Abstract
In Escherichia coli, the SecB/SecA branch of the Sec pathway and the twin-arginine translocation (Tat) pathway represent two alternative possibilities for posttranslational translocation of proteins across the cytoplasmic membrane. Maintenance of pathway specificity was analyzed using a model precursor consisting of the mature part of the SecB-dependent maltose-binding protein (MalE) fused to the signal peptide of the Tat-dependent TorA protein. The TorA signal peptide selectively and specifically directed MalE into the Tat pathway. The characterization of a spontaneous TorA signal peptide mutant (TorA*), in which the two arginine residues in the c-region had been replaced by one leucine residue, showed that the TorA*-MalE mutant precursor had acquired the ability for efficiently using the SecB/SecA pathway. Despite the lack of the “Sec avoidance signal,” the mutant precursor was still capable of using the Tat pathway, provided that the kinetically favored Sec pathway was blocked. These results show that the h-region of the TorA signal peptide is, in principle, sufficiently hydrophobic for Sec-dependent protein translocation, and therefore, the positively charged amino acid residues in the c-region represent a major determinant for Tat pathway specificity. Tat-dependent export of TorA-MalE was significantly slower in the presence of SecB than in its absence, showing that SecB can bind to this precursor despite the presence of the Sec avoidance signal in the c-region of the TorA signal peptide, strongly suggesting that the function of the Sec avoidance signal is not the prevention of SecB binding; rather, it must be exerted at a later step in the Sec pathway.
Proteins which have to be transported across the bacterial plasma membrane are, in most cases, initially synthesized as larger precursor proteins containing an N-terminal signal peptide (14, 30). The translocation of the vast majority of these proteins occurs in a more or less unfolded state and is mediated by the so-called Sec machinery, consisting of a protein conducting channel (SecYEG) and the SecA translocation ATPase, which couples the energy of ATP binding and hydrolysis to protein translocation (12).
In addition to Sec-mediated protein translocation, many bacteria possess a second protein export pathway for the translocation of a subset of precursor proteins. This Sec-independent mechanism has been designated Tat (for twin-arginine translocation) due to the fact that a characteristic amino acid sequence motif including two consecutive arginine residues can be identified in the signal peptides of the respective precursor proteins (1, 30, 33, 43). In marked contrast to the Sec system, which requires that its substrates be kept in a more or less unfolded state, the Tat pathway translocates its substrates in a fully folded or even oligomeric form across the membrane (18, 34, 36). Many of the Tat substrates are proteins that have to acquire a cofactor in the cytosol and, therefore, must fold before export (1, 2). In addition, some cofactorless proteins also use the Tat system, presumably because their rapid folding kinetics precludes transport via the Sec pathway (2, 39).
For Escherichia coli, four genes (tatA, tatB, tatC, and tatE) that encode components of the Tat export machinery have been identified (2, 33, 43). However, the molecular mechanism by which proteins are transported across the membrane by this machinery is completely unknown so far. Likewise, very little is known about the targeting of the Tat precursor proteins to the translocase. For the ΔpH import route into plant thylakoids, which is closely related to the bacterial Tat pathway, it had been shown that no soluble protein factors are required for transport across the membrane (29). It remains to be determined whether the same scenario (i.e., no requirement for a cytosolic factor) is true also for the bacterial Tat pathway.
Specificity of the precursor proteins for either the Sec pathway or the Tat pathway seems to lie predominantly in the nature of the respective signal peptides. Tat signal peptides possess a tripartite organization similar to that of Sec signal peptides: a positively charged N terminus (n-region) is followed by a core of hydrophobic amino acids (h-region) and a more polar C terminus (c-region) preceding the signal peptidase cleavage site (14, 30). In contrast to Sec signal sequences, Tat signal peptides contain an (S/T)-R-R-X-F-L-K consensus motif (where X is highly variable) at the boundary between the n- and the h-regions (1, 2). It is reasonable to assume that one or more of the components of the Tat pathway (but not of the Sec system) might recognize the precursor proteins by the presence of this motif. However, additional features of the signal peptides that go beyond the conserved sequence motif seem to be important for successful recognition of the precursor proteins by their respective translocase (3). Another important difference between Tat and Sec signal peptides seems to reside in the hydrophobicity of their h-regions, with Tat signal peptides being mostly less hydrophobic than their Sec counterparts (9). Likewise, the hydrophobicity of the n-region was reported to be another important factor determining Tat pathway exclusivity (20). Furthermore, in the c-region of Tat/ΔpH signal peptides, basic amino acids are frequently observed, whereas signal peptides of the Sec pathway show a strong bias against such residues near the signal peptidase cleavage site (2, 4). Together, these differences seem to prevent the mistargeting of stably folded Tat precursor proteins into the Sec pathway. At present, it is completely unknown at which step(s) of the export pathway this so-called “Sec avoidance” event takes place.
In this work, we have used the E. coli maltose-binding protein (MalE), which is the paradigm of a SecB-dependent precursor protein (14, 26), as a model protein for the analysis of the substrate specificity of the SecB/SecA and Tat pathways. Replacement of the pre-MalE signal peptide by the signal peptide of the Tat substrate TorA resulted in the selective and exclusive redirection of MalE into the Tat pathway. A spontaneous mutant of the TorA-MalE hybrid precursor, which had lost the positively charged amino acid residues in the c-region, gained the ability to use the Sec system. Furthermore, we could show that this mutant TorA-MalE precursor was still capable of using the Tat pathway, provided the kinetically favored Sec pathway was blocked. Our results clearly demonstrate that the nature of the signal peptide is crucial for the choice of the correct export pathway. Furthermore, our data strongly suggest that Tat signal peptides containing an h-region sufficiently hydrophobic for Sec-mediated protein export require an additional Sec avoidance activity, such as the presence of positively charged amino acid residues near the signal peptidase cleavage site, for the maintenance of Tat pathway specificity. Export of TorA-MalE was significantly retarded by SecB, suggesting that the Sec avoidance function of the positively charged amino acid residues in the c-region of the TorA signal peptide is not the prevention of SecB binding but rather is exerted at a later stage in the Sec pathway. Finally, since the TorA-MalE precursor is selectively targeted into the Tat pathway and since export of MalE into the periplasm is easily detectable in plate assays, this precursor represents an excellent reporter for the genetic analysis of Tat-dependent protein translocation in E. coli. The suitability of this reporter system was directly demonstrated by the isolation of mutants carrying a transposon insertion in known tat genes.
MATERIALS AND METHODS
Strains, plasmids, phages, and growth conditions.
The bacterial strains, plasmids, and phages used in this study are listed in Table 1. Bacterial strains were grown at 37°C in Luria-Bertani medium (28), minimal medium (41) supplemented with 0.4% maltose, 0.4% glucose, or 0.4% glycerol, or MacConkey medium (Difco) supplemented with 1% maltose. If required, isopropyl-β-d-thiogalactopyranoside was used at a 1 mM concentration. Antibiotic supplements were at the following concentrations: ampicillin, 100 μg/ml; tetracycline, 15 μg/ml; kanamycin, 50 μg/ml; chloramphenicol, 25 μg/ml. For phage P1-mediated transduction (28) of the ΔmalE444 deletion into various strains, E. coli MM129 (5) was used as a donor. The ΔmalE444 allele is an in-frame deletion which is nonpolar and lies entirely within malE, removing 765 bp of the gene (6). Tetracycline resistance in strain MM129 is conferred by Tn10, which is closely linked to the malEΔ444 deletion and leads to a maltose-independent expression of the lamB gene, which is necessary for phage λ infections (6). For TnphoA mutagenesis, λ::TnphoA-1 was used as described by Gutierrez et al. (17).
TABLE 1.
Bacterial strains, plasmids, and phages
| Strain, plasmid, or phage | Relevant propertiesa | Source or Reference |
|---|---|---|
| E. coli | ||
| LJ110 | Alias W3110, wild-type K-12, prototroph | J. Lengeler |
| MC4100 | F−araD139 Δ(argF-lac)U169 rpsL150 relA1 fruA25 | 7 |
| CK1953 | MC4100 secB::Tn5 | C. Kumamoto |
| DADE | MC4100 ΔtatA-tatD ΔtatE | 42 |
| MM129 | F−malT1c tsr-1 rpsL136 ΔmalE444 zjb729::Tn10 | 5 |
| GSJ100 | MC4100 × P1.MM129>Tcr ΔmalE444 zjb729::Tn10 | This study |
| GSJ101 | DADE × P1.MM129 > Tcr ΔmalE444 zjb729::Tn10 | This study |
| Plasmids | ||
| pMMB67EH | RSF1010-derivative, tacP, rrnB; Apr | 15 |
| pBBR1MCS-2 | mob, rep, lacZα+; Kmr | 25 |
| pBBR2MCS-2 | mob, rep, lacZα+; Apr | This study |
| pHSG575 | pSC101 replicon, lacZα+; Cmr | 40 |
| pBluescriptKS | lacZα+ cloning vector; Apr | Stratagene |
| pTatABC | pHSG575 derivative; carrying the tatABC operon of E. coli | This study |
| pMalE | pBBR2MCS-2 derivative; carrying the pre-malE gene | This study |
| pTorA-MalE | pBBR1MCS-2 derivative; carrying the torA-malE fusion gene | This study |
| pATorA-MalE | pBBR2MCS-2 derivative; carrying the torA-malE fusion gene | This study |
| pTorA*-MalE | pBBR1MCS-2 derivative; carrying the torA*-malE mutant fusion gene | This study |
| pATorA*-MalE | pBBR2MCS-2 derivative; carrying the torA*-malE mutant fusion gene | This study |
| pMTorA-MalE | pMMB67EH derivative; carrying the torA-malE fusion gene | This study |
| Phages | ||
| λTnphoA-1 | b221 cI857 Pam3::TnphoA Kmr | 17 |
| Plkc | Transducing phage | Laboratory stock |
Apr, ampicillin resistance; Kmr, kanamycin resistance; Cmr, chloramphenicol resistance; Tcr, tetracycline resistance.
DNA manipulations.
All DNA manipulations followed standard procedures (35). For the construction of the gene encoding the TorA-MalE fusion protein, a torA gene fragment was amplified by PCR using chromosomal DNA of E. coli LJ110 as a template and primers T-for (5′-TAA GGA TCC CGG CGA TAA GAA GGA AGA AAA-3′) and T-rev (5′-TTT GAA TTC GAC AGC GTC AGT CGC CGC TTG-3′). Via these primers, restriction sites were introduced immediately upstream of the torA ribosome binding site (BamHI) and downstream of the eighth codon of mature TorA (EcoRI). The amplified torA fragment was digested with BamHI and EcoRI and ligated into the BamHI/EcoRI-digested vector pBluescript KS, resulting in pBluetorA. A malE gene fragment encoding the mature MalE protein was amplified using chromosomal DNA of LJ110 as a template and primers M-for (5′-CTC TTT AAA ATC GAA GAA GGT AAA CTG G-3′) and M-rev (5′-GGC GTC GAC GCC GCA TCC GGC ATT TCA C-3′). Via these primers, restriction sites were introduced directly upstream of the region encoding the mature MalE (DraI) and downstream of the stop codon (SalI). The corresponding PCR fragment was digested with DraI and SalI and ligated into EcoRV/SalI-digested pBluetorA. The resulting torA-malE gene (encoding the signal peptide and eight amino acid residues of the mature part of TorA fused to a linker peptide of three amino acid residues and the mature part of MalE) (Fig. 1) was excised with SalI and XbaI, and the sticky ends were filled in with Klenow enzyme and ligated into the EcoRV site of the medium-copy-number vector pBBR1MCS-2 (25) or its Ampr derivative, pBBR2MCS-2 (see below). In the resulting plasmids (pTorA-MalE and pATorA-MalE), the torA-malE gene is placed under the regulatory control of the lac promoter. In addition, the torA-malE gene was also cloned into the medium-copy-number vector pMMB67EH (15) by transferring a 1.3-kb BamHI fragment from pTorA-MalE to BamHI-linearized pMMB67EH. In the resulting plasmid, pMTorA-MalE, the torA-malE gene is under the regulatory control of the tac promoter. For the construction of pBBR2MCS-2, a bla gene cassette was amplified by PCR using pBluescript as a template and primers amp-for (5′-AAG CTA GCG GCC ATC CTG ACG GAT GGC C-3′) and amp-rev (5′-CGC CAT GGT TAA TGC GCC GCT ACA GGG C-3′). The corresponding PCR product was phosphorylated with T4 polynucleotide kinase and ligated into the SmaI site of pBBR1MCS-2, resulting in pBBR2MCS-2. For the construction of pMalE, in which the gene encoding the authentic pre-MalE precursor is placed under the regulatory control of the lac promoter, the malE gene was amplified by PCR using chromosomal DNA of E. coli LJ110 as a template and primers MalE-for (5′-CAC GGG CCC TTC ACC AAC AAG GAC C-3′) and MalE-rev (5′-GGC GTC GAC GCC GCA TCC GGC ATT TCA C-3′). The resulting PCR product was digested with ApaI and SalI, and sticky ends were filled-in with the Klenow enzyme. After phosphorylation with T4 polynucleotide kinase, the DNA fragment was ligated into the EcoRV site of pBBR2MCS-2, resulting in the plasmid pMalE. For the construction of pTatABC, the tatABC genes were amplified by PCR using chromosomal DNA of E. coli MC4100 as a template and primers Tat-for (5′-CAG GAT CCA CAG AGG AAC ATG TAT GG-3′) and Tat-rev (5′-GCC CTGCAG GGC GGT TGA ATT TAT TC-3′). Via these primers, restriction sites were introduced immediately upstream of the ribosome binding site of the tatA gene (BamHI) and downstream of the stop codon of the tatC gene (PstI). The amplified fragment was cleaved with BamHI and PstI and ligated into the corresponding sites of the low-copy-number vector pHSG575 (40), thereby placing the tatABC genes under the regulatory control of the lac promoter. The correctness of all plasmid constructions was verified by DNA sequencing.
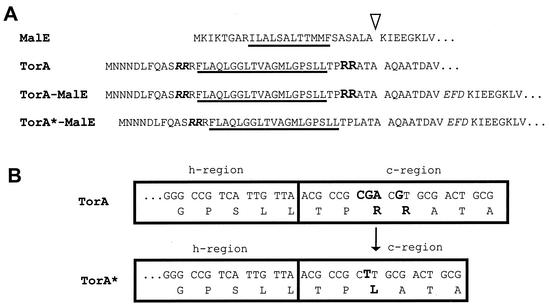
FIG. 1.
Signal peptides and early mature region of proteins used in this study. (A) The hydrophobic h-regions are underlined, and the signal peptidase cleavage site is indicated by an arrow. The RR residues of the twin-arginine motif are shown in bold and italics; the RR residues of the Sec avoidance signal are shown in bold. The mature regions of the TorA-MalE and TorA*-MalE fusion proteins consist of eight amino acid residues of the mature TorA protein followed by a linker sequence of three amino acid residues (shown in italics) and the mature MalE protein. (B) Nature of the spontaneous mutation that results in a replacement of the two arginine residues in the c-region of the TorA signal peptide by a leucine residue. The gene regions encoding the carboxyl-terminal parts of the h-regions and the c-regions of the wild-type (TorA) and mutant (TorA*) signal peptides are shown. Relevant nucleotide positions and amino acid residues are highlighted in bold.
Preparation of subcellular fractions.
Fractionation of cells into a fraction containing the cytosol and membranes (C/M) and a periplasmic fraction (P) was done by using an EDTA-lysozyme spheroplasting method (22) with slight modifications. E. coli strains were grown overnight in 10 ml of Luria-Bertani medium. The cells were harvested by centrifugation and washed once with ice-cold 30 mM Tris-HCl, pH 8.0. Subsequently, the cells were resuspended in 40 μl of ice-cold sucrose solution (20% sucrose, 30 mM Tris-HCl [pH 8.0]), after which 20 μl of ice-cold EDTA-lysozyme solution (1 mg of lysozyme/ml, 0.1 M EDTA) was added. After incubation on ice for 5 min, the resulting spheroplasts were spun down at 21,500 × g for 15 min at 4°C. After sampling of the supernatant (which represents the P fraction), the pellet was washed with sucrose solution, resuspended in 1 ml of ice-cold 30 mM Tris-HCl (pH 8.0), and disrupted in a bead mill by stirring with glass beads (0.1 to 0.25 mm in diameter). The glass beads were removed by centrifugation, and the supernatant (which corresponds to the C/M fraction) was sampled. C/M and P fractions corresponding to an identical number of cells were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting using anti-MalE antibodies as described earlier (18).
Pulse-chase experiments.
Pulse-chase experiments were performed as described earlier (3) with some minor variations. Cells were grown to an optical density at 600 nm of 0.4 in 6 ml of pulse-chase medium (minimal medium, according to the method of Tanaka et al. (41) containing 10 μg of thiamine/ml, 0.4% glycerol, and 0.5 mg of methionine assay medium (Difco)/ml. Two milliliters of the culture was labeled with 5 μl of l-[35S]methionine (10 μCi/μl; Amersham Life Science, Braunschweig, Germany) for 1 min, after which 52 μl of chase solution (45 mg of l-methionine/ml, 2 mg of chloramphenicol/ml) was added. If appropriate, sodium azide (final concentration, 3 mM) was added 5 min prior to labeling to inhibit Sec-dependent translocation. At the indicated chase time points, samples of 0.5 ml were withdrawn and precipitated overnight at 4°C with 150 μl of 40% trichloroacetic acid. After centrifugation, the precipitates were washed once with 1 ml of 80% acetone and dried for 5 min at 95°C. Subsequently, the precipitates were resuspended in 40 μl of lysis buffer I (2% SDS, 10 mM EDTA, 50 mM Tris-HCl [pH 8.0]) and incubated for 5 min at 95°C. After addition of 960 μl of lysis buffer II (2% Triton X-100, 0.15 M NaCl, 10 mM EDTA, 50 mM Tris-HCl [pH 8.0]), the samples were centrifuged and the supernatants were subjected to immunoprecipitation, SDS-PAGE, and fluorography as described previously (3). Fractionation of pulse-labeled cells into C/M and P fractions was done as described in the previous chapter. The resulting fractions were subsequently precipitated with trichloroacetic acid and further processed as described above.
RESULTS
Tat-dependent export of maltose-binding protein into the E. coli periplasm.
The periplasmic maltose-binding protein (MalE) is one of the model proteins that have extensively been used for the analysis of Sec-dependent protein translocation in E. coli (14, 26). MalE is part of the uptake system for maltose, and its presence in the periplasm is strictly required for growth of E. coli cells on minimal medium with maltose as the sole carbon source (10). In addition, colonies of E. coli show a red color (indicative of acid production) on MacConkey agar plates containing maltose only when MalE is translocated across the plasma membrane (13).
Proteins which are exported by the Tat pathway have to fold prior to membrane translocation (2). In contrast, MalE, which mainly uses the posttranslational mode of Sec-dependent translocation (23), has to be kept in a more or less unfolded, export-competent state by binding to the export-specific chaperone SecB (8). The signal peptide of MalE has been shown to slow down the folding of mature MalE (32), and it has been proposed in the so-called kinetic partitioning model that this slowing process allows SecB to discriminate between precursor proteins and other proteins in the cell (19). Despite these facts, we reasoned that it might be possible to export MalE via the Tat pathway, since previous results have shown that mature MalE (without its native Sec signal peptide) can rapidly fold into the native conformation also in the cytosol (27). Furthermore, we speculated that Tat signal peptides might exert reduced antifolding activity, or even lack antifolding activity, with respect to their mature proteins and, therefore, fusion of mature MalE to a Tat signal peptide might channel the normally SecB-dependent MalE protein into the Tat pathway.
To test this hypothesis, a gene was constructed that encodes a hybrid precursor protein consisting of the signal peptide and eight amino acid residues of the mature part of the strictly Tat-dependent trimethylamine N-oxide reductase (TorA) precursor protein (36) fused to a linker peptide of three amino acid residues and the mature part of MalE (Fig. 1A). The corresponding plasmid (pTorA-MalE) was transformed into E. coli GSJ100 (a malE-negative derivative of the tat wild-type strain MC4100) and GSJ101 (a malE-negative derivative of the tat deletion mutant DADE [42]), and the resulting strains GSJ100 (pTorA-MalE) and GSJ101 (pTorA-MalE) were streaked on minimal maltose and MacConkey maltose agar plates. As shown in Table 2, the presence of pTorA-MalE allowed growth of GSJ100 on maltose as the sole carbon source, and the colonies of this strain were red on MacConkey maltose indicator plates. In contrast, GSJ101 (pTorA-MalE) did not grow on minimal maltose plates and grew to pale colonies on MacConkey maltose agar plates. These results already suggested that the TorA signal peptide is able to target the mature part of MalE into the periplasmic space and that this targeting is dependent on the presence of the Tat protein export apparatus.
TABLE 2.
Phenotype of bacterial strains on MacConkey maltose and maltose minimal medium agar plates
| Bacterial strain | Growth on maltose minimal mediuma | Color of colonies on MacConkey maltosea |
|---|---|---|
| MC4100 | + | Red |
| GSJ100 | − | Pale |
| GSJ101 | − | Pale |
| GSJ100(pBBR1MCS-2) | − | Pale |
| GSJ101(pBBR1MCS-2) | − | Pale |
| GSJ100(pTorA-MalE) | + | Red |
| GSJ101(pTorA-MalE) | − | Pale |
| GSJ100(pTorA*-MalE) | + | Red |
| GSJ101(pTorA*-MalE) | + | Red |
Bacterial strains were streaked on minimal medium agar plates containing 0.4% maltose as a sole carbon source or on MacConkey agar plates containing 1 % maltose and incubated at 37°C. +, growth; −, no growth.
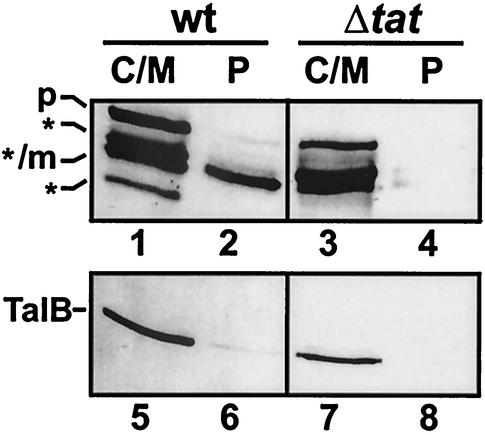
To more directly investigate the Tat-dependent export of MalE into the periplasm, the subcellular localization of MalE polypeptides was analyzed by EDTA-lysozyme spheroplasting (22). Spheroplasted cells were thus divided into a C/M fraction and a P fraction. The fractions were subsequently analyzed by SDS-PAGE and Western blotting using anti-MalE antibodies. As a control for the quality of the fractionation procedure, the distribution of the cytosolic enzyme transaldolase B (TalB) (38) was analyzed in parallel. As expected, the cytoplasmic TalB protein was found in the C/M fractions (Fig. 2, lanes 5 to 8). In the C/M fractions of GSJ100 (pTorA-MalE) and GSJ101 (pTorA-MalE), several MalE-derived polypeptides could be detected (Fig. 2, lanes 1 and 3). Besides a band corresponding in size to the unprocessed TorA-MalE precursor, several faster-migrating bands are present in the C/M fraction. Most likely, these smaller MalE polypeptides represent cytosolic degradation products of the TorA-MalE precursor. Importantly, in the periplasmic fraction of GSJ100 (pTorA-MalE), a protein corresponding in size to processed TorA-MalE can be identified (Fig. 2, lane 2). In contrast, no MalE-derived polypeptide was present in the periplasmic fraction of the tat mutant strain GSJ101 expressing the TorA-MalE hybrid protein (Fig. 2, lane 4).
FIG. 2.
Subcellular localization of MalE polypeptides. Cells of the tat wild-type (wt) strain GSJ100 containing pTorA-MalE and the tat deletion mutant (Δtat) GSJ101 containing pTorA-MalE were fractionated into a combined C/M fraction (lanes C/M) and a P fraction (lanes P) by EDTA-lysozyme spheroplasting as described in Materials and Methods. Samples were subjected to SDS-PAGE and Western blotting using anti-MalE antibodies (lanes 1 to 4) or, as a control, antibodies directed against the cytoplasmic protein TalB (lanes 5 to 8). p, TorA-MalE precursor; m, position of mature form of TorA-MalE in the P fraction; asterisks, positions of TorA-MalE degradation products in the C/M fraction.
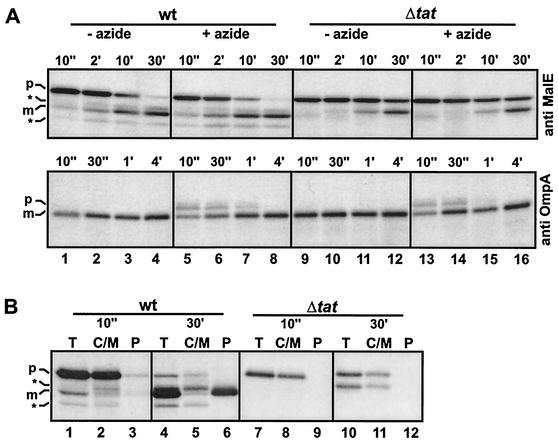
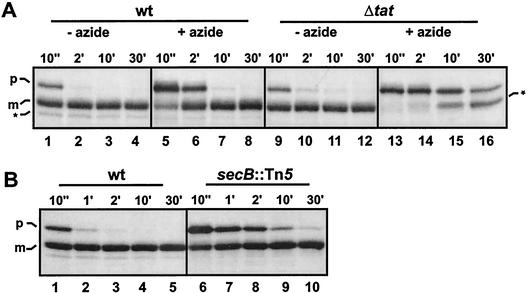
In addition to these Western blotting experiments, which represent a static situation, the membrane translocation and processing of the TorA-MalE precursor was analyzed by pulse-chase experiments. In GSJ100, the TorA-MalE precursor is almost completely converted into the periplasmic mature form within 30 min after the chase (Fig. 3A, upper part, lanes 1 to 4). In contrast, the majority of the TorA-MalE precursor is still present 30 min after the chase in the tat mutant GSJ101, and no polypeptide corresponding to the mature form can be detected. Instead, part of the precursor is degraded to a nonperiplasmic MalE polypeptide which is slightly larger than the mature exported form (Fig. 3A upper part, lanes 9 to 12). The mentioned subcellular localizations of the various MalE polypeptides were determined by fractionation of pulse-labeled cells sampled 10 s or 30 min after addition of the chase into C/M and P fractions (Fig. 3B) and were in full agreement with the subcellular localizations which had been deduced from the static Western blot experiments. Blocking of the Sec pathway by the addition of 3 mM sodium azide prior to labeling had no influence on the appearance of mature MalE in GSJ100 (Fig. 3A, upper part, lanes 5 to 8) but significantly retarded processing of the Sec-dependent precursor protein pre-OmpA (Fig. 3A, lower part, lanes 5 to 8). Taken together, our data clearly demonstrate that the TorA signal peptide can mediate export of MalE to the periplasm in a strictly Tat-dependent manner.
FIG. 3.
Processing of TorA-MalE. (A) Processing of TorA-MalE in the tat wild-type (wt) strain GSJ100 containing pTorA-MalE and the tat deletion mutant (Δtat) GSJ101 containing pTorA-MalE in the absence (−) or presence (+) of 3 mM sodium azide was analyzed by pulse-chase labeling and subsequent immunoprecipitation with either anti-MalE serum (upper panel) or anti-OmpA serum (lower panel), SDS-PAGE, and fluorography. Cells were labeled with l-[35S]methionine for 1 min prior to chase with an excess of nonradioactive methionine. Samples were withdrawn at the time points indicated above the respective lanes (e.g., 10", 10 s; 2′, 2 min). p, precursor form of TorA-MalE (upper panel) or OmpA (lower panel); m, mature form of TorA-MalE (upper panel) or OmpA (lower panel); asterisks, positions of TorA-MalE degradation products. (B) The subcellular localization of the MalE gene products observed in the pulse-chase experiments in wild-type (wt) or tat mutant (Δtat) cells was analyzed by fractionating samples withdrawn at 10 s or 30 min after the chase into a combined C/M fraction (lanes C/M) and a P fraction (P). Lanes T show the corresponding samples without fractionation. p, precursor form of TorA-MalE; m, mature form of TorA-MalE; asterisks, positions of TorA-MalE degradation products.
SecB retards the Tat-dependent membrane translocation of the TorA-MalE precursor.
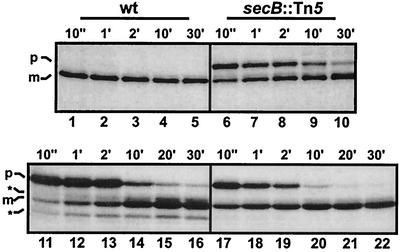
Since the possibility existed that signal peptides derived from Tat-dependent precursor proteins might differ from signal peptides derived from precursor proteins which use the SecB/SecA pathway with respect to their influence on the interaction of the attached mature parts with SecB, the export kinetics of TorA-MalE were analyzed by pulse-chase experiments with the secB wild-type strain MC4100 and its secB::Tn5 derivative, CK1953. As expected, the processing of the SecB-dependent authentic pre-MalE precursor is severely retarded for CK1953 (Fig. 4, lanes 6 to 10) compared to the very fast processing in MC4100 (Fig. 4, lanes 1 to 5). Interestingly, we found that the conversion of the TorA-MalE precursor to the mature form was significantly faster in the absence of SecB (Fig. 4, lanes 17 to 22) than in its presence (Fig. 4, lanes 11 to 16). Quantification of the bands from the sample taken 10 s after the chase revealed that 5% of the precursor had been converted to the mature form for the secB wild-type strain. In contrast, 25% of the precursor is converted to the mature protein in the absence of SecB within the same time period. Furthermore, for the secB mutant strain, hardly any nonexported degradation products of the TorA-MalE precursor can be detected, suggesting that the precursor which is bound to SecB is more sensitive to proteolytic attack. These results strongly suggest that SecB binds to at least a fraction of the TorA-MalE precursor, resulting in delayed export or partial degradation. In addition, these findings strongly suggest that the TorA signal peptide is not fundamentally different from the authentic pre-MalE signal peptide with respect to its effect on the kinetic partitioning of the attached mature MalE protein between binding to SecB and folding into the native conformation.
FIG. 4.
Processing of TorA-MalE is slowed down by SecB. Processing of authentic pre-MalE in the secB wild-type (wt) strain MC4100 containing pMalE (lanes 1 to 5) and the secB mutant (secB::Tn5) CK1953 containing pMalE (lanes 6 to 10) is shown. Processing of TorA-MalE in MC4100 (wt) containing pATorA-MalE (lanes 11 to 16) and CK1953 (secB::Tn5) containing pATorA-MalE (lanes 17 to 22) is shown. Cells were labeled with l-[35S]methionine for 1 min prior to chase with an excess of nonradioactive methionine. Samples were withdrawn at the time points indicated above the respective lanes (e.g., 10", 10 s; 1′, 1 min) and subjected to immunoprecipitation with anti-MalE serum, SDS-PAGE, and fluorography. p, precursor form of authentic pre-MalE (lanes 1 to 10) or TorA-MalE (lanes 11 to 22); m, mature form of pre-MalE (lanes 1 to 10) or TorA-MalE (lanes 11 to 22); asterisks, positions of TorA-MalE degradation products.
Positively charged amino acid residues in the c-region of the TorA signal peptide are required for Tat pathway specificity.
As indicated above, cells of the tat mutant strain GSJ101 expressing the TorA-MalE hybrid precursor appear pale on MacConkey maltose indicator plates due to the lack of exported MalE in the periplasm. However, during our experiments we repeatedly observed that single-mutant colonies with a red color arose spontaneously on the background of pale colonies. Since we had shown before that export of the TorA-MalE protein was strictly Tat dependent and since all the tat genes are deleted in GSJ101, the most likely explanation for the translocation of MalE into the periplasm of these spontaneous mutants would be that a mutation in the TorA-MalE precursor had occurred which allows this precursor to use the Sec pathway. From five independently isolated mutant colonies, the pTorA-MalE plasmid DNAs were isolated and retransformed into GSJ101. In all cases, the resulting strains gave red colonies on MacConkey maltose plates and were able to grow on minimal medium containing maltose (Table 2). This demonstrates that the respective mutations are indeed plasmid borne. DNA sequencing of the TorA-MalE region from the five plasmids showed that in all cases an identical mutation (CGA CGT to CTT; i.e., deletion of an entire codon plus a base substitution) had occurred which resulted in the replacement of the two arginine residues in the c-region of the TorA signal peptide (positions −4 and −5) by a leucine residue (Fig. 1B).
Export of the corresponding mutant precursor (designated TorA*-MalE) (Fig. 1) was analyzed by pulse-chase experiments with tat wild-type (GSJ100) and tat mutant (GSJ101) strains (Fig. 5A, lanes 1 to 4 and 9 to 12). In both strains, rapid processing of the TorA*-MalE precursor to the mature form occurred, clearly showing that, in contrast to the situation of the unaltered TorA-MalE precursor, translocation of TorA*-MalE across the plasma membrane is unaffected by the absence of functional Tat export machinery and, therefore, occurs in a Sec-dependent manner. In addition, the fast kinetics of TorA*-MalE processing, typical for Sec-dependent export, is in marked contrast to the slow processing kinetics of the TorA-MalE wild-type protein that is typical for Tat-dependent protein translocation (compare Fig. 5A, lanes 1 to 4, with Fig. 3A, upper part, lanes 1 to 4). Furthermore, our finding that processing of TorA*-MalE is sensitive to sodium azide is another strong indication for Sec-dependent export (see the following section). Since processing of the TorA*-MalE precursor was severely retarded for the secB mutant strain CK1953 (Fig. 5B, lanes 6 to 10) compared to the very fast processing in the secB wild-type strain, MC4100 (Fig. 5B, lanes 1 to 5), we conclude that the TorA*-MalE precursor is exported by the SecB/SecA branch of the Sec pathway. Taken together, these results clearly show that the two arginine residues in the c-region of the TorA signal peptide fulfill an important function in preventing the mistargeting of the attached mature protein into the Sec pathway.
FIG. 5.
Processing of TorA*-MalE. (A) Processing of TorA*-MalE in the tat wild-type (wt) strain GSJ100 containing pTorA*-MalE and the tat deletion mutant (Δtat) GSJ101 containing pTorA*-MalE in the absence (−) or presence (+) of 3 mM sodium azide was analyzed by pulse-chase labeling and subsequent immunoprecipitation with anti-MalE serum, SDS-PAGE, and fluorography. Cells were labeled with l-[35S]methionine for 1 min prior to chase with an excess of nonradioactive methionine. Samples were withdrawn at the time points indicated above the respective lanes (e.g., 10", 10s; 2′, 2 min). p, precursor form of TorA*-MalE; m, mature form of TorA*-MalE; asterisks, positions of TorA*-MalE degradation products. (B) Processing of TorA*-MalE in the secB wild-type (wt) strain MC41000 containing pATorA*-MalE (lanes 1 to 5) and the secB mutant (secB::Tn5) CK1953 containing pATorA*-MalE (lanes 6 to 10). Cells were labeled with l-[35S]methionine for 1 min prior to chase with an excess of nonradioactive methionine. Samples were withdrawn at the time points indicated above the respective lanes and subjected to immunoprecipitation with anti-MalE serum, SDS-PAGE, and fluorography. p, precursor form of TorA*-MalE; m, mature form of TorA*-MalE.
Translocation mediated by an ambivalent signal peptide via the Sec pathway is kinetically favored above Tat-dependent export.
To analyze the pathway specificity of the mutated TorA* signal peptide in more detail, export of the TorA*-MalE precursor was monitored by pulse-chase experiments with the strains GSJ100 and GSJ101 in the presence or absence of 3 mM sodium azide, respectively. As shown in Fig. 5A, lanes 5 to 8, azide significantly slows down processing of TorA*-MalE in GSJ100 compared to the situation without the inhibitor (Fig. 5A, lanes 1 to 4). Since azide is known to inhibit the SecA translocation ATPase (31), these results corroborate our notion that TorA*-MalE export occurs via the Sec pathway.
Surprisingly, a significant difference was observed when the processing kinetics of TorA*-MalE in the presence of azide were compared between the tat wild-type strain GSJ100 and the GSJ101 tat mutant derivative. In the absence of azide, the processing kinetics of TorA*-MalE are almost identical in GSJ100 (Fig. 5A, lanes 1 to 4) and GSJ101 (Fig. 5A, lanes 9 to 12). However, whereas nearly all of the precursor had been converted to the exported mature form in GSJ100 in the presence of azide after 30 min (Fig. 5A, lane 8), no mature form was detected under the same conditions for the tat mutant GSJ101 at this time point (Fig. 5A, lane 16). Instead, part of the precursor was degraded to a nonexported degradation product. An identical inhibition of pre-OmpA processing by azide was observed for both strains (data not shown). These results strongly suggest that the portion of the TorA*-MalE precursor which is still present at later time points (i.e., 10 and 30 min after the chase) for the GSJ101 tat mutant strain in the presence of azide is exported in the tat wild-type strain GSJ100. Since the only difference between the two strains is the presence or absence of a functional Tat system, the azide-insensitive export of this fraction of the TorA*-MalE precursor for GSJ100 must have occurred via the Tat pathway.
From our results we conclude that the TorA* signal peptide, which lacks the positively charged amino acid residues in the c-region, can in principle mediate protein translocation via both the Sec and the Tat pathways. Furthermore, in the presence of both pathways (i.e., in the absence of azide), the use of the Sec pathway is strongly favored above Tat-dependent export. Only in the case of a compromised Sec pathway is the Tat system used as an alternative.
The TorA-MalE hybrid precursor is an ideal reporter for the genetic analysis of Tat-dependent protein translocation.
Since we have found that export of the TorA-MalE precursor is strictly Tat dependent and since this export is easily detectable on MacConkey maltose indicator plates, we wanted to test whether these properties could be used as a genetic system to isolate mutations in the genes encoding known components of the Tat export apparatus. Accordingly, strain GSJ100 harboring pMTorA-MalE was subjected to transposon mutagenesis using the λTnPhoA-1 system (17). The mutagenized cells were plated on MacConkey maltose agar plates and inspected for the presence of pale colonies after overnight incubation. In fact, in a first experiment (without further optimization), pale mutant colonies appeared with a frequency of approximately 10−3 to 10−4. In order to test whether in some of the mutant colonies the transposon had inserted in one of the known tat genes, 11 randomly picked pale colonies were transformed with the plasmid pTatABC encoding the TatABC proteins, and the resulting transformants were tested again on MacConkey maltose agar plates. Six of the 11 isolates remained pale after transformation with pTatABC. Further characterization indicated that in these mutants the transposon had inserted in one of the genes involved in maltose uptake and metabolism (data not shown). In contrast, 5 of the 11 isolates became red again after transformation with pTatABC, strongly suggesting that in these mutants the transposon had inserted in either tatA, tatB, or tatC. To test this assumption, the transposon insertion site was rescued from the chromosome of one of the five mutant isolates. DNA sequencing showed that in fact, in the chosen mutant colony the transposon had inserted into the tatA gene at a position corresponding to amino acid residue 18 of the TatA protein.
These results clearly demonstrate that the TorA-MalE protein, besides being an excellent Tat-dependent model precursor protein, could be a powerful tool for the genetic analysis of Tat-dependent protein translocation in further experiments.
DISCUSSION
In the present work, we have shown that the replacement of the authentic signal peptide of the MalE precursor by the TorA signal peptide redirects this normally SecB-dependent protein selectively and exclusively into the Tat pathway. This result shows that the TorA signal peptide (i) is sufficient for directing MalE to the Tat translocase and (ii) does not allow translocation of MalE via the SecB/SecA pathway.
The main feature in the TorA signal peptide that is responsible for Tat pathway specificity (i.e., for avoiding channeling into the Sec pathway) seems to be the presence of two positively charged amino acid residues in the c-region. A spontaneous mutation, resulting in the replacement of the two arginine residues in the c-region (not to be confused with the two arginine residues of the twin-arginine motif in the n-region) by one leucine residue, allowed the mutant precursor (TorA*-MalE) to efficiently use the Sec pathway. These results strongly corroborate previous findings of Bogsch et al. (4) in the thylakoidal system. Using a ΔpH (Tat) pathway-specific 23K-PC hybrid precursor protein, these authors demonstrated that a positively charged lysine residue in the c-region of the 23K transfer signal is responsible for avoiding efficient interactions with the Sec-dependent import pathway. Based on these findings, it was proposed that, at least within the context of the 23K transfer peptide, the positively charged amino acid residue in the c-region constitutes a so-called Sec avoidance signal. For the bacterial system, Cristobal et al. (9) have found that removal of the positively charged amino acid residues in the c-region of a hybrid precursor protein consisting of the periplasmic P2 domain of leader peptidase fused to a TorA mutant signal peptide [TorA(10:10)], in which the central part of the h-region was replaced by a significantly more hydrophobic artificial h-region, resulted in an increased efficiency of Sec-dependent export of this model precursor. Although at first glance, the results of our study and the work of Cristobal et al. (9) seem to be very similar, an important difference exists. Since the TorA(10:10)-P2 precursor used by Cristobal et al. (9) was exported exclusively by the Sec pathway (but not the Tat pathway) irrespective of the presence or absence of the positively charged amino acid resides in the c-region, these results clearly showed that basic residues in the c-region negatively influence the efficiency of a Sec signal peptide. In contrast, the h-region of the TorA signal peptides used in our study remained unaltered, and, unlike the situation with the TorA(10:10)-P2 precursor, the removal of the arginine residues in the c-region resulted in the conversion of an exclusively Tat-dependent precursor into a Sec precursor protein. Our finding that TorA*-MalE is efficiently exported in a Sec-dependent manner clearly demonstrates that the hydrophobicity of the h-region of the TorA signal peptide is sufficiently high to allow productive interactions with the Sec machinery, and, therefore, the Sec-avoidance function of the two arginine residues in the c-region is absolutely required to prevent mistargeting into the Sec pathway, at least in the context of the TorA-MalE model precursor. In fact, the h-region of the TorA signal peptide, as defined by the criteria of Cristobal et al. (9), is only slightly less hydrophobic than the h-region of the authentic pre-MalE signal peptide (data not shown) (24). During the preparation of this report, a paper was published by Ize et al. (20) which showed that replacing the two arginine residues in the c-region of the TorA signal peptide by asparagine and glutamine allowed Sec-dependent export of two passenger proteins (colicin V [ColV] and green fluorescence protein [GFP]), whereas export of both proteins mediated by an unaltered signal peptide was exclusively Tat dependent. These results are in full agreement with our findings using the MalE reporter protein, both underscoring the importance of the Sec avoidance motif for maintenance of Tat pathway specificity.
Despite the lack of the Sec avoidance signal in the c-region, the TorA*-MalE precursor was still able to use the Tat pathway for membrane translocation. Inhibition of the Sec pathway by sodium azide resulted in a severe block of TorA*-MalE translocation for the tat deletion strain GSJ101, and no exported mature form was present 30 min after the chase. In contrast, the same precursor was converted almost completely into mature form after 30 min for the tat wild-type strain GSJ100, although the kinetics of processing was significantly slower in the presence of azide than in its absence. In the absence of the SecA inhibitor, rapid export of TorA*-MalE was observed for both strains. These results show that the use of the Sec pathway by a Tat signal peptide lacking the Sec avoidance signal is kinetically favored above the use of the Tat system and corroborate the findings of Ize et al. (20) showing that ColV and GFP fused to a TorA signal peptide lacking the Sec avoidance motif can be exported via the Tat pathway.
Our findings that TorA*-MalE, similar to the authentic pre-MalE precursor, is rapidly exported in the presence but not in the absence of SecB clearly show that the TorA* signal peptide allows efficient binding of SecB to the mature portion of the MalE protein and the subsequent targeting to the translocase by SecB-SecA interactions. Therefore, the TorA* mutant signal peptide must possess properties resembling those of the authentic pre-MalE signal peptide with respect to the kinetic partitioning of the corresponding precursor proteins between the binding to SecB and folding of the mature MalE into its stable tertiary structure. As mentioned before, according to the kinetic partitioning model an antifolding activity of the signal peptide is thought to favor SecB binding over folding of the mature part into a stable tertiary structure (19). However, it must be emphasized that the kinetic partitioning model has not remained uncontradicted (14), and therefore, alternative mechanisms might be responsible for SecB selectivity. Interestingly, the Tat-dependent export of the TorA-MalE precursor was found to be significantly slower for a SecB wild-type strain than for a SecB deficient mutant, indicating that this precursor also is able to interact with SecB. This result strongly suggests that the Sec avoidance function of the positively charged amino acid residues in the c-region of the TorA signal peptide is not the prevention of binding of the attached mature protein to SecB. Instead, other Sec avoidance events that prevent mistargeting of Tat precursor proteins into the Sec pathway, such as an interference of the positively charged amino acids in the vicinity of the signal peptidase cleavage site with proper channeling of the signal peptide into the SecYEG translocase (16), must be responsible for the maintenance of pathway specificity in the posttranslational modes of protein translocation. Recent results of Kebir and Kendall (24) showed that, surprisingly, the TorA signal peptide was able to interact with the SecA protein in vitro and to stimulate its ATPase activity, although to a relatively low extent. In line with these findings, the Sec avoidance step at the translocase level might occur after binding of the TorA signal peptide to SecA and, most likely, might be due to an unproductive interaction of the TorA signal peptide with the SecYEG components of the translocase.
The availability of various reporter proteins that confer an export-dependent phenotype has been of great value in genetic analyses of the Sec pathway (37). Only recently, suitable reporter proteins have been described for the Tat system (11, 21). The reporter system established by Ize et al. (21) is based on the bactericidal effect of ColV, which is exerted only when the colicin gains access to the inner membrane from the periplasmic side. Tat-dependent export of the ColV reporter results in the killing of the respective cells. Using a GFP-ssrA fusion as a reporter, DeLisa et al. (11) showed that export of the fusion by the Tat system rescues the reporter from degradation in the cytoplasm, and as a consequence, an increase in cellular fluorescence can be detected by fluorescence-activated cell sorter analysis. Due to the fact that the TorA-MalE hybrid precursor is specifically and exclusively exported via the Tat pathway without affecting the viability of the cells, and furthermore, since the presence or absence of MalE in the periplasm is easily detectable on MacConkey plates supplemented with maltose or on minimal medium containing maltose as the sole carbon source, the MalE protein possesses all the desirable properties that one would expect from a convenient reporter protein for the genetic analysis of Tat-dependent protein translocation. The feasibility of this system was tested in a transposon mutagenesis study. Without any further optimizations, transposon insertions in known tat genes could easily be isolated, clearly demonstrating the high potential of MalE as a reporter protein for the Tat pathway.
Acknowledgments
We thank M. Bagdasarian, W. Boos, C. Kumamoto, J. W. Lengeler, C. Manoil, M. Manson, T. Palmer, K.-L. Schimz, H. Schwarz, and Y.-D. Stierhoff for their generous gifts of bacterial strains, plasmids, phages, and antibodies. We thank A. Frerix for help with transposon mutagenesis, R. Halbach for excellent technical assistance, and H. Sahm for his continuous support.
This work was supported by grants Sp503/2-1 and Sp503/2-2 of the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Berks, B. C. 1996. A common export pathway for proteins binding complex redox cofactors? Mol. Microbiol. 22:393-404. [DOI] [PubMed] [Google Scholar]
- 2.Berks, B. C., F. Sargent, and T. Palmer. 2000. The Tat protein export pathway. Mol. Microbiol. 32:260-274. [DOI] [PubMed] [Google Scholar]
- 3.Blaudeck, N., G. A. Sprenger, R. Freudl, and T. Wiegert. 2001. Specificity of signal peptide recognition in Tat-dependent bacterial protein translocation. J. Bacteriol. 183:604-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogsch, E., S. Brink, and C. Robinson. 1997. Pathway specificity for a ΔpH-dependent precursor thylakoid lumen protein is governed by a ‘Sec-avoidance’ motif in the transfer peptide and a ‘Sec-incompatible’ mature protein. EMBO J. 16:3851-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brass, J. M., and M. D. Manson. 1984. Reconstitution of maltose chemotaxis in Escherichia coli by addition of maltose-binding protein to calcium-treated cells of maltose regulon mutants. J. Bacteriol. 157:881-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brass, J. M., M. D. Manson, and T. J. Larson. 1984. Transposon Tn10-dependent expression of the lamB gene in Escherichia coli. J. Bacteriol. 159:93-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casadaban, M. J. 1976. Transposition and fusion of the lac operon to selected promoters in Escherichia coli using bacteriophage λ and Mu. J. Mol. Biol. 104:541-555. [DOI] [PubMed] [Google Scholar]
- 8.Collier, D. N., V. A. Bankaitis, J. B. Weiss, and J. P. Bassford, Jr. 1988. The antifolding activity of SecB promotes the export of the E. coli maltose-binding protein. Cell 53:273-283. [DOI] [PubMed] [Google Scholar]
- 9.Cristobal, S., J.-W. de Gier, H. Nielsen, and G. von Heijne. 1999. Competition between Sec- and TAT-dependent protein translocation in Escherichia coli. EMBO J. 18:2982-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dassa, E., and P. Lambert. 1997. Activity of protein MalE (maltose-binding protein) fused to cytoplasmic and periplasmic regions of an Escherichia coli inner membrane protein. Res. Microbiol. 148:389-395. [DOI] [PubMed] [Google Scholar]
- 11.DeLisa, M. P., P. Samuelson, T. Palmer, and G. Georgiou. 2002. Genetic analysis of the twin arginine translocator secretion pathway in bacteria. J. Biol. Chem. 277:29825-29831. [DOI] [PubMed] [Google Scholar]
- 12.Driessen, A. J. M., P. Fekkes, and J. van der Wolk. 1998. The Sec system. Curr. Opin. Microbiol. 1:216-222. [DOI] [PubMed] [Google Scholar]
- 13.Duplay, P., H. Bedouelle, A. Fowler, I. Zabin, W. Saurin, and M. Hofnung. 1984. Sequences of the malE gene and of its product, the maltose-binding protein of Escherichia coli K12. J. Biol. Chem. 259:10606-10613. [PubMed] [Google Scholar]
- 14.Fekkes, P., and A. M. M. Driessen. 1999. Protein targeting to the bacterial cytoplasmic membrane. Microbiol. Mol. Biol. Rev. 63:161-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fürste, J. P., W. Pansegrau, R. Frank, H. Blöcker, P. Scholz, M. Bagdasarian, and E. Lanka. 1986. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene 48:119-131. [DOI] [PubMed] [Google Scholar]
- 16.Geller, B., H.-Y. Zhu, S. Cheng, A. Kuhn, and R. E. Dalbey. 1993. Charged residues render pro-OmpA potential dependent for initiation of membrane translocation. J. Biol. Chem. 268:9442-9447. [PubMed] [Google Scholar]
- 17.Gutierrez, C., J. Barondess, C. Manoil, and J. Beckwith. 1987. The use of transposon TnphoA to detect genes for cell envelope proteins subject to a common regulatory stimulus. Analysis of osmotically regulated genes in Escherichia coli. J. Mol. Biol. 195:289-297. [DOI] [PubMed] [Google Scholar]
- 18.Halbig, D., T. Wiegert, N. Blaudeck, R. Freudl, and G. A. Sprenger. 1999. The efficient export of NADP-containing glucose-fructose oxidoreductase to the periplasm of Zymomonas mobilis depends both on an intact twin-arginine motif in the signal peptide and on the generation of a structural export signal induced by cofactor binding. Eur. J. Biochem. 263:543-551. [DOI] [PubMed] [Google Scholar]
- 19.Hardy, S. J. S., and L. L. Randall. 1991. A kinetic partitioning model of selective binding of nonnative proteins by the bacterial chaperone SecB. Science 251:439-443. [DOI] [PubMed] [Google Scholar]
- 20.Ize, B., F. Gérard, and L.-F. Wu. 2002. In vivo assessment of the Tat signal peptide specificity in Escherichia coli. Arch. Microbiol. 178:548-553. [DOI] [PubMed] [Google Scholar]
- 21.Ize, B., F. Gérard, M. Zhang, A. Chanal, R. Volhoux, T. Palmer, A. Filloux, and L.-F. Wu. 2002. In vivo dissection of the Tat translocation pathway in Escherichia coli. J. Mol. Biol. 317:327-335. [DOI] [PubMed] [Google Scholar]
- 22.Jensch, T., and B. Fricke. 1997. Localization of alanyl aminopeptidase and leucyl aminopeptidase in cells of Pseudomonas aeruginosa by application of different methods for periplasm release. J. Basic Microbiol. 37:115-128. [DOI] [PubMed] [Google Scholar]
- 23.Josefsson, L.-G., and L. L. Randall. 1981. Different exported proteins in E. coli show differences in the temporal mode of processing in vivo. Cell 25:151-157. [DOI] [PubMed] [Google Scholar]
- 24.Kebir, M. O., and D. A. Kendall. 2002. SecA specificity for different signal peptides. Biochemistry 41:5573-5580. [DOI] [PubMed] [Google Scholar]
- 25.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 26.Kumamoto, C. A. 1991. Molecular chaperones and protein translocation across the Escherichia coli inner membrane. Mol. Microbiol. 5:19-22. [DOI] [PubMed] [Google Scholar]
- 27.Liu, G., T. B. Topping, and L. L. Randall. 1989. Physiological role during export for the retardation of folding by the leader peptide of maltose-binding protein. Proc. Natl. Acad. Sci. USA 86:9213-9217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 29.Mori, H., and K. Cline. 2001. Post-translational protein translocation into thylakoids by the Sec- and delta-pH-dependent pathways. Biochim. Biophys. Acta 1541:80-90. [DOI] [PubMed] [Google Scholar]
- 30.Müller, M., H.-G. Koch, K. Beck, and U. Schäfer. 2001. Protein traffic in bacteria: multiple routes from the ribosome to and across the membrane. Progr. Nucleic Acid Res. Mol. Biol. 66:107-157. [DOI] [PubMed] [Google Scholar]
- 31.Oliver, D. B., R. J. Cabelli, K. M. Dolan, and G. P. Jarosik. 1990. Azide-resistant mutants of Escherichia coli alter the SecA protein, an azide-sensitive component of the protein export machinery. Proc. Natl. Acad. Sci. USA 87:8227-8231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park, S., G. Liu, T. B. Topping, W. H. Cover, and L. L. Randall. 1988. Modulation of folding pathway of exported proteins by the leader sequence. Science 239:1033-1035. [DOI] [PubMed] [Google Scholar]
- 33.Robinson, C., and A. Bolhuis. 2001. Protein targeting by the twin-arginine translocation pathway. Nat. Rev. Mol. Cell Biol. 2:350-358. [DOI] [PubMed] [Google Scholar]
- 34.Rodrigue, A., A. Chanal, K. Beck, M. Müller, and L.-F. Wu. 1999. Co-translocation of a periplasmic enzyme complex by a hitchhiker mechanism through the bacterial Tat pathway. J. Biol. Chem. 274:13223-13228. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Santini, C.-L., B. Ize, A. Chanal, M. Müller, G. Giordano, and L.-F. Wu. 1998. A novel Sec-independent periplasmic protein translocation pathway in Escherichia coli. EMBO J. 17:101-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schatz, P. J., and J. Beckwith. 1990. Genetic analysis of protein export in Escherichia coli. Annu. Rev. Genet. 24:215-248. [DOI] [PubMed] [Google Scholar]
- 38.Sprenger, G. A., U. Schörken, G. Sprenger, and H. Sahm. 1995. Transaldolase B of Escherichia coli K-12: cloning of its gene, talB, and characterization of the enzyme from recombinant strains. J. Bacteriol. 177:5930-5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stanley, N. R., T. Palmer, and B. C. Berks. 2000. The twin arginine consensus motif of Tat signal peptides is involved in Sec-independent protein targeting in Escherichia coli. J. Biol. Chem. 275:11591-11596. [DOI] [PubMed] [Google Scholar]
- 40.Takeshita, S., M. Sato, M. Toba, W. Masahashi, and T. Hashimoto-Gotoh. 1987. High-copy-number and low-copy-number plasmid vectors for lacZα-complementation and chloramphenicol- or kanamycin-resistance selection. Gene 61:63-74. [DOI] [PubMed] [Google Scholar]
- 41.Tanaka, H., S. A. Lerner, and E. C. C. Lin. 1967. Replacement of a phosphoenolpyruvate-dependent phosphotransferase by a nicotinamide adenine dinucleotide-linked dehydrogenase for the utilization of mannitol. J. Bacteriol. 93:642-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wexler, M., F. Sargent, R. L. Jack, N. R. Stanley, E. G. Bogsch, C. Robinson, B. C. Berks, and T. Palmer. 2000. TatD is a cytoplasmic protein with DNase activity. No requirement for TatD family proteins in Sec-independent protein export. J. Biol. Chem. 275:16717-16722. [DOI] [PubMed] [Google Scholar]
- 43.Wu, L.-F., B. Ize, A. Chanal, Y. Quentin, and G. Fichant. 2000. Bacterial twin-arginine signal peptide-dependent protein translocation pathway: evolution and mechanism. J. Mol. Microbiol. Biotechnol. 2:179-189. [PubMed] [Google Scholar]