Abstract
To facilitate analysis of plant cell wall polysaccharide structure and composition, we cloned 74 genes encoding polysaccharide-degrading enzymes from Aspergillus nidulans, Aspergillus fumigatus, and Neurospora crassa and expressed the genes as secreted proteins with C-terminal Myc and 6× His tags. Most of the recombinant enzymes were active in enzyme assays, and optima for pH and temperature were established. A subset of the enzymes was used to fragment polysaccharides from the irregular xylem 9 (irx9) mutant of Arabidopsis. The analysis revealed a decrease in the abundance of xylan in the mutant, indicating that the IRX9 gene, which encodes a putative family 43 glycosyltransferase, is required for xylan synthesis.
Keywords: Arabidopsis, fingerprinting, hydrolase, mutant, xylan
Plant cell wall polysaccharides are the most abundant organic compounds found in nature. Their structures are very complex, and their exact nature and relative abundances vary from one plant species to another and from tissue to tissue within a plant. They are conventionally divided into three groups: cellulose, hemicellulose (e.g., xyloglucans, xylans, and mannans), and pectins. Analysis of polysaccharide structure is a very challenging task and requires specialized techniques that, in most cases, differ widely from those used for the characterization of other types of biological macromolecules. In general, sequential extraction of individual polymers from the alcohol-insoluble residue of the plant material is used as a first step (1–3). Crude extracts are frequently fractionated by various size-exclusion methods (4–6). The positions of the linkages between the sugars in the polysaccharides can be determined by methylation analysis or reductive cleavage (7, 8), but finding the sequence and arrangement of the sugars usually involves a detailed characterization of smaller fragments. Fragmentation of the polymers can be achieved by fairly selective chemical methods such as partial acid hydrolysis and Smith degradation or by endo-hydrolase enzymes (9). The resulting oligomers can be separated from each other with techniques such as HPLC or capillary electrophoresis (10–13). For structure elucidation of purified oligosaccharides, one- and two-dimensional NMR spectroscopy has been used (10, 14) as well as mass spectrometry using various ionization techniques such as electrospray ionization (ESI) or MALDI (3, 15, 16).
The most crucial step of the analysis is the partial degradation of polysaccharides into smaller oligomers that can be structurally characterized. Chemical cleavages such as partial acid hydrolysis are not completely specific for any one glycosidic linkage in the polymer and so produce a complex mixture of products that can be difficult to purify and can lead to low yields of individual oligomers. An alternative to acid cleavage is the use of enzymes that hydrolyze the polysaccharides much more specifically and under less drastic conditions. However, pure enzymes for this purpose are available only for some commercially important polysaccharides, such as starch, and for substrates under intense study (e.g., proteoglycans).
Pure carbohydrate-degrading enzymes are an extremely useful tool for analyzing plant cell wall polysaccharides and for profiling of polymers in WT and mutant cell walls. To be useful for structural work, such enzymes must be completely pure or at least free from undesirable activity. Most plant saprophytes and parasites secrete a suite of enzymes for degrading plant polysaccharides; thus, isolation of pure enzymes from them involves extensive purification. For this reason, the only feasible way to obtain a reproducible access to enzymes with properties required is to clone the genes into a suitable host. Here, we report the production of a set of 74 plant cell wall hydrolytic enzymes from Aspergillus nidulans (72 enzymes), Aspergillus fumigatus (1 enzyme), and Neurospora crassa (1 enzyme), which are freely available to the research community. Because these enzymes can be readily produced without contaminating activities, they are very useful for the accurate analysis of most polysaccharides that are the constituents of the plant cell wall. By using these enzymes, we were able to identify cell wall differences in the Arabidopsis thaliana irx9 mutant.
Results and Discussion
As saprophytic or pathogenic organisms, fungi are capable of degrading a wide variety of different polysaccharides (17). Although genes for many polysaccharide-degrading enzymes have been cloned, we were unable to obtain many of the clones for production of research grade enzymes. Therefore, we have developed a comprehensive suite of publicly available clones for useful enzymes. This research was facilitated by the observation that homologs of many polysaccharide hydrolytic enzymes could be identified in the genome sequence that was recently obtained for the saprophytic fungus, A. nidulans (18). Several enzymes active on xylan (19–24), pectate (25), arabinan (26–28), glucans (29, 30), and other polysaccharides (31, 32) have been cloned or isolated previously. Nevertheless, these enzymes cannot be obtained without laborious steps of purification. In addition, the need for an easy access to a range of different enzymes demands the creation of an enzyme collection ideally covering every linkage involved in plant cell wall polysaccharides.
Sequences of polysaccharide-degrading enzymes in the genome of A. nidulans were identified by performing a BLASTp search with representatives of all known fungal polysaccharide degrading enzymes from the Carbohydrate-Active Enzymes (CAZy) database (33). Genes for 72 selected cell wall-degrading enzymes were obtained either from genomic DNA (for intron-free genes) or from cDNA generated from fungus grown on pectin, xylan, or gum arabic. Additionally, a xylogalacturonase and an endo-β-(1, 6)-galactanase were cloned from A. fumigatus and N. crassa, respectively. The coding sequence of each gene was modified by addition of yeast α-factor signal peptide at the N terminus and a Myc tag and a 6× His tag at the C terminus. Pichia pastoris is capable of producing and secreting large amounts of recombinant proteins and normally secretes very little additional protein into the medium, making it ideal for production of recombinant proteins (34). Because of the 6× His tag, enzymes can be easily purified from culture filtrates by using affinity chromatography (35).
The recombinant genes were placed under control of a methanol-inducible promoter and introduced into the P. pastoris genome. Clones that exhibited satisfactory levels of expression of the recombinant enzyme were identified by screening the media of small cultures by dot-blots using an anti-Myc antibody. The most productive clones have been deposited in the Fungal Genetics Stock Center (FGSC) at the University of Missouri (Kansas City, MO), and the empirically determined sequences of the genes were deposited in GenBank.
Table 1 shows the list of enzymes cloned and basic information about the activities of many of them. More detailed information is available in Table 2, which is published as supporting information on the PNAS web site. Enzymes are now available for the cleavage of backbones and side chains of most common plant cell wall polysaccharides such as cellulose, hemicelluloses (glucans, xyloglucans, xylans, and mannans), and pectins (polygalacturonic acid, xylogalacturonans, and rhamnogalacturonan I). We envision that some of the enzymes might also act on the complex pectic polysaccharide rhamnogalacturonan II and on the glycan moiety of arabinogalactan proteins (AGP). We have included enzymes for which we could not detect activity although recombinant protein was expressed. It is quite likely that a suitable substrate has not yet been identified for them. Some of the enzymes from the list exhibit a strong preference for certain structures. Oligoxyloglucan-reducing end-specific xyloglucanobiohydrolase (OREX, AN1542.2) acts on oligoxyloglucan fragments only if the second xylose from the reducing end is not further substituted (35). Another example is the α-rhamnosidase (AN10277.3). This enzyme does not hydrolyze 4-nitrophenyl-α-l-rhamnopyranoside or naringin or hesperidin (in which α-l-rhamnopyranose is linked to glucose via α(1→2)- and α(1→6)-linkages, respectively), but it releases a terminal α(1→4)-linked l-rhamnopyranose from the nonreducing end of rhamnogalacturonan oligomers.
Table 1.
List of enzymes (including a brief description of activity) expressed in P. pastoris ordered by activity toward respective polysaccharides
| Enzyme | Accession no. | pHopt | Topt, °C | Activity* | Known substrates |
|---|---|---|---|---|---|
| Active on glucans | |||||
| Endo-β(1,4)-glucanase | AN1602.2 | <6 | ND | 1.12 | Soluble CMC† |
| Endo-β(1,4)-glucanase | AN5214.2 | 4.0 | 52 | 2.85 | Soluble CMC,† cellooligos Glc4 (slow)/Glc5/Glc6 |
| Endo-β(1,4)-glucanase | AN1285.2 | 4.0 | 57 | 5.30 | Soluble CMC,† cellooligos Glc4/Glc5/Glc6, barley β-glucan and lichenan |
| Endo-β(1,4)-glucanase | AN3418.2 | 5.5 | 42 | 1.85 | CMC,† cellooligos Glc4/Glc5/Glc6, barley β-glucan and lichenan and tamarind XG |
| Cellobiohydrolase | AN0494.2 | ND | ND | 1.82 | Soluble CMC,† cellooligos Glc3 (slow)/Glc4/Glc5/Glc6 |
| Cellobiohydrolase | AN5282.2 | 5.5 | 57 | 2.40 | Soluble CMC,† cellooligos Glc3/Glc4/Glc5/Glc6, barley β-glucan and lichenan, avicel |
| Cellobiohydrolase | AN5176.2 | ND | ND | ND | ND |
| β-Glucosidase | AN2227.2 | ND | ND | ND | ND |
| β-Glucosidase | AN2612.2 | ND | ND | ND | ND |
| β-Glucosidase | AN0712.2 | 5.5 | 52 | A | PNP-β-glucoside† |
| β-Glucosidase | AN1551.2 | ND | ND | ND | Not active on PNP-β-glucoside |
| β-Glucosidase | AN1804.2 | 6.0 | 52 | 87.84 | PNP-β-glucoside† |
| α-/β-Glucosidase | AN7345.2 | ND | ND | 1.66 | PNP-β-glucoside† and PNP-α-glucoside |
| Mixed-linked glucanase | AN2385.2 | ND | ND | 2.33 | Laminarin,† lichenan, soluble CMC, not on pustulan |
| Endo-β(1,3)-glucanase | AN4700.2 | ND | ND | 1.35 | Laminarin† lichenan |
| Exo-β(1,3)-glucanase | AN7533.2 | ND | ND | 0.58 | Lichenan† |
| Endo-β(1,3)-glucanase | AN7950.2 | ND | ND | ND | ND |
| β-(1,6)-Glucanase | AN3777.2 | ND | ND | 0.15 | Pustulan (β-1,6-glucan)† |
| Active on xyloglucans | |||||
| XG-specific endoglucanase | AN0452.2 | 6.5 | 47 | 1.97 | Tamarind XG† |
| Oligoxyloglucan reducing | AN1542.2 | 3.0 | 42 | 0.08 | Tamarind XG,† tamarind XG oligomers |
| End-specific xyloglucanase | |||||
| α-Fucosidase | AN8149.2 | ND | ND | A | Cotton XG oligomers, not active on PNP-fucoside |
| α-Xylosidase | AN7505.2 | ND | ND | 0.21 | PNP-α-xyloside† |
| Active on xylans | |||||
| Endo-β(1,4)-xylanase | AN1818.2 | 4.9 | 52 | 51.16 | LWX†, BWX, RAX, OSX, Xyl4 and Xyl6 |
| Endo-β(1,4)-xylanase | AN3613.2 | 5.4 | 52 | 18.83 | LWX†, BWX, RAX, OSX, Xyl4 and Xyl6 |
| β-Xylosidase | AN2359.2 | 5.1 | 52 | 29.00 | PNP-β-xyloside,† Xyl6 |
| β-Xylosidase/α-arabinosidase | AN8401.2 | 4.4 | 48 | 0.28 | RAX† and Xyl6, not on PNP-β-xyloside |
| Acetylxylan esterase | AN3294.2 | ND | ND | 2.33 | Naphthyl acetate, PNP-acetate† |
| Acetylxylan esterase | AN6093.2 | 7.5 | 49 | 33.33 | Naphthyl acetate, PNP-acetate† |
| Ferulic acid esterase | AN5267.2 | 6.1 | 37 | 0.03 | PNP-acetate, methyl ferulate,† wheat arabinoxylan |
| α-Glucuronidase | AN9286.2 | 4.0 | 30 | 0.02 | LWX,† APTS labeled 4-O-methyl glucuronosyl Xyl3 |
| Active on mannans | |||||
| Endo-β(1,4)-mannanase | AN3297.2 | ND | ND | 0.67 | LBG,† GG |
| Endo-β(1,4)-mannanase | AN3358.2 | 5.5 | 52 | 2.13 | LBG,† GG |
| Endo-β(1,4)-mannanase | AN6427.2 | ND | ND | 2.05 | LBG,† GG |
| Endo-β(1,4)-mannanase | AN7413.2 | ND | ND | ND | ND |
| β-Mannosidase | AN3368.2 | ND | ND | 3.22 | PNP-β-mannoside† |
| α-Galactosidase | AN7152.2 | 5.0 | 52 | 1.50 | PNP-α-galactoside†; not on raffinose, LBG, or GG |
| α-Galactosidase | AN7624.2 | ND | ND | ND | Not active on PNP-α-galactoside, raffinose |
| α-Galactosidase | AN8138.2 | 3.5 | 52 | 42.17 | PNP-α-galactoside,† raffinose, LBG, GG |
| α-Galactosidase | AN9035.2 | ND | ND | ND | Not active on PNP-α-galactoside, raffinose |
| Active on pectin | |||||
| Pectin lyase | AN2331.2 | 7.0 | ND | 3.50 | Citrus pectin† |
| Pectin lyase | AN2569.2 | ND | ND | 5.00 | Citrus pectin† |
| Pectate lyase | AN0741.2 | 8.5 | 37 | 0.67 | Pectic acid† |
| Pectate lyase | AN3337.2 | 9.2 | 37 | 11.33 | Pectic acid† |
| Pectate lyase | AN7646.2 | 8.5 | 22 | 1.17 | Pectic acid† |
| Pectate lyase | AN8453.2 | 7.8 | 22 | 1.83 | Pectic acid† |
| Endo-polygalacturonase | AN4372.2 | 5.1 | 38 | 21.42 | Pectic acid,† less active on citrus pectin |
| Endo-polygalacturonase | AN8327.2 | 4.8 | 48 | 18.58 | Pectic acid,† less active on citrus pectin |
| Exo-polygalacturonase | AN8761.2 | 4.4 | 48 | 25.00 | Pectic acid,† less active on citrus pectin |
| Exo-polygalacturonase | AN9045.2 | ND | ND | A | GalA oligomers |
| Pectin methyl esterase | AN3390.2 | 8.0 | 30 | 10.00 | Citrus pectin† |
| Rhamnogalacturonase | AN9134.2 | ND | ND | 0.58 | Linseed RG† |
| Rhamnogalacturonan lyase | AN6395.2 | ND | ND | 1.15 | Linseed RG† |
| Rhamnogalacturonan lyase | AN7135.2 | ND | ND | 0.55 | Linseed RG† |
| Rhamnogalacturonan | AN2528.2 | ND | ND | 11.67 | PNP-acetate,† naphthyl acetate |
| Acetylesterase α-l-rhamnosidase | AN10277.3 | 5.0 | ND | A | RG oligomers, not on PNP-α-L-rhamnoside, not on naringin or hesperidin |
| Endo-α(1,5)-arabinosidase | AN6352.2 | ND | ND | 0.17 | Debranched arabinan† |
| Endo-α(1,5)-arabinosidase | AN8007.2 | ND | ND | 0.02 | Debranched arabinan† |
| Endo-α(1,5)-arabinosidase | AN3044.2 | ND | ND | ND | ND |
| α-l-arabinofuranosidase | AN1571.2 | 4.8 | 65 | 23.50 | PNP-α-arabinofuranoside,† sugar beet arabinan, Ara7 |
| α-l-arabinofuranosidase | AN7908.2 | 5.4 | 47 | 7.83 | PNP-α-arabinofuranoside,† sugar beet arabinan, RAX |
| α-l-arabinofuranosidase | AN1277.2 | ND | ND | A | Ara7 |
| Endo-β(1,4)-galactanase | AN5727.2 | 5.0 | ND | 66.58 | Potato pectic galactan† |
| Xylogalacturonase | Afu8g06890 | ND | ND | A | Water melon xylogalacturonan |
| β-Galactosidase | AN3201.2 | ND | ND | ND | ND |
| Miscellaneous | |||||
| β-Glucuronidase | AN5361.2 | ND | ND | ND | ND |
| Cutinase | AN7541.2 | ND | ND | 304.17 | PNP-butyrate† |
| Cutinase | AN7180.2 | ND | ND | 1.33 | PNP-butyrate† |
| α-Glucosidase | AN0941.2 | 5.5 | 52 | 0.22 | PNP-α-glucoside† |
| α-Glucosidase | AN4843.2 | ND | ND | ND | Not active on PNP-α-glucoside |
| Endo-β-(1,6)-galactanase | NCU09702.1 | ND | ND | ND | ND |
| α-1,2-Mannosidase | AN0787.2 | ND | ND | ND | Not active on PNP-α-mannoside |
| α-1,2-Mannosidase | AN3566.2 | ND | ND | ND | Not active on PNP-α-mannoside |
| α-1,3-Glucanase (mutanase) | AN7349.2 | ND | ND | 0.40 | α-1,3-Glucan (mutan)† from A. nidulans |
| N,O-diacetylmuramidase | AN6470.2 | 4.0 | 24 | 4510.0‡ | Dried micrococcus cells (bacterial cells)† |
A, activity observed but specific activity not determined; ND, not determined; Glc, glucose; Xyl, xylose; Ara, arabinose; GalA, galacturonic acid (subscript indicates a linear oligosaccharide chain, e.g., Xyl6 = xylohexaose); CMC, carboxymethyl cellulose; XG, xyloglucan; LWX, larch wood xylan; BWX, birch wood xylan; RAX, rye arabinoxylan; OSX, oat spelt xylan; LBG, locust bean gum; GG, gum guar; RG, rhamnogalacturonan; PNP, para-nitrophenyl; APTS, aminopyrene trisulfonic acid; NR, nonreducing. Additional information is presented in Table 2.
*Unless otherwise mentioned, the activity (nKat/ml culture) was tested at a temperature of 37°C (hydrolases) or 30°C (lyases) and at optimum pH based on the reported properties of similar enzymes.
†The substrate used for measuring the activity, expressed in nKat/ml (nmol/s/ml) of culture filtrate after 72 h of methanol induction.
‡The activity (1 unit) is expressed as decrease in turbidity (absorbance at 450 nm) by 0.001 absorbance.
We have observed that, because the enzymes described here can be obtained without any major contaminating activities (36), it is possible to allow digestions of cell wall material to go to completion without concern that unexpected hydrolysis will occur from contaminating activities. Because many enzymes lose activity very rapidly at higher temperatures, we recommend using a temperature of 37°C or lower for long-term incubations (e.g., overnight). This condition also applies to enzymes with optimum temperatures of >50°C.
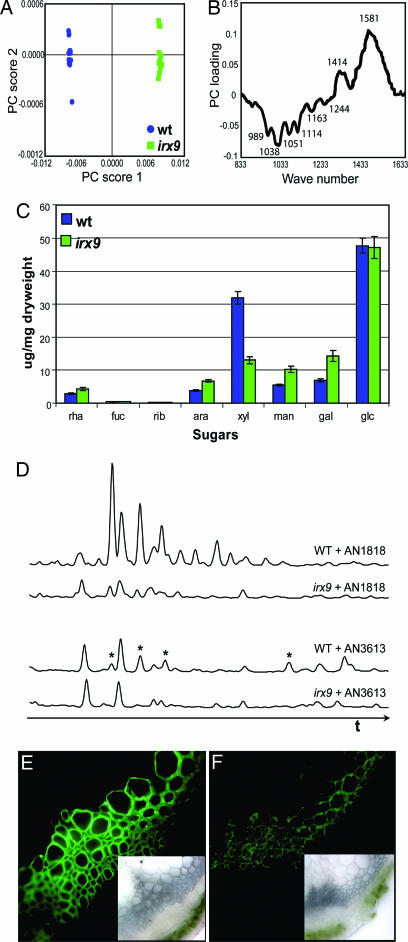
We envision these enzymes to be very valuable in analyses of cell wall mutants. For instance, in previous studies, the known Arabidopsis xyloglucan mutants mur1, mur2, and mur3 could be easily distinguished by the different oligosaccharide pattern generated by xyloglucanase treatment (37). To illustrate the utility of the enzymes, we have applied purified enzymes from Table 1 to characterize the cell wall defect of the Arabidopsis irregular xylem 9 (irx9) mutant. The IRX9 gene (AT2G37090), which encodes a putative family 43 glycosyl transferase, was selected for mutant analysis following the observation that it was coordinately expressed with the cellulose synthase subunits during secondary cell wall formation (38). None of the other family 43 members from Arabidopsis have been characterized. The mutant has a dwarfed phenotype, exhibits a collapsed xylem, and has a 20% decreased cellulose content (39). We applied Fourier transform infrared (FTIR) spectroscopy of ball-milled stem cell walls in combination with principal component analysis to detect differences between WT and irx9 spectra (Fig. 1). Based on principal component (PC) 1, the spectra show a clear separation (Fig. 1A). These differences are also seen in the PC1 loading plot in the carbohydrate fingerprint regions (1,200–800 cm−1) that contains information characteristic of polysaccharides (Fig. 1B). Obvious differences in cellulose (1,059 cm−1 and 1,033 cm−1) were not observed. However, the positive correlation of the WT in this region (e.g., 1,038 cm−1, 1,051 cm−1, and 1,114 cm−1) indicates that there are potential alterations in noncellulosic polymers, presumably in the hemicelluloses.
Fig. 1.
Cell wall analysis of A. thaliana WT and irx9 stems. (A and B) Principal component plots for WT vs. mutant irx9 spectra. Spectra show a clear separation based on principal component (PC) 1. Spectra show differences in the carbohydrate fingerprint regions that correspond to the deformations of cellulosic and noncellulosic polymers. (C) Monosaccharide composition of WT and irx9 as determined by gas chromatography of alditol acetate derivatives of total polysaccharides from stems. (D) Capillary electrophoresis electropherograms of WT and irx9 after endo-xylanase AN1818.2 (Upper) and AN3613.2 (Lower) treatment. (E and F) Indirect immunofluorescence and differential interference contrast micrographs of LM11 antibody binding to transverse sections of WT (E) and irx9 (F).
A difference in cell wall composition of mutant and WT was confirmed by analysis of the monosaccharide composition of stems. The xylose content of the irx9 mutant is reduced by >50% compared with the WT (Fig. 1C). Xylose is a significant component of xylan, xyloglucan, and xylogalacturonan. Therefore, we infer a significant decrease in the amount of one or more of these polymers in the mutant. To obtain additional information, cell wall material from stems of irx9 and WT was incubated with a variety of enzymes from Table 1 (pectin lyases, pectate lyases, polygalacturonases, mannanases, xyloglucanase, xylogalacturonase, xylanases, endo-glucanases, and endo-galactanase), and the resulting oligosaccharides were separated by capillary electrophoresis after 8-aminopyrene-1,3,6-trisulfonate (APTS) labeling. The only difference between irx9 and WT resulted from incubation with the two endo-xylanases (AN1818.2 and AN3613.2). After digesting with endo-xylanase AN1818.2 (Fig. 1D Upper), peaks in WT are more intense and show a different fingerprint pattern compared with the irx9 mutant. The differences after incubation with endo-xylanase AN3613.2 were not as obvious as with AN1818.2. However, asterisks mark peaks that were significantly more intense in the WT (Fig. 1D Lower). We believe that the basis for this effect is that the two xylanases have slightly different modes of action: AN3613.2 requires a higher number of unsubstituted backbone residues than AN1818.2 and therefore is less active on more highly branched xylans.
In combination with the results of the monosaccharide composition, we conclude that the more abundant peaks in the WT after endo-xylanase digestion result from a higher xylan content. Interestingly, another irx mutant, irx7 also referred to as fra8, displayed a very similar sugar composition as irx9 and was shown to be involved in biosynthesis of glucuronoxylan (40). The lower xylan content in irx9 was also confirmed by immunolabeling of stem sections with monoclonal antibodies LM10 and LM11, which specifically bind to low-substituted (LM10) and low- and higher-substituted xylans (LM11) (41). Whereas we did not see significant differences using LM10 (Fig. 2, which is published as supporting information on the PNAS web site), the immunofluorescence of WT (Fig. 1E) after incubation with LM11 was more intense than in the irx9 mutant (Fig. 1F), indicating a reduced amount of higher-substituted xylan in this mutant, consistent with the results of the enzymatic digestion. Thus, we infer that IRX9 may be a xylan synthase. Additional studies of the catalytic properties of the protein will be necessary to test this hypothesis.
We envision that the enzymes described here will be a powerful tool in the analysis of WT and mutant cell walls, complementing the commonly used techniques such as IR-spectroscopy, monosaccharide analysis, and immunolabeling. By using the enzymes to hydrolyze specific polysaccharides, it is possible to selectively investigate one type of polymer in a cell wall at a time. Reproducible selective cleavage of specific polymers will facilitate the use of mass spectrometry and NMR for structural analysis of cell walls. The recombinant Pichia strains have been deposited in the FGSC. We encourage researchers to share their experience with these enzymes by sending annotations to the online database maintained at the Carnegie Institution.
Materials and Methods
Chemicals.
Unless otherwise stated, chemicals and substrates were purchased from Sigma. Unless indicated otherwise, polysaccharides were purchased from Megazyme (Bray, County Wicklow, Ireland). Methyl ferulate (MFA) was prepared according to the method described in Borneman et al. (42). Commercially unavailable oligosaccharides were generated by partial acid or enzymic hydrolysis of polysaccharides followed by purification by ion-exchange and/or size exclusion chromatography (43). Oligosaccharides were reductively aminated with 8-aminopyrene-1,3,6-trisulfonate (APTS) or 8-aminonaphthalene-1,3,6-trisulfonic acid (ANTS) and purified by gel filtration (13).
Construction of Expression Plasmid.
A. nidulans FGSC A4 (Glasgow WT), A. fumigatus Af293, and N. crassa (74-OR23-1VA) were obtained from the FGSC (University of Missouri, Kansas City, MO) (44). A. nidulans and A. fumigatus were grown in complete medium (minimal medium supplemented with 0.5% yeast extract, 1% peptone, 2% glucose, and Hutner’s trace elements, pH 4.5) at 37°C (45). N. crassa was grown in yeast extract/peptone/dextrose (YPD) medium. Chromosomal DNA was extracted according to protocol 12 in ref. 46. For induction of enzymes, mycelium of A. nidulans and A. fumigatus was sterile-filtered (miracloth) and transferred into media containing minimal medium (pH 4.5), Hutner’s trace elements, and 0.5% pectin (Sigma), 0.5% larch wood xylan (Sigma), or 2% gum arabic (Sigma), respectively. RNA was extracted by using Trizol (Invitrogen), and first-strand cDNA synthesis was performed by using SuperScript reverse transcriptase (Invitrogen) and oligo(dT) primer. DNA was amplified by PCR by using specific primers (including restriction sites) designated in Table 3, which is published as supporting information on the PNAS web site. The amplified fragments were cloned at the corresponding sites into vector pPICZαC (Invitrogen) or pPICZαA (for xylogalacturonase Afu8g06890) and transformed into P. pastoris X-33 (Invitrogen) as described (35). Screening of recombinant clones, expression optimization, and purification of enzymes were done as described (35).
Enzyme Assays.
A pH-dependent assay was performed at 37°C with buffers in the range of pH 1–10 (47). For temperature optimum determinations, the assays were performed at the optimum pH at temperatures in the range of 16–70°C for 15 min.
Assay mixtures contained substrate and suitably diluted enzyme in 50 mM buffer of optimum pH. The mixture was incubated either at 37°C (for hydrolases) or at 30°C (for lyases and esterases) for 10–60 min, and the reaction was terminated by heating at 100°C for 5 min. Enzyme activity was determined spectrophotometrically by measuring (i) the release of reducing groups from respective polysaccharides (exo-polygalacturonase, endo-polygalacturonase, xylogalacturonase, endo-galactanase, rhamnogalacturonase, endo-arabinanase, endo-glucanase, cellobiohydrolase, endo-xylanase, endo-mannanase, and mutanase), (ii) the generation of unsaturated galacturonides (pectate lyases, pectin lyases, and RG lyases), (iii) the release of p-nitrophenol (pNP) from respective pNP-substrates (xylosidase, glucosidase, arabinofuranosidase, mannosidase, galactosidase, RG acetylesterase, and cutinase), (iv) release of naphthol from naphthyl acetate (acetyl xylan esterases), (v) release of methanol from pectin (pectin methyl esterase), (vi) conversion of methyl ferulate to ferulic acid (ferulic acid esterases), or (vii) decrease in the turbidity (N,O-diacetylmuramidase).
Reducing groups were determined by using the 2-cyanoacetamide method (48), whereas the unsaturated galacturonides were estimated by increase in absorbance at A235, after acidification to pH 3.7 with acetate buffer (49). The release of p-nitrophenol was quantified spectrophotometrically at A420, after addition of 0.5 M sodium carbonate, pH 11.5 (50). The release of naphthol was measured spectrophotometrically at A560 after incubation with Fast Garnet GBC sodium salt (51), whereas the methanol was oxidized to formaldehyde for reaction with acetylacetone, and measuring at A412 (52). The decrease in the absorbance at A369, measured at pH 10.0 (for methyl ferulate) was used to quantify the activity of ferulic acid esterase (53), whereas the decrease in the absorbance at A450 (turbidity) was used to assay N,O-diacetylmuramidase (54). One katal of enzyme activity is defined as the amount of the enzyme required to release 1 mol of product per second under the standard assay conditions. For product characterization, the assay reaction mixtures were withdrawn at different time intervals, heated at 100°C for 5 min, and then either derivatized with 8-aminonaphthalene-1,3,6-trisulfonic acid or 8-aminopyrene-1,3,6-trisulfonate by reductive amination for capillary electrophoresis (13) or desalted by Dowex beads for MALDI-TOF-MS analysis (55).
Plant Material and Genetic Analysis.
A. thaliana plants were grown as described in ref. 38. Similarly, insertion lines corresponding to At2g37090 (SALK_058238 [irx9-1] and SALK_057033 [irx9-2]) were obtained and screened as described in ref. 38. Primer sequences for genomic screens were 5′-CTTAGAATGTATTTGACCGCCC-3′ (forward) and 5′-CCTATTAGCCGATAACAATGCC-3′ (reverse) for irx9-1 and 5′-GCTCCAATCTGGTTTAGTGCTC-3′ (forward) and 5′-CCTTCAACTTTGTATCGTCCTCC-3′ (reverse) for irx9-2. RT-PCR was used to confirm disruption of mRNA production. Primer sequences used for RT-PCR were 5′-TTCCTTGAAGAGAGGGTTATGGG-3′ (forward) and 5′-TCTTGGAACAATCTTGTGCCG-3′ (reverse).
Cell Wall Analysis.
Ball-milled cell wall material (1 mg) each from WT and irx9 was incubated in triplicate in respective buffer (25 mM, 400 μl) with 0.1 unit of purified enzymes of endo-mannanase (AN3358.2, pH 5.5), endo-glucanase (AN1285.2, pH 4.0), pectin lyases (AN2331.2 or AN2569.2, pH 7.0), pectate lyase (AN3337.2, pH 7.0), endo-polygalacturonases (AN4372.2 or AN8327.2, pH 4.0), xyloglucanase (AN0452.2, pH 6.5), endo-xylanases (AN1818.2 or AN3613.2, pH 5.5), endo-galactanase (AN5727.2, pH 5), and xylogalacturonase (Afu8g06890, pH 5.5) in an orbital shaker at 37°C for 12 h. Capillary electrophoresis analysis was performed as described in ref. 35.
Fourier transform infrared analyses were performed as described in ref. 38. The neutral sugar composition was determined by gas chromatography (56).
Gas Chromatography.
An Agilent gas chromatograph (6890N) with splitless injection (220°C) was equipped with a Supelco SP2330 capillary column (30 m × 0.25 mm, 0.2 μm film thickness). Column temperatures were as follows: 11 min isotherm at 180°C, then with 2°C/min to 220°C, 15 min isothermal at 220°C, then with 20°C/min to 240°C, 1 min isothermal at 240°C. Injection volumes were as follows: 1 μl, carrier gas helium 3.4 ml/min (constant flow), hydrogen 40 ml/min, air 250 ml/min, helium make-up 45 ml/min, detector temperature 250°C.
Immunolabeling.
Hand-cut stem sections (≈200 μm in thickness) from 7-week-old plants were fixed for 1 h at room temperature in PEM buffer (50 mM piperazine-N-N′-bis [2-ethane-sulfonic acid]/5 mM MgSO4/5 mM EGTA, pH 6.9), containing 4% paraformaldehyde. After rinsing three times with PBS buffer (0.1 M sodium phosphate, 0.9% NaCl, pH 7.0), sections were blocked with 3% BSA in PBS buffer for 1 h. Sections were rinsed three times in PBS buffer and incubated with primary antibodies [LM10 and LM11; diluted 1:20 (41)] at 4°C overnight, subsequently rinsed three times with PBS buffer and incubated with goat anti-rat IgG antibody (Molecular Probes) conjugated to the fluorophore Alexa Fluor 488 (diluted 1:1,000). Sections were rinsed three times with PBS, mounted in PBS buffer, and photographed by using a Leica (Deerfield, IL) D500 camera attached to a Leica DMRB compound microscope using epifluorescent filters (57). Sections were counterstained with calcofluor to visualize cellulose depositions. Sections probed only with secondary antibodies were used as controls.
Supplementary Material
Acknowledgments
We thank Rolf Prade, David Pesta, and Patricia Ayoubi for performing the BLAST search. This work was supported in part by Grant DE-FG02-03ER15444 from the U.S. Department of Energy Basic Energy Sciences Division. S.B. received fellowships from the Josef-Schormüller-Gedächtnisstiftung and the Deutsche Forschungsgemeinschaft (DFG). S.P. was a recipient of Swedish Research Council Fellowship 623-2004-4254.
Abbreviation
- FGSC
Fungal Genetics Stock Center.
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. DQ490466–DQ490522) and the FGSC database, www.fgsc.net (accession nos. 10061–10134).
References
- 1.Coimbra M. A., Delgadillo I., Waldron K. W., Selvendran R. R. In: Modern Methods of Plant Analysis: Plant Cell Wall Analysis. Linskens H. F., Jackson J. F, editors. Vol. 17. Berlin: Springer; 1996. pp. 19–44. [Google Scholar]
- 2.MacDougall A. J., Rigby N. M., Ring S. G. Plant Physiol. 1997;114:353–362. doi: 10.1104/pp.114.1.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strasser G. R., Amado R. Carbohydr. Polymers. 2002;48:263–269. [Google Scholar]
- 4.Deery M. J, Stimson E, Chappell C. G. Rapid Commun. Mass Spectrom. 2001;15:2273–2283. doi: 10.1002/rcm.458. [DOI] [PubMed] [Google Scholar]
- 5.Cohen A., Schagerlof H., Nilsson C., Melander C., Tjerneld F., Gorton L. J. Chromatogr. A. 2004;1029:87–95. doi: 10.1016/j.chroma.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 6.Teleman A., Nordström M., Tenkanen M., Jacobs A., Dahlman O. Carbohydr. Res. 2003;338:525–534. doi: 10.1016/s0008-6215(02)00491-3. [DOI] [PubMed] [Google Scholar]
- 7.Björndal H., Hellerquist C. G., Lindberg B., Svensson S. Angew. Chem. 1970;82:643–674. [Google Scholar]
- 8.Rolf D., Gray G. R. J. Am. Chem. Soc. 1982;104:3539–3541. [Google Scholar]
- 9.Mort A. J., Pierce M. L. In: Carbohydrate Analysis: High Performance Liquid Chromatography and Capillary Electrophoresis. El Rassi Z., editor. Amsterdam: Elsevier; 1994. pp. 3–37. [Google Scholar]
- 10.Broberg A., Thomsen K. K., Duus J. O. Carbohydr. Res. 2000;328:375–382. doi: 10.1016/s0008-6215(00)00108-7. [DOI] [PubMed] [Google Scholar]
- 11.Stroop C. J. M., Bush C. A., Marple R. L., LaCourse W. R. Anal. Biochem. 2002;303:176–185. doi: 10.1006/abio.2002.5582. [DOI] [PubMed] [Google Scholar]
- 12.Ray B., Loutelier-Bourhis C., Lange C., Condamine E., Driouich A., Lerouge P. Carbohydr. Res. 2004;339:201–208. doi: 10.1016/j.carres.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 13.Evangelista R. A., Liu M., Chen F. C. Anal. Chem. 1995;67:2239–2245. [Google Scholar]
- 14.Cardoso S. M., Silva A. M. S., Coimbra M. A. Carbohydr. Res. 2002;337:917–924. doi: 10.1016/s0008-6215(02)00082-4. [DOI] [PubMed] [Google Scholar]
- 15.Vierhuis E., York W. S., Kolli V. S., Vincken J., Schols H. A., Van Alebeek G. W., Voragen A. G. Carbohydr. Res. 2001;332:285–297. doi: 10.1016/s0008-6215(01)00096-9. [DOI] [PubMed] [Google Scholar]
- 16.Harvey D. J. Intern. J. Mass Spectrom. 2003;226:1–35. [Google Scholar]
- 17.Vries R. P., Visser J. Microbiol. Mol. Biol. Rev. 2001;65:497–522. doi: 10.1128/MMBR.65.4.497-522.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galagan J. E., Calvo S. E., Cuomo C., Ma L.-J., Wortman J. R., Batzoglou S., Lee S.-I., Bastürkmen M., Spevak C. C., Clutterbuck J., et al. Nature. 2005;438:1105–1115. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- 19.Fernández-Espinar M. T., Ramón D., Piñaga F., Vallés S. FEMS Microbiol. Lett. 1992;91:91–97. doi: 10.1111/j.1574-6968.1994.tb06622.x. [DOI] [PubMed] [Google Scholar]
- 20.Fernández-Espinar M. T., Piñaga F., Sanz P., Ramón D., Vallés S. FEMS Microbiol. Lett. 1993;113:223–228. doi: 10.1111/j.1574-6968.1994.tb06622.x. [DOI] [PubMed] [Google Scholar]
- 21.Fernández-Espinar M. T., Piñaga F., de Graaf L. H., Visser J., Ramón D., Vallés S. Appl. Environ. Microbiol. 1994;42:555–562. [Google Scholar]
- 22.Kumar S., Ramón D. FEMS Microbiol. Lett. 1996;135:287–293. [Google Scholar]
- 23.Pérez-González J. A., de Graaf L. H., Visser J., Ramón D. Appl. Environ. Microbiol. 1996;62:2179–2182. doi: 10.1128/aem.62.6.2179-2182.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pérez-González J. A., van Peij N. N. M. E., Bezoen A., MacCabe A. P., Ramón D., de Graaf L. H. Appl. Environ. Microbiol. 1998;64:1412–1419. doi: 10.1128/aem.64.4.1412-1419.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ho M.-C., Whitehead M. P., Cleveland T. E., Dean R. A. Curr. Genet. 1995;27:142–149. doi: 10.1007/BF00313428. [DOI] [PubMed] [Google Scholar]
- 26.Ramón D., v. d. Veen P., Visser J. FEMS Microbiol. Lett. 1993;113:15–22. doi: 10.1111/j.1574-6968.1993.tb06481.x. [DOI] [PubMed] [Google Scholar]
- 27.Fernández-Espinar M. T., Peña J. L., Piñaga F., Vallés S. FEMS Microbiol. Lett. 1994;115:107–112. doi: 10.1111/j.1574-6968.1994.tb06622.x. [DOI] [PubMed] [Google Scholar]
- 28.Gielkens M., González-Candelas L., Sánchez-Torres P., van de Vondervoort P., de Graaf L., Visser J., Ramón D. Microbiology. 1999;145:735–751. doi: 10.1099/13500872-145-3-735. [DOI] [PubMed] [Google Scholar]
- 29.Chikamatsu G., Shirai K., Kato M., Kobayashi T., Tsukagoshi N. FEMS Microbiol. Lett. 1999;175:239–245. doi: 10.1111/j.1574-6968.1999.tb13626.x. [DOI] [PubMed] [Google Scholar]
- 30.Lockington R. A., Rodbourn L., Barnett S., Carter C. J., Kelly J. M. Fungal Genet. Biol. 2002;37:190–196. doi: 10.1016/s1087-1845(02)00504-2. [DOI] [PubMed] [Google Scholar]
- 31.Ríos S., Pedregosa A. M., Monistrol I. F., Laborda F. FEMS Microbiol. Lett. 1993;112:35–42. doi: 10.1111/j.1574-6968.1993.tb06420.x. [DOI] [PubMed] [Google Scholar]
- 32.Eades C. J., Hintz W. E. Gene. 2000;255:25–34. doi: 10.1016/s0378-1119(00)00298-5. [DOI] [PubMed] [Google Scholar]
- 33.Coutinho P. M., Deleury E., Davies G. J., Henrissat B. J. Mol. Biol. 2003;328:307–317. doi: 10.1016/s0022-2836(03)00307-3. [DOI] [PubMed] [Google Scholar]
- 34.Cereghino G., Cereghino J., Ilgen C., Cregg J. Curr. Opin. Biotechnol. 2002;13:329–332. doi: 10.1016/s0958-1669(02)00330-0. [DOI] [PubMed] [Google Scholar]
- 35.Bauer S., Vasu P., Mort A. J., Somerville C. R. Carbohydr. Res. 2005;340:2590–2597. doi: 10.1016/j.carres.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 36.Fu J., Prade R., Mort A. Carbohydr. Res. 2001;330:73–81. doi: 10.1016/s0008-6215(00)00268-8. [DOI] [PubMed] [Google Scholar]
- 37.Lerouxel O., Choo T. S., Séveno M., Usadel B., Faye L., Lerouge P., Pauly M. Plant Physiol. 2002;130:1754–1763. doi: 10.1104/pp.011965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Persson S., Wei H., Milne J., Page G. P., Somerville C. R. Proc. Natl. Acad. Sci. USA. 2005;102:8633–8638. doi: 10.1073/pnas.0503392102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brown D. M., Zeef L. A., Ellis J., Goodacre R., Turner S. R. Plant Cell. 2004;17:2281–2295. doi: 10.1105/tpc.105.031542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhong R., Peña M. J., Zhou G.-K., Nairn C. J., Wood-Jones A., Richardson E. A., Morrison W. H., III, Darvill A. G., York W. S., Ye Z.-H. Plant Cell. 2005;17:3390–3408. doi: 10.1105/tpc.105.035501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCartney L., Marcus S. E., Knox J. P. J. Histochem. Cytochem. 2005;53:543–546. doi: 10.1369/jhc.4B6578.2005. [DOI] [PubMed] [Google Scholar]
- 42.Borneman W. S., Hartley R. D., Morrison H., Akin D. E., Ljungdahl L. G. Appl. Microbiol. Biotechnol. 1990;33:345–351. [Google Scholar]
- 43.Zhan D., Janssen P., Mort A. J. Carbohydr. Res. 1998;308:373–380. doi: 10.1016/s0008-6215(98)00096-2. [DOI] [PubMed] [Google Scholar]
- 44.McCluskey K. Adv. Appl. Microbiol. 2003;52:245–262. doi: 10.1016/s0065-2164(03)01010-4. [DOI] [PubMed] [Google Scholar]
- 45.Barratt R. W., Johnson G. B., Ogata W. N. Genetics. 1965;52:233–246. doi: 10.1093/genetics/52.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sambrook J., Russell D. W. Molecular Cloning: A Laboratory Manual. 3rd Ed. Vol. 1. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 2000. pp. 4.70–4.71. [Google Scholar]
- 47.Gomori G. Methods Enzymol. 1955;1:138–145. [Google Scholar]
- 48.Honda S., Nishimura Y., Takahashi M., Chiba H., Kakehi K. Anal. Biochem. 1982;119:194–199. doi: 10.1016/0003-2697(82)90685-6. [DOI] [PubMed] [Google Scholar]
- 49.Benen J. A. E., Kester H. C. M., Parenicova L., Visser J. Biochemistry. 2000;39:15563–15569. doi: 10.1021/bi000693w. [DOI] [PubMed] [Google Scholar]
- 50.Gilead S., Shoham Y. Appl. Environ. Microbiol. 1995;61:170–174. doi: 10.1128/aem.61.1.170-174.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hespell R. B., O’Bryan-Shah P. J. Appl. Environ. Microbiol. 1988;54:1917–1922. doi: 10.1128/aem.54.8.1917-1922.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wood P. J., Siddiqui I. R. Anal. Biochem. 1971;39:418–428. doi: 10.1016/0003-2697(71)90432-5. [DOI] [PubMed] [Google Scholar]
- 53.Faulds C. B., Williamson G. Microbiology. 1994;140:779–787. [Google Scholar]
- 54.Croux C., Canard B, Goma G., Soucaille P. Appl. Environ. Microbiol. 1992;58:1075–1081. doi: 10.1128/aem.58.4.1075-1081.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Körner R., Limberg G., Mikkelsen J. D., Roepstorff P. J. Mass Spectr. 1998;33:836–842. doi: 10.1002/(SICI)1096-9888(199809)33:9<836::AID-JMS694>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 56.Blakeney A. B., Harris P. J., Henry R. J., Stone B. A. Carbohydr. Res. 1983;113:291–299. [Google Scholar]
- 57.Adam L., Somerville S. Plant J. 1996;9:341–356. doi: 10.1046/j.1365-313x.1996.09030341.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.