in fresh 7H9 medium, and ≈106 cells were added to clear-bottom 96-well microtiter plates. Outer-perimeter wells were filled with sterile water to prevent dehydration. TRIM was dissolved in dimethyl sulfoxide, and 2-fold serial dilutions of the drug were made in 0.1 ml of 7H9 in microtiter plates. Wells containing drug only and no Msm cells were the autofluorescence controls. Additional controls consisted of wells containing cells and medium only. Plates were incubated for 12 h, after which 30 μl of AB (Biosource International, Camarillo, CA) solution (1:1 dilution in Middlebrook 7H9-Tween medium) was added to wells containing cells only, further incubated, and observed for the appearance of a pink color. AB was added to each well after the emergence of a pink color in the “cells-only” wells. Plates were further incubated at 37°C, and the result was recorded after 6 h. Fluorescence intensity was measured in a Cytofluor II microplate fluorometer (PerSeptive Biosystems, Framingham, MA) in bottom-reading mode with excitation at 530 nm and emission at 590 nm. Msm transformants containing empty vectors hsp60[F1,2]/hsp60[F3] show a low level of background fluorescence.
yed Graph Editor.
The network of proteins that associate with the Cfp-10 was generated by using yed graph editor software (Tübingen, Germany).
Supplementary Material
Acknowledgments
We thank John Trombley, Andrea Stanus, Olga Danielchanka, and Enatra Hale for excellent technical assistance and members of the Steyn laboratory, Mary Hondalus, Michael Niederweis, and Miriam Braunstein, for critical review of this manuscript. This work was supported by the University of Alabama at Birmingham Center for Aids Research (A.J.C.S.) and National Institutes of Health Grant AI058131 (to A.J.C.S.). A.S. is a recipient of a postdoctoral fellowship from the Heiser Foundation.
Abbreviations
- Mtb
Mycobacterium tuberculosis
- M-PFC
mycobacterial protein fragment complementation
- Msm
Mycobacterium smegmatis
- mDHFR
murine dihydrofolate reductase
- SRP
signal-recognition pathway
- Y2H
yeast two hybrid
- TRIM
trimethoprim
- KAN
kanamycin
- HYG
hygromycin
- AB
Alamar blue.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Tekaia F., Gordon S. V., Garnier T., Brosch R., Barrell B. G., Cole S. T. Tuber. Lung Dis. 1999;79:329–342. doi: 10.1054/tuld.1999.0220. [DOI] [PubMed] [Google Scholar]
- 2.Cohen B. D., Lowy D. R., Schiller J. T. Mol. Cell. Biol. 1993;13:6462–6468. doi: 10.1128/mcb.13.10.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li B., Fields S. FASEB J. 1993;7:957–963. doi: 10.1096/fasebj.7.10.8344494. [DOI] [PubMed] [Google Scholar]
- 4.Remy I., Michnick S. W. Proc. Natl. Acad. Sci. USA. 2001;98:7678–7683. doi: 10.1073/pnas.131216098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rain J. C., Selig L., De Reuse H., Battaglia V., Reverdy C., Simon S., Lenzen G., Petel F., Wojcik J., Schachter V., et al. Nature. 2001;409:211–215. doi: 10.1038/35051615. [DOI] [PubMed] [Google Scholar]
- 6.Steyn A. J., Collins D. M., Hondalus M. K., Jacobs W. R., Jr., Kawakami R. P., Bloom B. R. Proc. Natl. Acad. Sci. USA. 2002;99:3147–3152. doi: 10.1073/pnas.052705399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steyn A. J., Joseph J., Bloom B. R. Mol. Microbiol. 2003;47:1075–1089. doi: 10.1046/j.1365-2958.2003.03356.x. [DOI] [PubMed] [Google Scholar]
- 8.Bystroff C., Kraut J. Biochemistry. 1991;30:2227–2239. doi: 10.1021/bi00222a028. [DOI] [PubMed] [Google Scholar]
- 9.Pelletier J. N., Campbell-Valois F. X., Michnick S. W. Proc. Natl. Acad. Sci. USA. 1998;95:12141–12146. doi: 10.1073/pnas.95.21.12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Appleman J. R., Howell E. E., Kraut J., Kuhl M., Blakley R. L. J. Biol. Chem. 1988;263:9187–9198. [PubMed] [Google Scholar]
- 11.Braunstein M., Griffin T. I., Kriakov J. I., Friedman S. T., Grindley N. D., Jacobs W. R., Jr. J. Bacteriol. 2000;182:2732–2740. doi: 10.1128/jb.182.10.2732-2740.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harth G., Lee B. Y., Horwitz M. A. Infect Immun. 1997;65:2321–2328. doi: 10.1128/iai.65.6.2321-2328.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Renshaw P. S., Panagiotidou P., Whelan A., Gordon S. V., Hewinson R. G., Williamson R. A., Carr M. D. J. Biol. Chem. 2002;277:21598–21603. doi: 10.1074/jbc.M201625200. [DOI] [PubMed] [Google Scholar]
- 14.Roberts D. M., Liao R. P., Wisedchaisri G., Hol W. G., Sherman D. R. J. Biol. Chem. 2004;279:23082–23087. doi: 10.1074/jbc.M401230200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McNerney R., Kiepiela P., Bishop K. S., Nye P. M., Stoker N. G. Int. J. Tuberc. Lung Dis. 2000;4:69–75. [PubMed] [Google Scholar]
- 16.Stanley S. A., Raghavan S., Hwang W. W., Cox J. S. Proc. Natl. Acad. Sci. USA. 2003;100:13001–13006. doi: 10.1073/pnas.2235593100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guinn K. M., Hickey M. J., Mathur S. K., Zakel K. L., Grotzke J. E., Lewinsohn D. M., Smith S., Sherman D. R. Mol. Microbiol. 2004;51:359–370. doi: 10.1046/j.1365-2958.2003.03844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okkels L. M., Muller E. C., Schmid M., Rosenkrands I., Kaufmann S. H., Andersen P., Jungblut P. R. Proteomics. 2004;4:2954–2960. doi: 10.1002/pmic.200400906. [DOI] [PubMed] [Google Scholar]
- 19.Daugelat S., Kowall J., Mattow J., Bumann D., Winter R., Hurwitz R., Kaufmann S. H. Microbes Infect. 2003;5:1082–1095. doi: 10.1016/s1286-4579(03)00205-3. [DOI] [PubMed] [Google Scholar]
- 20.Fortune S. M., Jaeger A., Sarracino D. A., Chase M. R., Sassetti C. M., Sherman D. R., Bloom B. R., Rubin E. J. Proc. Natl. Acad. Sci. USA. 2005;102:10676–10681. doi: 10.1073/pnas.0504922102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brodin P., Majlessi L., Marsollier L., de Jonge M. I., Bottai D., Demangel C., Hinds J., Neyrolles O., Butcher P. D., Leclerc C., et al. Infect Immun. 2006;74:88–98. doi: 10.1128/IAI.74.1.88-98.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pym A. S., Brodin P., Brosch R., Huerre M., Cole S. T. Mol. Microbiol. 2002;46:709–717. doi: 10.1046/j.1365-2958.2002.03237.x. [DOI] [PubMed] [Google Scholar]
- 23.Keenan R. J., Freymann D. M., Stroud R. M., Walter P. Annu. Rev. Biochem. 2001;70:755–775. doi: 10.1146/annurev.biochem.70.1.755. [DOI] [PubMed] [Google Scholar]
- 24.Yamane K., Bunai K., Kakeshita H. Biosci. Biotechnol. Biochem. 2004;68:2007–2023. doi: 10.1271/bbb.68.2007. [DOI] [PubMed] [Google Scholar]
- 25.Urbanus M. L., Scotti P. A., Froderberg L., Saaf A., de Gier J. W., Brunner J., Samuelson J. C., Dalbey R. E., Oudega B., Luirink J. EMBO Rep. 2001;2:524–529. doi: 10.1093/embo-reports/kve108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bolhuis A., Matzen A., Hyyrylainen H. L., Kontinen V. P., Meima R., Chapuis J., Venema G., Bron S., Freudl R., van Dijl J. M. J. Biol. Chem. 1999;274:24585–24592. doi: 10.1074/jbc.274.35.24585. [DOI] [PubMed] [Google Scholar]
- 27.Porankiewicz J., Wang J., Clarke A. K. Mol. Microbiol. 1999;32:449–458. doi: 10.1046/j.1365-2958.1999.01357.x. [DOI] [PubMed] [Google Scholar]
- 28.Nielsen E., Akita M., Davila-Aponte J., Keegstra K. EMBO J. 1997;16:935–946. doi: 10.1093/emboj/16.5.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pan Q., Losick R. J. Bacteriol. 2003;185:5275–5278. doi: 10.1128/JB.185.17.5275-5278.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sassetti C. M., Boyd D. H., Rubin E. J. Mol. Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.