Abstract
After synaptic vesicle fusion, vesicle proteins must be segregated from plasma membrane proteins and recycled to maintain a functional vesicle pool. We monitored the distribution of synaptobrevin, a vesicle protein required for exocytosis, in Caenorhabditis elegans motor neurons by using a pH-sensitive synaptobrevin GFP fusion protein, synaptopHluorin. We estimated that 30% of synaptobrevin was present in the plasma membrane. By using a panel of endocytosis and exocytosis mutants, we found that the majority of surface synaptobrevin derives from fusion of synaptic vesicles and that, in steady state, synaptobrevin equilibrates throughout the axon. The surface synaptobrevin was enriched near active zones, and its spatial extent was regulated by the clathrin adaptin AP180. These results suggest that there is a plasma membrane reservoir of synaptobrevin that is supplied by the synaptic vesicle cycle and available for retrieval throughout the axon. The size of the reservoir is set by the relative rates of exo- and endocytosis.
Keywords: AP180, endocytosis, pHluorin, synaptic vesicle
Neurotransmitter released at synapses originates from a recycling pool of synaptic vesicles (SVs) (1–3). Several processes are required for neurotransmitter secretion, including biogenesis of SVs, docking with the plasma membrane, ATP-dependent priming of SVs to make them fusion-competent, calcium-evoked fusion, and endocytic recycling (4–6). The fusion step is believed to be mediated by the SNARE complex, a four-helix coiled-coil structure consisting of a vesicle SNARE (v-SNARE), synaptobrevin/VAMP, and two target membrane SNARE (t-SNARE) proteins, syntaxin 1 and SNAP-25, on the plasma membrane (7–10).
Accurate sorting of SNAREs to vesicle and plasma membranes is critical for the coordination of SV fusion (11, 12). The t-SNAREs syntaxin and SNAP-25 are abundant in the plasma membrane but are excluded from recycling SVs (13, 14), although syntaxin has been found in some intracellular compartments (13, 15–17). Several studies have documented a significant fraction of endogenous synaptobrevin (14) or synaptobrevin GFP (18, 19) in the plasma membrane. Surface synaptobrevin could be derived from fusion of SV precursors undergoing anterograde transport via the constitutive secretory pathway (20–23). Alternatively, surface synaptobrevin could reflect diffusion within the plasma membrane after vesicle fusion (19) or “stranded” vesicles that fail to undergo endocytosis (24). Finally, some authors have argued that surface synaptobrevin results from missorting, particularly in cases in which synaptobrevin is overexpressed (22).
Several questions remain concerning the surface pool of synaptobrevin. Does this pool arise from v-SNAREs that escape retrieval after exocytosis? Which endocytic pathways regulate this pool of synaptobrevin? Is the spatial distribution of surface synaptobrevin restricted in some manner? To address these questions, we used synaptopHluorin (SpH), a pH-sensitive variant of GFP fused to the luminal domain of synaptobrevin (25–29), to analyze genes that regulate the surface pool of v-SNAREs in the nematode Caenorhabditis elegans.
Results
To develop an optical reporter for surface synaptobrevin, we expressed SpH in cholinergic motor neurons. Fluorescence was observed both in cell bodies and in axonal processes in the ventral and dorsal nerve cords. We used two criteria to determine whether SpH behaves like endogenously expressed synaptobrevin. First, SpH required the unc-104 KIF1A motor protein for its synaptic localization, as is the case for endogenous synaptobrevin (data not shown) (30, 31). Second, pan-neuronal expression of SpH rescued the locomotion and sensitivity to the cholinesterase inhibitor aldicarb defects of snb-1 synaptobrevin mutants (data not shown) (32, 33). Thus, SpH functionally replaced synaptobrevin at the neuromuscular junction.
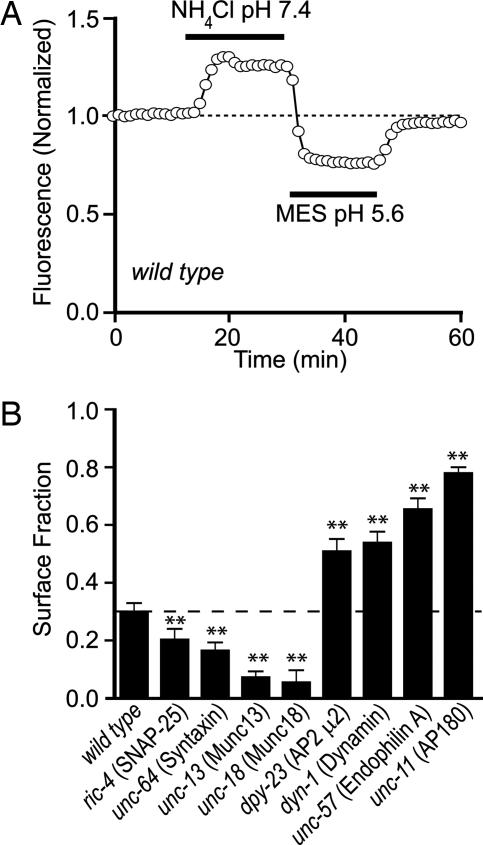
SpH is highly pH-sensitive such that fluorescence is expected to be quenched in the acidic environment of the SV lumen, whereas SpH residing on the plasma membrane will be unquenched, producing a 20-fold increased fluorescence per SpH molecule (25). Individual animals were dissected, and axonal fluorescence from a 20- to 100-μm region of the dorsal nerve cord was focused onto a photodiode. To determine the surface fraction of axonal SpH, we measured SpH fluorescence changes caused by neutralizing intracellular compartments (with pH 7.4 NH4Cl) and by quenching surface SpH (with pH 5.6 Mes) (Fig. 1A and Fig. 6, which is published as supporting information on the PNAS web site). On the basis of these measurements, we estimate that 30% of axonal SpH resides on the cell surface. This amount was similar to the surface abundance previously determined in cultured hippocampal neurons (25) and in Torpedo axons (14). The surface fraction calculation assumes that SVs are acidic, and it may underestimate the true surface percentage if SVs are closer to a neutral pH, as has been observed in Drosophila terminals (34). However, the ratiometric approach used here gave surface percentage estimates that were largely independent of vesicle pH (see Supporting Methods and Fig. 7, which are published as supporting information on the PNAS web site).
Fig. 1.
Measuring surface and vesicular SpH in cholinergic motor neurons. (A) A representative experiment in a wild-type animal. Photodiode current measurements from the fluorescent spot were made every 15 sec while the extracellular solution was exchanged as indicated (see Methods). Note that there is a large contribution of background light pooled onto the photodetector (see Supporting Methods for details). (B) Summary of SpH surface ratios in a panel of synaptic mutants: wild-type nuIs122, ric-4 SNAP-25, unc-64 syntaxin, unc-13 Munc13, unc-18 Munc18, dpy-23 μ2 AP2, dyn-1 dynamin, unc-57 endophilin A, and unc-11 AP180. See Methods for the calculation of surface ratios. Data are shown as mean ± SEM. ∗∗, P < 0.01 by Student’s t test, compared with wild type.
To determine whether the surface SpH pool depended on synaptic transmission, we repeated the measurements in various SV exocytosis and endocytosis mutants. For exocytosis mutants, we examined SpH fluorescence in mutants lacking the t-SNAREs ric-4 SNAP-25 or unc-64 syntaxin 1, as well as in mutants lacking two syntaxin-binding proteins, unc-13 Munc13 and the Sm protein unc-18 Munc18/nSec1 (35–39). All four mutants significantly reduced the surface pool of SpH, with 75% and 94% reductions observed in unc-13 and unc-18 mutants, respectively (Fig. 1B). More modest reductions in SpH surface ratios were observed in t-SNAREs mutants, likely because hypomorphic alleles were analyzed, inasmuch as null alleles cause a lethal phenotype (36, 40).
The clathrin adaptins unc-11 AP180 and dpy-23 AP2 μ2 and the endocytic proteins unc-57 endophilin A and dyn-1 dynamin are required for SV endocytosis (41–46). In all four endocytic mutants studied, surface SpH increased significantly (70–160%) (Fig. 1B).
To confirm that the surface fraction of SpH was altered in these mutants, we examined the rate of SpH bleaching in wild-type, unc-18, and unc-11 mutants. A shift toward more quenched SpH (e.g., an exocytosis mutant) would be predicted to slow the bleach rate, whereas the opposite shift should accelerate the bleach rate. This prediction was confirmed experimentally (see Fig. 8, which is published as supporting information on the PNAS web site), thereby providing further evidence that vesicle pH differed appreciably from extracellular pH.
These experiments suggest that a significant fraction of SpH resides on the plasma membrane. This surface pool is supplied by the exocytosis of SVs and recycled by clathrin-mediated endocytosis. In addition, even if the absolute surface fraction was underestimated by assuming an acidic SV lumen, the relative shifts in surface SpH observed across the panel of synaptic mutants are robust.
Spatial Organization of Vesicular and Surface Synaptobrevin.
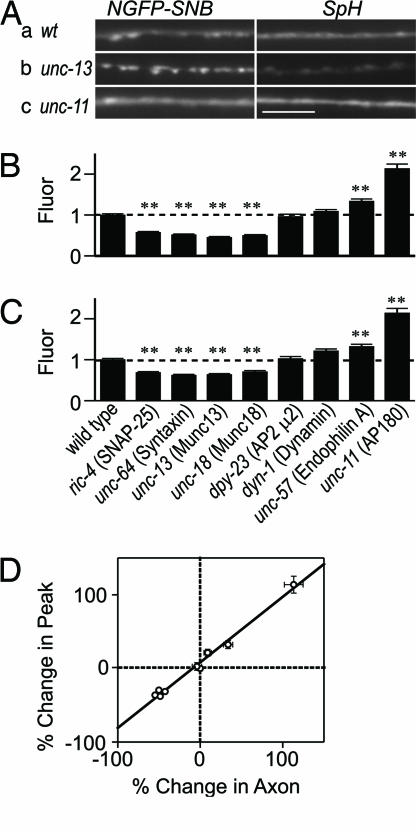
GFP-tagged synaptobrevin has been widely used as a presynaptic marker, and its enrichment at synapses is thought to correspond to the SV pool. We therefore wondered whether surface synaptobrevin might also be enriched near synapses. To detect the spatial features of in vivo synaptobrevin distribution, we imaged two distinct forms of GFP-tagged synaptobrevin in intact animals. First, to analyze the distribution of total (i.e., surface and internal) synaptobrevin, we imaged an N-terminal GFP-tagged synaptobrevin (NGFP-SNB) in the motor axons of intact animals. In NGFP-SNB, the GFP is appended to the cytoplasmic domain of synaptobrevin; consequently, NGFP-SNB molecules in SVs and those in the plasma membrane are equally fluorescent. NGFP-SNB fluorescence is markedly punctate, but we also observed a small amount of diffuse axonal fluorescence between puncta, likely representing surface synaptobrevin (Fig. 2Aa).
Fig. 2.
Imaging synaptobrevin in intact animals. (Aa) The N-terminal tag places GFP (NGFP-SNB) in the cytoplasm. (Ab) Motor neurons expressing a C-terminal pHluorin-tagged synaptobrevin (SpH) that places the fluorophore in the vesicle lumen. (Scale bars, 5 μm.) (Ba) Expression of SpH in the ventral cord. Neuromuscular junctions are located on the bottom cord (∗). (Bb) Expression of UNC-10 RIM1::RFP in the ventral cord. (Bc) Colocalization of SpH (green) and UNC-10 (red). (C) A line scan of SpH in a wild-type animal. Peaks (red arrowheads) and baseline (dashed line) were located by using automated software, as described in Supporting Methods.
We found that 70–90% of SpH fluorescence originates from the unquenched surface pool, assuming a vesicle pH between 5.6 and 6.3 (see Supporting Methods, Eq. 11); consequently, SpH images primarily reflect surface synaptobrevin. In wild-type animals, SpH fluorescence was moderately punctate, indicating that surface SpH was spatially restricted (Fig. 2Ab). These SpH puncta colocalized with the presynaptic active zone marker unc-10 Rim1 (47, 48), suggesting that a local pool of surface SpH is sequestered near active zones (Fig. 2B). This surface cluster of SpH may correspond to a specialized endocytic zone where v-SNAREs are recycled to the SV pool, as has been described in other synapses (49–51).
Total and surface synaptobrevin distributions were measured in exocytosis (unc-13 Munc13) and endocytosis (unc-11 AP180) mutants by quantitative fluorescence microscopy (Fig. 2C, and see Methods). In unc-13 mutants, NGFP-SNB punctal fluorescence increased 40% (Fig. 3Ab Left). The increased punctal fluorescence is indicative of an increased SV pool because previous ultrastructural studies have shown that unc-13 mutants have a 74% increase in the number of cholinergic SVs found at neuromuscular junctions (35). Decreased SV exocytosis should also cause a reduction in surface synaptobrevin, which was evident in our experiments. The unc-13 mutants had decreased NGFP-SNB axonal fluorescence, and SpH fluorescence was nearly eliminated, particularly in the axons between puncta (Fig. 4Ab Right). These findings are consistent with the decreased SpH surface ratio measured in dissected unc-13 mutants (Fig. 1B).
Fig. 3.
Effects of synaptic mutants on synaptobrevin distribution in vivo. (A) Representative image of the dorsal cord in wild-type (a), unc-13 Munc13 (b), and unc-11 AP180 (c) mutant animals. (Left) NGFP-SNB. (Right) SpH. (B) Peak absolute SpH fluorescence across the nine strains: nuIs122, ric-4, unc-64, unc-13, unc-18, dpy-23, dyn-1, unc-57, and unc-11. (C) Axon absolute fluorescence. Individual images are normalized to fluorescent bead standards (see Methods). (D) Plot of percentage change in peak fluorescence vs. percentage change in axon fluorescence for each of the nine strains measured in B and C. A linear regression of the data gave a slope of 0.9, y intercept of 8.2%, and Pearson’s R2 of 0.99 (P < 0.001). All data are normalized to wild type; bars are mean ± SEM. ∗∗, P < 0.01 by Student’s t test, compared with wild type. Note that some error bars are too small to be visible on this scale.
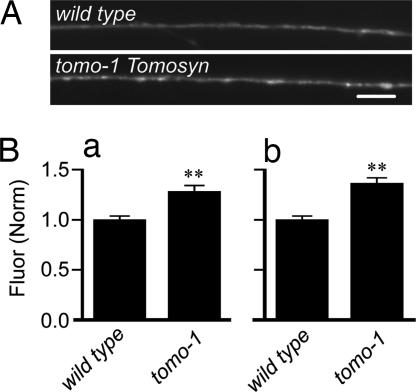
Fig. 4.
Loss of tomosyn increases surface synaptobrevin abundance. (A) Representative images of the dorsal cord from the wild-type strain (Upper) and from the tomosyn mutant tomo-1 (Lower). (Scale bar, 5 μm.) (Ba) Absolute peak fluorescence (relative to a fluorescent bead standard, see Methods) for wild type and tomo-1. (Bb) Absolute axonal fluorescence. Data are normalized to wild type; bars are mean ± SEM. ∗∗, P < 0.001 by Student’s t test, compared with wild type.
Conversely, disruption of endocytosis in unc-11 mutants significantly increased axonal fluorescence of both NGFP-SNB and SpH (Fig. 3Ac), consistent with the increased surface SpH ratio (160%) observed in dissected animals (Fig. 1B). Punctal SpH fluorescence was also increased by a similar degree, indicating that perisynaptic surface clusters could persist despite a large increase in total surface abundance. Thus, large bidirectional changes in plasma membrane SpH in intact animals were observed when exocytosis and endocytosis were disrupted, consistent with the pHluorin measurements in dissected animals (Fig. 1).
Perisynaptic SpH Is in Equilibrium with Axonal SpH.
If SV components are recycled at specialized endocytic zones, we might expect that the recycling of perisynaptic and axonal SpH would be differentially regulated. To test this idea, we quantified perisynaptic and axonal SpH across eight exocytic and endocytic mutants (Fig. 3 B and C). Axonal and perisynaptic surface SpH were generally affected equally across the mutant panel. This correlation was quantified by plotting the relative changes in peak vs. axonal fluorescence for each mutant (Fig. 3D). Perisynaptic and axonal surface SpH were tightly correlated (R = 0.99), with a regression slope of 0.9, suggesting that an equilibrium was established between the two surface pools under steady-state conditions.
Tomosyn Regulates Surface Synaptobrevin Abundance.
In all of the mutants examined thus far, steady-state surface synaptobrevin abundance was altered by disruption of vesicle fusion or endocytosis. If the surface abundance of synaptobrevin is activity-dependent, we would expect that mutants that have increased rates of vesicle fusion would also have altered surface synaptobrevin levels. To test this idea, we imaged tomo-1 mutants, which lack tomosyn, a highly conserved 130-kDa cytoplasmic protein initially found as a binding partner of syntaxin (52, 53). The carboxyl-terminal SNARE motif of tomosyn forms a core complex with SNAP-25 and syntaxin and has been proposed to act as a competitive inhibitor of synaptobrevin. Overexpression of tomosyn in neuroendocrine cells inhibits vesicle fusion (53, 54), and in C. elegans, mutations in the tomosyn gene, tomo-1, enhance acetylcholine release at the neuromuscular junction (55). Both peak and axonal surface SpH increased significantly in tomo-1 mutants (Fig. 4). Thus, the surface pool of synaptobrevin can be augmented by increasing the rate of delivery and by decreasing the rate of retrieval.
AP180 Regulates the Size of Perisynaptic SpH Clusters.
We examined the effects of mutations that disrupt endocytosis on the spatial restriction of surface synaptobrevin by measuring the width of perisynaptic SpH puncta (Fig. 5). The average punctal width was ≈1 μm in wild-type animals and was not significantly altered in dpy-23 AP2, dyn-1 dynamin, or unc-57 endophilin A mutants (all P > 0.05). In unc-11 AP180 mutants, the average SpH puncta width was increased by 35% (Fig. 5 A and B and Fig. 9, which is published as supporting information on the PNAS web site), and the entire distribution of punctal widths was shifted toward larger values (Fig. 5C) (P < 10−10). This change in SpH punctal widths is unlikely to be caused by a change in the geometry of unc-11 AP180 synapses because the SpH puncta are >1 μm wide and are consequently not diffraction-limited; similar puncta widths were measured with confocal microscopy (data not shown). We cannot exclude the possibility that geometric changes contribute to the increased brightness of synaptic varicosities; however, prior ultrastructural studies (44) did not observe large changes in the geometry of synaptic membranes in unc-11 mutants. Transgenic animals expressing 50% less SpH displayed identical puncta widths (see Fig. 10, which is published as supporting information on the PNAS web site). Finally, internal postendocytic compartments are unlikely to contribute to increased puncta width because most of the SpH fluorescence in unc-11 mutants could be quenched with acidic saline (Fig. 11, which is published as supporting information on the PNAS web site). These results suggest that the increased SpH puncta widths observed in unc-11 mutants are unlikely to be caused by changes in SpH abundance or in the distribution of intracellular organelles. Therefore, we propose that AP180 may play an important role in restricting a pool of surface synaptobrevin near active zones.
Fig. 5.
Effects of endocytosis mutants on surface synaptobrevin puncta width. (A) Summary of endocytosis mutant effects on SpH punctal width for the following strains (number of animals): wild type, dpy-23 μ2 AP2, dyn-1 dynamin, unc-57 endophilin A, and unc-11 AP180. (B) Assortment of representative puncta from wild-type (Upper) and unc-11 AP180 mutant (Lower) animals. (Scale bar, 1 μm.) (C) Histogram of punctal widths (measured as full width at half maximum, see Methods) for wild-type (black) and unc-11 (red) animals. Widths are binned in 0.2-μm intervals and normalized to the wild-type peak. Data are from 1,672 puncta (90 animals) for wild type and 414 puncta (35 animals) for unc-11. All data are normalized to wild type; bars are mean ± SEM. ∗∗, P < 0.01 by Student’s t test, compared with wild type.
Discussion
Our results lead to four primary conclusions. First, there is a sizeable pool of synaptobrevin on the plasma membrane of cholinergic motor axons in C. elegans. This surface synaptobrevin is derived from the SV pool, and it clusters near active zones in a manner regulated by AP180. Second, the surface pool of synaptobrevin is largely recycled by clathrin- and dynamin-dependent endocytosis. Third, perisynaptic surface synaptobrevin is in equilibrium with nonsynaptic axonal surface synaptobrevin. Fourth, increasing the rate of secretion (in tomo-1 mutants) results in a net increase of the surface pool.
Sources of Plasma Membrane Synaptobrevin.
Synaptobrevin has been observed in the plasma membrane in multiple neuronal cell types by using immunofluorescence, electron microscopy, biochemical fractionation, and GFP fusion proteins (14, 18, 19, 56). In principle, there are two likely sources for this plasma membrane pool: (i) diffusion from fused SVs and (ii) exocytosis of SV precursors undergoing anterograde transport to nerve terminals. Our results favor the former hypothesis. For surface synaptobrevin to accumulate from SVs, some fraction of vesicular protein must escape retrieval during endocytosis and diffuse away from perisynaptic endocytic zones, as has been observed in cultured hippocampal neurons (19, 56). Surface synaptobrevin was nearly eliminated in mutants lacking UNC-13 and UNC-18, proteins that are critical for SV exocytosis at active zones but are not required for the constitutive secretion pathway. Enhanced synaptic acetylcholine secretion in tomo-1 tomosyn mutants increased the surface abundance of synaptobrevin, further corroborating the synaptic origins of this surface component. Thus, the surface pool of synaptobrevin derives largely from SV fusion at nerve terminals.
Routes of Synaptobrevin Retrieval.
Several endocytic pathways have been proposed to mediate v-SNARE recapture. On a time scale of 1–30 sec, SV endocytosis is thought to occur at specialized endocytic zones that neighbor active zones. The perisynaptic enrichment of surface synaptobrevin observed here could reflect the spatial extent of these endocytic zones. On slower time scales, constitutive endocytosis and pinocytosis of nonsynaptic membrane will capture this axonal synaptobrevin and, therefore, will likely contribute to recycling throughout the axon (50, 57, 58). We observed a tight correlation between the amounts of SpH in perisynaptic surface clusters vs. nonsynaptic axonal SpH over a 5-fold change in surface SpH concentration under steady-state conditions. This correlation is probably the result of equilibration between the two compartments. Inasmuch as we observed parallel effects on surface synaptobrevin both locally (perisynaptic membrane) and globally (axonal membrane) at steady state, all of these endocytic routes are likely to contribute to regulation of the surface synaptobrevin pool. The sorting of plasma membrane synaptobrevin to SVs depends on a targeting sequence found in its cytoplasmic domain (22). This sorting signal can act autonomously to direct synaptobrevin to SVs, regardless of the synaptobrevin’s initial location (20, 21, 59, 60). These studies suggest that nonsynaptic synaptobrevin can be efficiently sorted into SVs, thereby providing a means of coupling surface abundance to replenishment of the vesicle pool and enhancing the recycling efficiency of the vesicle cycle.
The equilibration of surface synaptobrevin across the axon, as observed here, is consistent with several prior studies (19, 56). Allersma et al. (61) reported that in <1 sec after vesicle fusion, GFP-tagged synaptobrevin equilibrated over a 1-μm2 region of membrane. Rapid forms of endocytosis such as “kiss-and-run” events may limit lateral diffusion of synaptobrevin, whereas complete collapse of the vesicle membrane followed by clathrin-mediated endocytosis could result in relatively greater lateral diffusion. Rapid endocytic events occurring as soon as 1 sec after exocytosis have been reported at a mammalian central synapse (62). On this time scale, endogenous synaptobrevin would be expected to diffuse ≈3 μm, assuming a diffusion coefficient of 2.5 μm2/sec (56, 63). However, after more intense periods of activity, recycling times slowed to >20 sec, theoretically allowing a synaptobrevin molecule to diffuse >10–15 μm from its release site. Because three to four synapses are found per 10 μm in the worm nerve cords, these results support our conclusion that surface synaptobrevin derived from SV exocytosis rapidly equilibrates throughout the axon.
Maintenance of a Surface v-SNARE Pool.
The steady-state abundance of plasma membrane synaptobrevin results from a balance between the rate of insertion by means of SV fusion and removal by endocytosis. In hippocampal neurons, increasing the rate of exocytosis (beyond a threshold level of 2 Hz at room temperature) overwhelmed retrieval mechanisms and resulted in a parallel increase in surface synaptobrevin (64). We observed a similar phenomenon (a 30–35% increase in surface SpH) when exocytosis was increased in mutants lacking the inhibitory SNARE tomosyn. Because all mutants tested in this study affected the ratio of surface-to-vesicular synaptobrevin, we conclude that SV exocytosis is not inextricably coupled to endocytosis. Taken together, these results suggest that the surface pool of synaptobrevin is determined by the rates of SV exocytosis and endocytosis, and consequently correlates with recent synaptic activity.
Role of AP180 in Recycling of Surface Synaptobrevin.
AP180 localizes to presynaptic endocytic zones and is thought to regulate local curvature of the lipid bilayer during assembly of the clathrin lattice (43, 65, 66). Loss of AP180 resulted in a 35% widening of perisynaptic surface SpH clusters. Nonet et al. (44) reported that AP180 mutants selectively disrupted retrieval of synaptobrevin from the plasma membrane, suggesting a role for AP180 as a synaptobrevin chaperone. AP180 also regulates synaptic localization of synaptotagmin and cysteine-string protein, two additional SV-associated proteins (43). Our observations provide evidence that AP180 functions to concentrate surface synaptobrevin near active zones. However, AP180 is not essential for maintenance of a steady-state gradient; we still detected localized SpH peaks in unc-11 AP180 mutants.
Functional Significance of Surface v-SNAREs.
We propose that the plasma membrane pool of synaptobrevin is an integral component of the SV cycle. Several results are consistent with this idea. First, we and others have shown that a significant fraction of total synaptobrevin is found at the cell surface (56). Further, the surface pool of synaptobrevin is derived from SV exocytosis and is equilibrated throughout the axon. In cultured rodent neurons, synaptobrevin released from one synapse can be recycled at a neighboring synapse to replenish its vesicle pool (19). In these examples, surface abundance of synaptobrevin is regulated by synaptic activity, and the plasma membrane pool provides a source of v-SNAREs during recovery from bouts of exocytosis (67). Taken together, these results suggest that surface synaptobrevin is an important component of the SV cycle. Further experiments will be necessary to determine whether the abundance of surface synaptobrevin regulates synaptic efficacy.
Methods
Strains and Plasmids.
Strain maintenance and genetic manipulation were performed as described in ref. 68. Animals were cultivated at 20°C on agar nematode growth media seeded with HB101 bacteria. Strains used in this work are listed in Supporting Methods. Strain KP#557 encodes a pHluorin-tagged worm synaptobrevin (SNB-1) in which superecliptic pHluorin was inserted at the C terminus expressed under the snb-1 promoter. KP#558 is the same SNB-1::pHluorin construct expressed under the acr-2 promoter. KP#704 encodes a GFP-tagged SNB-1 in which GFP was inserted at the N terminus (NGFP-SNB) expressed under the acr-2 promoter. KP#930 encodes unc-10 cDNA tagged with a tandem repeat of monomeric red fluorescent protein (mRFP) expressed under the acr-2 promoter.
In Vivo Microscopy and Image Analysis.
GFP and pHluorin-expressing animals were mounted on agarose pads and viewed on a Zeiss Axiovert microscope with an Olympus PlanApo 100× NA 1.4 objective, as described in ref. 69. Images were captured with a Hamamatsu Photonics ORCA digital camera, and line scans were analyzed with custom software in IGOR Pro (WaveMetrics, Lake Oswego, OR). Images of 500-nm fluorescein-conjugated beads (Molecular Probes) were captured during each imaging session to provide a fluorescence standard for comparing absolute fluorescence levels between animals. Background signal (charge-coupled device dark current and slide autofluorescence) was subtracted before analysis. Automated line scan analysis is described in Supporting Methods.
Worm Dissection and Fluorometric Microscopy.
Saline solution recipes are listed in Supporting Methods. SpH-expressing animals were glued to Sylgard-coated coverslips (Dow-Corning), dissected, and placed in a perfusion chamber for gravity-fed superfusion of extracellular saline solutions. A 20- to 100-μm region of the dorsal nerve cord was excited under epifluorescence illumination by a xenon arc lamp, and the emission fluorescence was focused onto a custom-built photodiode to establish the baseline fluorescence value. The photodiode current was sampled for 30 msec every 15 sec, digitized at 10 kHz (National Instruments, Austin, TX), digitally filtered, and analyzed with custom software using IGOR Pro (WaveMetrics). For most experiments, the application of NH4Cl followed by Mes pH 5.6 was repeated at least once and the results were averaged. In some experiments, the order of application was reversed, with similar results. NH4Cl and Mes solutions were applied until a stable level of fluorescence was observed (at least 5 min). Derivation of the surface fraction calculations is provided in Supporting Methods.
Supplementary Material
Acknowledgments
We thank G. Garriga (University of California, Berkeley, CA) and the C. elegans Genetic Stock Center for strains, J. Rothman (Columbia University, New York, NY) for reagents, J. Madison for advice, J. Bai and S. Cotman for assistance with the confocal imaging, and members of the Kaplan laboratory for comments on this manuscript. This work was supported by National Institutes of Health Research Grant GM54728 (to J.M.K.) and by a Damon Runyon postdoctoral fellowship (to J.S.D.).
Abbreviations
- SV
synaptic vesicle
- v-SNARE
vesicle SNARE
- t-SNARE
target membrane SNARE
- SpH
synaptopHluorin
- NGFP-SNB
N-terminal GFP-tagged synaptobrevin.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Betz W. J., Angleson J. K. Annu. Rev. Physiol. 1998;60:347–363. doi: 10.1146/annurev.physiol.60.1.347. [DOI] [PubMed] [Google Scholar]
- 2.Schneggenburger R., Sakaba T., Neher E. Trends Neurosci. 2002;25:206–212. doi: 10.1016/s0166-2236(02)02139-2. [DOI] [PubMed] [Google Scholar]
- 3.von Gersdorff H., Matthews G. Annu. Rev. Physiol. 1999;61:725–752. doi: 10.1146/annurev.physiol.61.1.725. [DOI] [PubMed] [Google Scholar]
- 4.Calakos N., Scheller R. H. Physiol. Rev. 1996;76:1–29. doi: 10.1152/physrev.1996.76.1.1. [DOI] [PubMed] [Google Scholar]
- 5.Jahn R., Südhof T. C. Annu. Rev. Biochem. 1999;68:863–911. doi: 10.1146/annurev.biochem.68.1.863. [DOI] [PubMed] [Google Scholar]
- 6.Murthy V. N., De Camilli P. Annu. Rev. Neurosci. 2003;26:701–728. doi: 10.1146/annurev.neuro.26.041002.131445. [DOI] [PubMed] [Google Scholar]
- 7.Sollner T., Rothman J. E. Trends Neurosci. 1994;17:344–348. doi: 10.1016/0166-2236(94)90178-3. [DOI] [PubMed] [Google Scholar]
- 8.Rothman J. E., Warren G. Curr. Biol. 1994;4:220–233. doi: 10.1016/s0960-9822(00)00051-8. [DOI] [PubMed] [Google Scholar]
- 9.Fasshauer D., Sutton R. B., Brunger A. T., Jahn R. Proc. Natl. Acad. Sci. USA. 1998;95:15781–15786. doi: 10.1073/pnas.95.26.15781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sutton R. B., Fasshauer D., Jahn R., Brunger A. T. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 11.Jahn R., Lang T., Südhof T. C. Cell. 2003;112:519–533. doi: 10.1016/s0092-8674(03)00112-0. [DOI] [PubMed] [Google Scholar]
- 12.Südhof T. C. Annu. Rev. Neurosci. 2004;27:509–547. doi: 10.1146/annurev.neuro.26.041002.131412. [DOI] [PubMed] [Google Scholar]
- 13.Mitchell S. J., Ryan T. A. J. Neurosci. 2004;24:4884–4888. doi: 10.1523/JNEUROSCI.0174-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taubenblatt P., Dedieu J. C., Gulik-Krzywicki T., Morel N. J. Cell Sci. 1999;112:3559–3567. doi: 10.1242/jcs.112.20.3559. Pt 20. [DOI] [PubMed] [Google Scholar]
- 15.Salem N., Faundez V., Horng J. T., Kelly R. B. Nat. Neurosci. 1998;1:551–556. doi: 10.1038/2787. [DOI] [PubMed] [Google Scholar]
- 16.Otto H., Hanson P. I., Jahn R. Proc. Natl. Acad. Sci. USA. 1997;94:6197–6201. doi: 10.1073/pnas.94.12.6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kretzschmar S., Volknandt W., Zimmermann H. Neurosci. Res. 1996;26:141–148. doi: 10.1016/s0168-0102(96)01086-3. [DOI] [PubMed] [Google Scholar]
- 18.Ahmari S. E., Buchanan J., Smith S. J. Nat. Neurosci. 2000;3:445–451. doi: 10.1038/74814. [DOI] [PubMed] [Google Scholar]
- 19.Li Z., Murthy V. N. Neuron. 2001;31:593–605. doi: 10.1016/s0896-6273(01)00398-1. [DOI] [PubMed] [Google Scholar]
- 20.Sampo B., Kaech S., Kunz S., Banker G. Neuron. 2003;37:611–624. doi: 10.1016/s0896-6273(03)00058-8. [DOI] [PubMed] [Google Scholar]
- 21.Grote E., Hao J. C., Bennett M. K., Kelly R. B. Cell. 1995;81:581–589. doi: 10.1016/0092-8674(95)90079-9. [DOI] [PubMed] [Google Scholar]
- 22.Grote E., Kelly R. B. J. Cell Biol. 1996;132:537–547. doi: 10.1083/jcb.132.4.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsukita S., Ishikawa H. J. Cell Biol. 1980;84:513–530. doi: 10.1083/jcb.84.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gandhi S. P., Stevens C. F. Nature. 2003;423:607–613. doi: 10.1038/nature01677. [DOI] [PubMed] [Google Scholar]
- 25.Sankaranarayanan S., De Angelis D., Rothman J. E., Ryan T. A. Biophys. J. 2000;79:2199–2208. doi: 10.1016/S0006-3495(00)76468-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miesenbock G., De Angelis D. A., Rothman J. E. Nature. 1998;394:192–195. doi: 10.1038/28190. [DOI] [PubMed] [Google Scholar]
- 27.Samuel A. D., Silva R. A., Murthy V. N. J. Neurosci. 2003;23:373–376. doi: 10.1523/JNEUROSCI.23-02-00373.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poskanzer K. E., Marek K. W., Sweeney S. T., Davis G. W. Nature. 2003;426:559–563. doi: 10.1038/nature02184. [DOI] [PubMed] [Google Scholar]
- 29.Li Z., Burrone J., Tyler W. J., Hartman K. N., Albeanu D. F., Murthy V. N. Proc. Natl. Acad. Sci. USA. 2005;102:6131–6136. doi: 10.1073/pnas.0501145102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hall D. H., Hedgecock E. M. Cell. 1991;65:837–847. doi: 10.1016/0092-8674(91)90391-b. [DOI] [PubMed] [Google Scholar]
- 31.Yonekawa Y., Harada A., Okada Y., Funakoshi T., Kanai Y., Takei Y., Terada S., Noda T., Hirokawa N. J. Cell Biol. 1998;141:431–441. doi: 10.1083/jcb.141.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller K. G., Alfonso A., Nguyen M., Crowell J. A., Johnson C. D., Rand J. B. Proc. Natl. Acad. Sci. USA. 1996;93:12593–12598. doi: 10.1073/pnas.93.22.12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rand J., Nonet M. In: C. elegans II. Riddle D., Blumenthal T., Meyer B., Priess J., editors. Woodbury, NY: Cold Spring Harbor Lab. Press; 1997. pp. 1049–1052. [Google Scholar]
- 34.Sturman D. A., Shakiryanova D., Hewes R. S., Deitcher D. L., Levitan E. S. Biophys. J. 2006;90:L45–L47. doi: 10.1529/biophysj.106.080978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richmond J. E., Davis W. S., Jorgensen E. M. Nat. Neurosci. 1999;2:959–964. doi: 10.1038/14755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saifee O., Wei L., Nonet M. L. Mol. Biol. Cell. 1998;9:1235–1252. doi: 10.1091/mbc.9.6.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Augustin I., Rosenmund C., Südhof T. C., Brose N. Nature. 1999;400:457–461. doi: 10.1038/22768. [DOI] [PubMed] [Google Scholar]
- 38.Weimer R. M., Richmond J. E. Curr. Top. Dev. Biol. 2005;65:83–113. doi: 10.1016/S0070-2153(04)65003-4. [DOI] [PubMed] [Google Scholar]
- 39.Verhage M., Maia A. S., Plomp J. J., Brussaard A. B., Heeroma J. H., Vermeer H., Toonen R. F., Hammer R. E., van den Berg T. K., Missler M., et al. Science. 2000;287:864–869. doi: 10.1126/science.287.5454.864. [DOI] [PubMed] [Google Scholar]
- 40.Nonet M. L., Saifee O., Zhao H., Rand J. B., Wei L. J. Neurosci. 1998;18:70–80. doi: 10.1523/JNEUROSCI.18-01-00070.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takei K., Mundigl O., Daniell L., De Camilli P. J. Cell Biol. 1996;133:1237–1250. doi: 10.1083/jcb.133.6.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang B., Koh Y. H., Beckstead R. B., Budnik V., Ganetzky B., Bellen H. J. Neuron. 1998;21:1465–1475. doi: 10.1016/s0896-6273(00)80664-9. [DOI] [PubMed] [Google Scholar]
- 43.Bao H., Daniels R. W., Macleod G. T., Charlton M. P., Atwood H. L., Zhang B. J. Neurophysiol. 2005;94:1888–1903. doi: 10.1152/jn.00080.2005. [DOI] [PubMed] [Google Scholar]
- 44.Nonet M. L., Holgado A. M., Brewer F., Serpe C. J., Norbeck B. A., Holleran J., Wei L., Hartwieg E., Jorgensen E. M., Alfonso A. Mol. Biol. Cell. 1999;10:2343–2360. doi: 10.1091/mbc.10.7.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schuske K. R., Richmond J. E., Matthies D. S., Davis W. S., Runz S., Rube D. A., van der Bliek A. M., Jorgensen E. M. Neuron. 2003;40:749–762. doi: 10.1016/s0896-6273(03)00667-6. [DOI] [PubMed] [Google Scholar]
- 46.Harris T. W., Schuske K., Jorgensen E. M. Traffic. 2001;2:597–605. doi: 10.1034/j.1600-0854.2001.002009597.x. [DOI] [PubMed] [Google Scholar]
- 47.Koushika S. P., Richmond J. E., Hadwiger G., Weimer R. M., Jorgensen E. M., Nonet M. L. Nat. Neurosci. 2001;4:997–1005. doi: 10.1038/nn732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Betz A., Thakur P., Junge H. J., Ashery U., Rhee J. S., Scheuss V., Rosenmund C., Rettig J., Brose N. Neuron. 2001;30:183–196. doi: 10.1016/s0896-6273(01)00272-0. [DOI] [PubMed] [Google Scholar]
- 49.Roos J., Kelly R. B. Curr. Biol. 1999;9:1411–1414. doi: 10.1016/s0960-9822(00)80087-1. [DOI] [PubMed] [Google Scholar]
- 50.Teng H., Wilkinson R. S. J. Neurosci. 2000;20:7986–7993. doi: 10.1523/JNEUROSCI.20-21-07986.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Teng H., Cole J. C., Roberts R. L., Wilkinson R. S. J. Neurosci. 1999;19:4855–4866. doi: 10.1523/JNEUROSCI.19-12-04855.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pobbati A. V., Razeto A., Boddener M., Becker S., Fasshauer D. J. Biol. Chem. 2004;279:47192–47200. doi: 10.1074/jbc.M408767200. [DOI] [PubMed] [Google Scholar]
- 53.Hatsuzawa K., Lang T., Fasshauer D., Bruns D., Jahn R. J. Biol. Chem. 2003;278:31159–31166. doi: 10.1074/jbc.M305500200. [DOI] [PubMed] [Google Scholar]
- 54.Yizhar O., Matti U., Melamed R., Hagalili Y., Bruns D., Rettig J., Ashery U. Proc. Natl. Acad. Sci. USA. 2004;101:2578–2583. doi: 10.1073/pnas.0308700100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dybbs M., Ngai J., Kaplan J. M. PLoS Genet. 2005;1:6–16. doi: 10.1371/journal.pgen.0010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sankaranarayanan S., Ryan T. A. Nat. Cell Biol. 2000;2:197–204. doi: 10.1038/35008615. [DOI] [PubMed] [Google Scholar]
- 57.Holt M., Cooke A., Wu M. M., Lagnado L. J. Neurosci. 2003;23:1329–1339. doi: 10.1523/JNEUROSCI.23-04-01329.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meshul C. K., Pappas G. D. Brain Res. 1984;290:1–18. doi: 10.1016/0006-8993(84)90730-3. [DOI] [PubMed] [Google Scholar]
- 59.West A. E., Neve R. L., Buckley K. M. J. Cell Biol. 1997;139:917–927. doi: 10.1083/jcb.139.4.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martinez-Arca S., Arold S., Rudge R., Laroche F., Galli T. Traffic. 2004;5:371–382. doi: 10.1111/j.1398-9219.2004.00180.x. [DOI] [PubMed] [Google Scholar]
- 61.Allersma M. W., Wang L., Axelrod D., Holz R. W. Mol. Biol. Cell. 2004;15:4658–4668. doi: 10.1091/mbc.E04-02-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu W., Xu J., Wu X. S., Wu L. G. J. Neurosci. 2005;25:11676–11683. doi: 10.1523/JNEUROSCI.2972-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bacia K., Schuette C. G., Kahya N., Jahn R., Schwille P. J. Biol. Chem. 2004;279:37951–37955. doi: 10.1074/jbc.M407020200. [DOI] [PubMed] [Google Scholar]
- 64.Fernandez-Alfonso T., Ryan T. A. Neuron. 2004;41:943–953. doi: 10.1016/s0896-6273(04)00113-8. [DOI] [PubMed] [Google Scholar]
- 65.Yao P. J., Petralia R. S., Bushlin I., Wang Y., Furukawa K. J. Comp. Neurol. 2005;481:58–69. doi: 10.1002/cne.20362. [DOI] [PubMed] [Google Scholar]
- 66.McMahon H. T. Curr. Biol. 1999;9:R332–R335. doi: 10.1016/s0960-9822(99)80206-1. [DOI] [PubMed] [Google Scholar]
- 67.Kaneda Y., Grote E., Bonzelius F., Desnos C., Herman G., Clift-O’Grady L., Kelly R. B. Cold Spring Harbor Symp. Quant. Biol. 1995;60:379–387. doi: 10.1101/sqb.1995.060.01.042. [DOI] [PubMed] [Google Scholar]
- 68.Brenner S. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Burbea M., Dreier L., Dittman J. S., Grunwald M. E., Kaplan J. M. Neuron. 2002;35:107–120. doi: 10.1016/s0896-6273(02)00749-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.