Abstract
UDP-GlcNAc acyltransferase (LpxA) catalyzes the first step of lipid A biosynthesis, the transfer of the R-3-hydroxyacyl chain from R-3-hydroxyacyl acyl carrier protein (ACP) to the glucosamine 3-OH group of UDP-GlcNAc. LpxA is essential for the growth of Escherichia coli and related Gram-negative bacteria. The crystal structure of the E. coli LpxA homotrimer, determined previously at 2.6 Å in the absence of substrates or inhibitors, revealed that LpxA contains an unusual, left-handed parallel β-helix fold. We now present the crystal structure at 1.8 Å resolution of E. coli LpxA in a complex with a pentadecapeptide, peptide 920. Three peptides, each of which adopts a β-hairpin conformation, are bound per LpxA trimer. The peptides are located at the interfaces of adjacent subunits in the vicinity of the three active sites. Each peptide interacts with residues from both adjacent subunits. Peptide 920 is a potent inhibitor of E. coli LpxA (Ki = 50 nM). It is competitive with respect to acyl-ACP but not UDP-GlcNAc. The compact β-turn structure of peptide 920 bound to LpxA may open previously uncharacterized approaches to the rational design of LpxA inhibitors with antibiotic activity.
Keywords: antibiotic, crystal structure, Escherichia coli, inhibitor, outer membrane
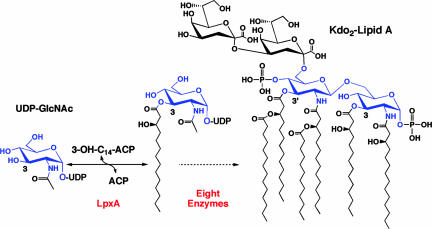
Lipopolysaccharide constitutes the outer monolayer of the outer membrane in Gram-negative bacteria (1–3). Lipopolysaccharide maintains the integrity of the outer membrane, which functions as a barrier to molecules >500 Da (4). Lipid A (endotoxin) is the hydrophobic moiety that anchors lipopolysaccharide into the outer membrane (1, 2). It is a potent activator of the human innate immune system via Toll-like receptor 4 (TLR4) (5–7). The minimal lipopolysaccharide required for growth usually consists of two 3-deoxy-d-manno-octulosonic acid (Kdo) residues attached to lipid A (Fig. 1; ref. 2). Because lipid A biosynthetic enzymes have no mammalian counterparts, they are attractive targets for the design of new antibiotics (10, 11).
Fig. 1.
Function of LpxA in lipid A biosynthesis. LpxA catalyzes the first step of lipid A biosynthesis. It transfers the R-3-hydroxyacyl moiety from R-3-hydroxyacyl-ACP to the 3-position of UDP-GlcNAc. E. coli LpxA is highly selective for 14 carbons (8). The glucosamine ring is colored blue, and the 3-position is indicated. Kdo2-lipid A is the minimal lipopolysaccharide that supports growth in most bacteria (9). The acyl chains at the 3 and 3′ positions of Kdo2-lipid A are derived from LpxA.
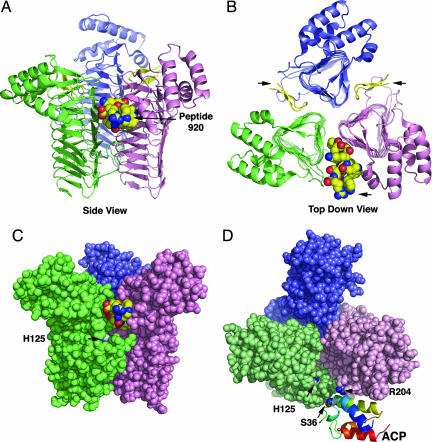
Kdo2-lipid A is synthesized by a conserved pathway consisting of nine enzymes (1, 2). LpxA catalyzes the first step (Fig. 1), the thermodynamically unfavorable 3-O-acylation of UDP-GlcNAc (Keq = 0.01) (12–14). The crystal structure of Escherichia coli LpxA, previously determined at 2.6 Å (15), revealed that the enzyme is a homotrimer. It adopts a distinctive, left-handed parallel β-helix fold (Fig. 2A and B), which is specified by the presence of 24 complete and six partial hexad repeats (15). Three contiguous hexad repeats (18-aa residues) fold into one coil of the β-helix (Fig. 2 A and B). Many other bacterial acyl and acetyl transferases later were shown to possess this fold (17–22).
Fig. 2.
Crystal structure of the LpxA–peptide 920 complex at 1.8 Å. (A) In this side view, the individual LpxA subunits of the homotrimer are colored pink, green, and blue. The LpxA N termini, located at the bottom, form the start of the β-helix domain of each subunit. Peptide 920 (yellow) is in a β-hairpin conformation with its N and C termini protruding. In the space-filling model, peptide 920 carbons are yellow, nitrogens blue, and oxygens red. (B) Top-down view of LpxA with bound peptide 920. The threefold symmetry and precise stacking of the 18-aa residues in each β-helix coil are visualized clearly. Arrows point to protruding peptide 920. (C) Side view of a space-filling model of LpxA, highlighting the relative positions of H125 and peptide 920, which are not in direct contact. (D) Top-down view of free LpxA, docked computationally to butyryl-ACP based on NMR constraints (35). The positions of H125, R204 (a proposed contact site between LpxA and ACP), and S36 of ACP are indicated. The location of the butyryl phosphopantetheine moiety, which is attached to S36, was not included in the published model (16).
Structures of LpxA have not been solved in the presence of substrates or inhibitors. Mutagenesis suggests that the active site is located in a large cleft between adjacent subunits (8, 23). This region contains several conserved basic residues (K76, H122, H125, H144, H160, and R204) (23). H125 (Fig. 2 C and D) may be the catalytic base (Fig. 1B), because, of the conserved residues in the cleft, only the H125A substitution abolishes LpxA activity completely (23). The structure of Helicobacter pylori LpxA, solved with a bound 1-S-octyl-β-d-thioglucoside molecule (24), is consistent with a catalytic role for H125, because N-δ1 of H121 (equivalent to E. coli H125) is in close proximity to the 1-S-octyl-β-d-thioglucoside 3-OH group.
Recently, a pentadecapeptide (termed peptide 920) with high affinity for LpxA was discovered by using phage display (25). When expressed in E. coli as a fusion protein with GST, bacterial growth was inhibited. Specificity for LpxA was inferred from the resistance of cells overexpressing LpxA to killing by peptide 920 (25). Inhibition of LpxA in vitro was not investigated. We now present the 1.8-Å crystal structure of E. coli LpxA with bound peptide 920, which adopts a compact β-turn conformation. The peptide occupies part of the proposed active site region inferred from mutagenesis studies (8, 23). Inhibition by peptide 920 is competitive with respect to R-3-hydroxymyristoyl-ACP (Ki = 50 nM) but not UDP-GlcNAc, suggesting that peptide 920 contacts LpxA in a region that overlaps or occludes the R-3-hydroxymyristoyl-ACP-binding site.
Results
Inhibition of LpxA by Peptide 920.
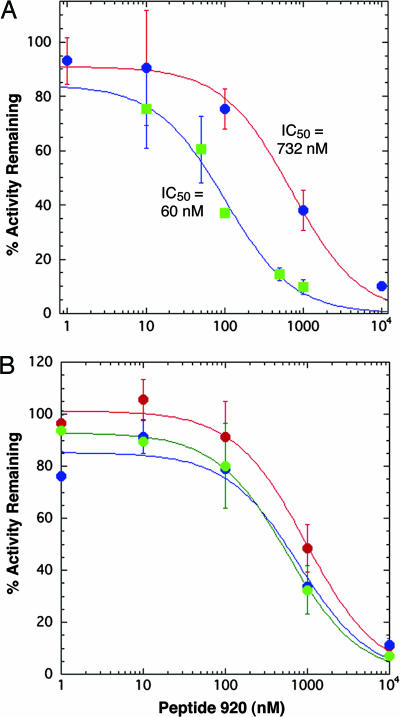
Peptide 920 (NH2-SSGWMLDPIAGKWSR-COOH) is a potent inhibitor of E. coli LpxA with an IC50 of 60 ± 9 nM when assayed with 1 μM UDP-GlcNAc and 1 μM R-3-hydroxymyristoyl-ACP (Fig. 3A). Competitive inhibitors often are substrate analogues, but peptide 920 displays no similarity to either ACP or UDP-GlcNAc. To test whether peptide 920 competes with R-3-hydroxymyristoyl-ACP, the UDP-GlcNAc concentration was held at 1 μM, and the R-3-hydroxymyristoyl-ACP was varied (1, 25, 50, or 100 μM). The IC50 of peptide 920 was determined under each condition. Increasing the acyl-ACP concentration from 1 to 100 μM shifted the IC50 from 60 ± 9 nM to 730 ± 107 nM (Fig. 3A), consistent with Eq. 3, which predicts a 12-fold shift. At 25 and 50 μM R-3-hydroxymyristoyl-ACP, the IC50 values were 390 ± 18 nM and 536 ± 87 nM, respectively. The average calculated Ki was 50 ± 13 nM with Km set at 5 μM for acyl-ACP.
Fig. 3.
Inhibition of LpxA activity by peptide 920. Peptide 920 at the indicated concentrations was preincubated in the assay mixture at 30°C for 3 min. The reaction was initiated by the addition of 1 nM LpxA. The data were fit to Eqs. 1 or 2 for IC50 determination. (A) The ≈12-fold shift in the IC50 indicates that peptide 920 is competing with acyl-ACP. The green squares show that the IC50 is 60 ± 9 nM when acyl-ACP is 1 μM. The blue circles show a shift in IC50 to 730 ± 107 nM when acyl-ACP is 100 μM. In both cases, UDP-GlcNAc was 1 μM. The initial rate that corresponds to 100% at 1 μM acyl-ACP is 2.8 ± 0.4 nmol·min−1mg−1. At 100 μM acyl-ACP, it is 6.6 ± 1.3 nmol·min−1mg−1. (B) Peptide 920 does not compete effectively with UDP-GlcNAc, because increasing UDP-GlcNAc from 1 μM (green) to 500 μM (blue), or even to 5 mM (red), does not change the IC50 significantly with acyl-ACP held at 50 μM. The initial rate that corresponds to 100% at 1 μM UDP-GlcNAc is 7.1 ± 0.8 nmol·min−1mg−1. At 500 μM UDP-GlcNAc, it is 4.4 × 102 ± 1.4 × 102 nmol·min−1mg−1. At 5 mM UDP-GlcNAc, it is 2.3 × 103 ± 4.9 × 102 nmol·min−1mg−1. Error bars show SDs of triplicates.
To test whether peptide 920 competes with UDP-GlcNAc (Km = 0.8 mM) (23), the R-3-hydroxymyristoyl-ACP was held at 50 μM, and the IC50 for peptide 920 was determined at 5 mM, 500 μM, or 1 μM UDP-GlcNAc. The IC50 was similar at all three concentrations (923 ± 230 nM, 760 ± 220 nM, and 700 ± 65 nM, respectively) (Fig. 3B). If peptide 920 were competing with UDP-GlcNAc, a 7-fold shift in the IC50 would be expected. The combined data suggest that peptide 920 contacts LpxA in a region that overlaps mainly with the R-3-hydroxymyristoyl-ACP substrate-binding site.
Crystal Structure of the Peptide 920–LpxA Complex.
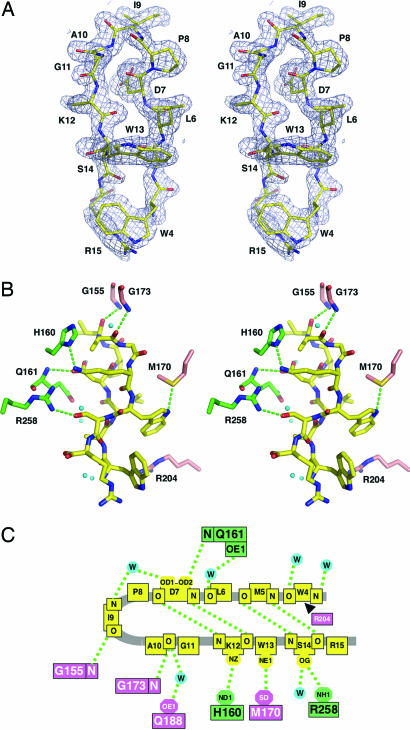
No crystal structures of LpxA with bound acyl-ACP or UDP-GlcNAc have been reported. To determine how peptide 920 inhibits LpxA, LpxA was crystallized in the presence of a 25-fold molar excess of peptide. Crystals of the complex diffracted to 1.8-Å resolution and were solved by molecular replacement by using the previously determined structure of free LpxA (PDB ID code 1LXA) (15) as the search model. The refinement converged to Rwork and Rfree values of 0.194 and 0.220, respectively (Table 1), with good geometry and low clash scores (26). All 262 residues of the enzyme with the exception of the N-terminal methionine side chain were visible in the electron density map. Peptide 920 was well resolved, except for the three residues (SSG) at its N-terminus and its M5 side chain (Fig. 4A). A truncated analogue of peptide 920, lacking the three N-terminal amino acid residues not seen in the crystal structure, inhibits LpxA with the same IC50 as the pentadecapeptide (data not shown).
Table 1.
Data collection and refinement statistics
| Measurement | Value |
|---|---|
| Source/detector | RU200/Raxis-IV |
| Space group | P213 |
| A = b= c, Å | 96.73 |
| Wavelength, Å | 1.5418 |
| Resolution range,* Å | 18–1.8 |
| Last shell (Å) | 1.86–1.80 |
| Unique reflections | 26,451 |
| Completeness, % (last shell) | 99.8 (100) |
| Average I/σ(I) (last shell) | 25.8 (3.9) |
| Redundancy (last shell) | 10.4 (10.3) |
| Rmerge,† % | 7.3 |
| Nonsolvent atoms | |
| Protein | 2,056 |
| Peptide | 100 |
| Solvent atoms | 410 |
| Phosphate ions | 5 |
| DMSO | 22 |
| Water | 297 |
| rmsd from ideality | |
| Bond lengths, Å | 0.0061 |
| Bond angles, ° | 1.56 |
| R value‡ (F > 2σF) | 19.4 (18.6) |
| R free‡ (F > 2σF) | 22.0 (21.9) |
| All-atom clash score | 6.42 |
*Resolution limit was defined as the highest resolution shell where the average I/σI was 2.
†Rmerge = ΣhklΣi| Ii(hkl) − 〈I(hkl)〉|/ΣhklΣI(hkl).
‡R = Σ|Fo − Fc|/ΣFo. Ten percent of reflections were used to calculate Rfree.
Fig. 4.
Noncovalent interactions between peptide 920 and LpxA. (A) Stereoview of the final 2 Fo − Fc electron density map contoured at 1 σ around peptide 920. The peptide is shown as a stick model with carbons in yellow. (B) Stereoview of important protein–peptide 920 interactions. The peptide 920 carbons are colored yellow, whereas the LpxA subunit carbons are either green or pink, according to the coloring scheme in Fig. 2. All of the N and O atoms are blue and red, respectively. Hydrogen bonds are green dashed lines. Water molecules are cyan. Hydrogen bonds involving water are not included for clarity. Only one of two possible conformations is shown for the R258 and M170 side chains. The peptide is rotated ≈90° clockwise along its vertical axis when compared with A. (C) Schematic drawing of key interactions between peptide 920 and LpxA. Hydrogen bonds are shown as green dashes. A van der Waals interaction is indicated by the black arrowhead. Water molecules are cyan balls.
Although the crystal structure of LpxA-peptide 920 contains a single enzyme–peptide complex in the asymmetric unit, three asymmetric units associate around a crystallographic 3-fold symmetry axis to form the LpxA homotrimer with three bound peptides (Fig. 2 A and B). This result is consistent with the expectation that there are three active sites per functional trimer (23).
The left-handed parallel β-helix conformation present in free LpxA (15) is not perturbed in the LpxA–peptide 920 complex (Fig. 2 A and B). Similarly, comparison of peptide-bound LpxA with unbound LpxA shows that the presence of the inhibitory peptide does not alter the structure of the proposed active cleft of LpxA. The average rmsd between the peptide-bound and unbound LpxA is just 0.34 Å for all Cα pairs of the backbone. Similarly, the side chains of the enzyme that interact with the peptide (Fig. 4 B and C) do not move appreciably from their unbound conformations with rmsds for all atoms in each side-chain pair of only 0.1 Å. LpxA thus appears to be a rigid structure, and any deformations that may arise during peptide binding likely involve adaptation of the peptide conformation rather than LpxA.
Conformation of Peptide 920 and Noncovalent Interactions with LpxA.
LpxA-bound peptide 920 is folded into a β-hairpin conformation (Fig. 4A). The hairpin is inserted loop-first, leaving the N- and C-termini of the peptide exposed to solvent (Figs. 2 A and B and 4B). Peptide 920 was derived from a library in which the P8 residue was fixed (25). In the structure of the complex, P8 (Fig. 4 A and B) adopts a trans-configuration at the start of the β-turn. However, P8 does not appear to stabilize the hairpin bend of the peptide, because when this residue is changed to A, the IC50 for inhibition is unchanged (data not shown). This result is consistent with the fact that P8 of peptide 920 does not have any significant interactions with LpxA (Fig. 4 B and C). At the end of a very tight β-turn, G11 of peptide 920 may provide conformational flexibility (Fig. 4 A and B). Although G11 does not make any significant contacts with LpxA, this residue may be essential for β-hairpin formation.
Each peptide makes extensive contacts with the enzyme, burying ≈1,400 Å2 of surface area from the solvent. Electron density for most of the peptide (Fig. 4A) and all of the surrounding LpxA residues (data not shown) was excellent. The peptide does not disrupt any of the contacts between the adjacent subunits of the trimeric enzyme but rather supplements them with bridging peptide–enzyme contacts (Fig. 4B). Three direct hydrogen bonds are formed between peptide 920 and each of the adjacent subunits (Fig. 4 B and C). In addition, a water-mediated hydrogen bond is formed with each subunit (Fig. 4C). At least six water molecules are hydrogen-bonded directly to the peptide (Fig. 4 B and C). Residues involved in direct hydrogen bonding with the peptide are H160, Q161, and R258 from the green chain and G173, M170, and G155 from the pink chain (Fig. 4 B and C). Residues that make water-mediated interactions with the peptide are Q161 from green chain and Q188 from the pink chain (Fig. 4C). Four additional hydrogen bonds within the peptide and a possible van der Waals attraction with R204 are shown in Fig. 4C.
Peptide 920 Binding in Relation to Residues Within the LpxA Active Site.
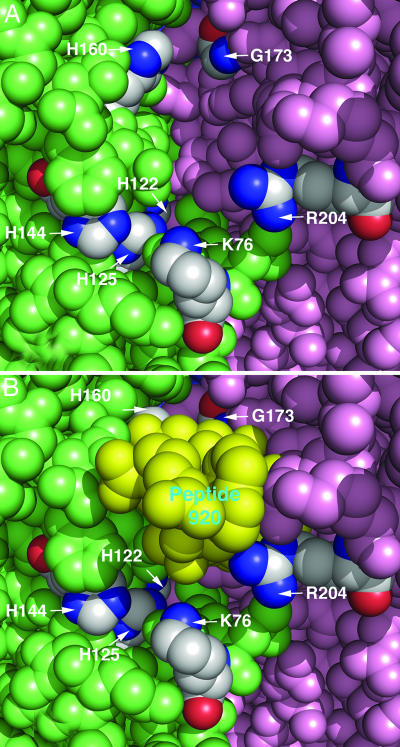
Mutagenesis experiments have implicated specific LpxA residues as being critical for substrate binding (K76, H122, H144, and H160), acyl chain-length recognition (G173), and catalysis (H125) (refs. 8 and 23; Fig. 5A). R204 (Fig. 5A), which is likewise essential for catalytic activity (23), may be involved in docking ACP (Fig. 2D). Three of these residues (H160, G173, and R204) interact directly with peptide 920 (Figs. 4 B and C and 5B), whereas the others are partially occluded (Fig. 5B). H125, the proposed catalytic base (23), exposes only a single imino nitrogen to the active site cleft, and access to this atom is partially blocked by the bound peptide (Fig. 5B). H160 hydrogen bonds to K12 of peptide 920 (Fig. 4 B and C) and is rendered inaccessible upon inhibitor binding (Fig. 5B). K76 and R204 are located at the mouth of the proposed active site cleft (Fig. 5 A and B). R204 is in position to form a van der Waals contact with the peptide (Fig. 4 B and C). Interestingly, the A10 carbonyl group of peptide 920 forms a hydrogen bond with the backbone nitrogen of G173, the residue that is involved in determining acyl chain-length selectivity (8).
Fig. 5.
Location of peptide 920 and conserved residues implicated in catalysis. (A) View into one of the three peptide-binding cavities of the LpxA trimer in the absence of peptide 920. LpxA subunits are colored pink, green, or blue (not visible) as in Fig. 2. Key residues implicated in catalysis or substrate binding by mutagenesis are colored according to element with carbons in gray. (B) Same view as above but with peptide 920 present in yellow. Conserved LpxA residues are labeled in white. There is some space between peptide 920 and the conserved residues H125, H144, H122, and K76. Because of the hydrogen bonding of peptide 920 with G173 and H160, these residues are largely hidden from view. In the complete model, a few water molecules are located in the space beneath peptide 920 (data not shown).
Discussion
Infections caused by Gram-negative bacteria remain a serious health concern (27, 28). Many pathogens, such as strains of Pseudomonas aeruginosa in cystic fibrosis patients (29, 30), now are resistant to commercially available antibiotics. Therefore it is necessary to develop new antibacterial agents that target previously unexploited systems, such as the enzymes that assemble the lipid A component of lipopolysaccharide (2, 8, 9, 31–35). The cytosolic acyltransferase LpxA catalyzes the first step of the lipid A pathway (Fig. 1) and is essential for growth in almost all Gram-negative pathogens (2). E. coli LpxA, which contains a glycine residue at position 173, is highly selective for a 14-carbon R-3-hydroxyacyl chain activated on ACP, whereas P. aeruginosa LpxA, which contains a methionine residue at the equivalent position, is selective for 10 carbons (8, 36). Indeed, the G173M mutation converts E. coli LpxA into a C10 enzyme, whereas the reciprocal M169G substitution converts P. aeruginosa LpxA to a C14 acyltransferase (8). LpxA is therefore also an interesting system for studying lipid protein recognition at the atomic level. The previous 2.6-Å crystal structure of E. coli LpxA revealed the first example of a left-handed parallel β-helical secondary structure (Fig. 2 A and B; ref. 15), but it did not include substrates, inhibitors, or solvent molecules. The more recent Helicobacter pylori LpxA structure (24) was similar in its overall fold to E. coli LpxA.
We now have cocrystallized E. coli LpxA and the antibacterial inhibitor, peptide 920, and have solved its structure to 1.8 Å resolution. Our structure includes 297 water, 22 DMSO, and 5 phosphate molecules per monomer (data not shown) that could not be seen in the previous analysis of E. coli LpxA at 2.6 Å. The overall LpxA/peptide 920 complex is remarkably similar to free LpxA (15). Peptide 920 binds to LpxA in the general region of the proposed active site cleft (Fig. 5), which was anticipated by mutagenesis of conserved residues (23). Because inhibition by peptide 920 is competitive with respect to acyl-ACP, it is likely that peptide 920 overlaps or occludes at least a portion of the R-3-hydroxymyristoyl-ACP binding region of LpxA, blocking productive interaction and catalysis. Some of the conserved residues implicated in substrate binding (H160, R204) (16, 23) and acyl chain-length selectivity (G173) (8) interact directly with peptide 920 (Figs. 4 and 5). Interestingly, the residual activity of the H160A LpxA variant is two orders of magnitude less sensitive to inhibition by peptide 920 than wild-type LpxA (data not shown), consistent with the fact that H160 hydrogen bonds K12 of the peptide (Fig. 4 B and C). The fact that peptide 920 interacts directly with G173, the acyl chain ruler (Fig. 4 B and C), further supports the idea that peptide 920 binds to same region of the LpxA active site that engages the R-3-hydroxymyristoyl chain.
No crystal structures of LpxA with bound acyl-ACP or UDP-GlcNAc have been reported. Modeling studies, by using the coordinates from the butyryl-ACP crystal structure (37) in conjunction with residual dipolar couplings and chemical shift perturbations of 15N-labeled ACP in the presence of E. coli LpxA (36), suggest that three acyl-ACPs can bind to the LpxA homotrimer. Each acyl-ACP is thought to dock in the vicinity of the basic cleft located between adjacent LpxA subunits (36), near R204 (Fig. 2D). This residue, which is essential for activity (23), contacts W13 of peptide 920 (Fig. 4 B and C). It is not far removed from H125 (the catalytic base) (Fig. 5A) and other conserved active-site residues implicated in LpxA function by site-directed mutagenesis (23). With the exception of R204, there is apparently no overlap between the LpxA-binding sites for peptide 920 and ACP (Figs. 2D, 4 B and C, and 5B; ref. 16). However, the location of the acyl-phosphopantetheine moiety, which is covalently linked to S36 of ACP, was not included in the docking model (Fig. 2D; ref. 16). The docking model therefore supports the idea that peptide 920 and, specifically, the acyl-phosphopantetheine portion of R-3-hydroxymyristoyl-ACP might be competing for the same or overlapping sites on LpxA.
Acyl-ACPs participate in diverse biochemical pathways, including fatty acid (38), phospholipid (39), polyketide (40, 41), and nonribosomal peptide biosynthesis (42). Structures of long-chain acyl-ACPs have resisted structure determination, and many of the enzymes that use these acyl-ACPs as substrates are either membrane-bound or large modular proteins with multiple active sites. Despite many unsuccessful attempts in the past, a crystal structure of LpxA with bound R-3-hydroxymyristoyl-ACP remains an important objective, because it would provide significant insights into the LpxA mechanism and the acyl chain-length selection process. Furthermore, crystallization of LpxA with the product UDP-3-O-R-3-hydroxymyristoyl-GlcNAc has not been investigated. Because LpxA efficiently catalyzes the reverse reaction (Fig. 1; refs. 14 and 23), the crystal structure of LpxA with bound UDP-3-O-R-3-hydroxymyristoyl-GlcNAc would be especially informative.
Based on genetic evidence (43) and the toxicity associated with peptide 920 expression in living cells (25), E. coli LpxA is a validated antibiotic target. Peptide inhibitors are often poor drug candidates because they do not cross membranes and are subject to proteolysis. The peptide 920–LpxA complex nevertheless provides an interesting starting point for further inhibitor development. It should be possible to design more potent, cyclic analogues of peptide 920 (44) or modified polyketides (40, 42) that would not be susceptible to proteolysis and could cross membranes.
Materials and Methods
Sample Preparation and Crystallization.
E. coli LpxA was overexpressed as described in refs. 8 and 23. The purification scheme consists of Green19 agarose (Sigma) affinity chromatography (pH 7.4), followed by Source Q ion-exchange chromatography (pH 8) and Superdex 200 gel filtration chromatography in 10 mM potassium phosphate buffer (pH 7) containing 250 mM NaCl (8, 23). Purity was assessed by using SDS/PAGE, LpxA activity assays (8, 23), and electrospray ionization mass spectrometry (45). Peptide 920 (NH2-SSGWMLDPIAGKWSR-COOH), a pentadecapeptide (25), was prepared at the University of North Carolina Peptide Synthesis Facility (Chapel Hill). Before crystallization, peptide 920 was added to a concentrated LpxA solution (20 mg/ml) at a 25-fold molar excess (12.5 mM) and incubated overnight at 4°C. Crystals of the LpxA–peptide 920 complex were grown at 18°C by using the hanging drop vapor diffusion method (46). Droplets contained 2 μl of the LpxA–peptide 920 mixture and 2 μl of 0.8–1.8 M phosphate buffer (pH 6.3–6.9) obtained by mixing concentrated stocks of NaH2PO4 and K2HPO4, and 30–35% DMSO. Crystalline cubes appeared after 48 h and grew to ≈0.6 mm after 2 weeks.
Data Collection.
Crystals of the LpxA–peptide 920 complex were cryoprotected in 1.4 M Na/K phosphate, pH 8.2/45% DMSO and then were flash cooled in liquid nitrogen. Diffraction data were collected on an R-Axis IV image plate detector. Typical crystals diffract to 1.8 Å and belong to space group P213 (a = b = c = 96.74 Å). The asymmetric unit contains one LpxA monomer in complex with one peptide 920, and the biologically functional trimer lies on the crystallographic 3-fold axis.
Structure Determination and Refinement.
Diffraction data were reduced and scaled by using hkl2000 (47). Initial phases were obtained by molecular replacement with the program molrep (48). The search model consisted of a single LpxA monomer from the earlier structure determination at 2.6 Å (PDB ID code 1LXA) (15). Model building was performed iteratively in o (49) with rounds of simulated annealing, energy minimization, and B factor refinement in CNS (50). The programs reduce and probe (51) were used to highlight steric clashes in the model for correction. Identification of nonwater solvent molecules derived from the crystallization medium (phosphate ions or DMSO) was aided by the use of anomalous difference maps. The quality of the final model was assessed by using molprobity and procheck (26, 52). The figures were drawn by using pymol (DeLano Scientific, San Carlos, CA). Data collection and refinement statistics are presented in Table 1. Atomic coordinates for the complex have been deposited with the Protein Data Bank (ID code 2AQ9).
Inhibition of LpxA Activity by Peptide 920.
LpxA assay conditions were similar to those published in ref. 14 with minor modifications. The LpxA reaction monitors the conversion of [α-32P]UDP-GlcNAc to [α-32P]UDP-3-O-(R-3-hydroxymyristoyl)-GlcNAc. The assay components are 40 mM Hepes (pH 8), 1 mg/ml BSA, 1–100 μM R-3-hydroxymyristoyl-ACP, and 1–5,000 μM [α-32P]UDP-GlcNAc (2 × 106 cpm/nmol) (53), as indicated. Peptide 920 was dissolved in DMSO and preincubated with the reaction mixture at 30°C for 3 min in the absence of enzyme at concentrations ranging from 1 nM to 10,000 nM. For all in vitro assays, the final concentration of DMSO was adjusted to 10% to match the peptide solvent.
The reactions were initiated by the addition of LpxA to 1 nM and incubated at 30°C for various times up to 10 min. Reactions were terminated by spotting 1-μl portions onto a silica TLC plate (53). The spots were air dried for 10 min before developing in chloroform/methanol/water/acetic acid (25:15:4:2, vol/vol). The plates were exposed to PhosphorImager screens overnight, and the data were evaluated with Molecular Dynamics PhosphorImager equipped with imagequant software.
Inhibition of LpxA by peptide 920 was analyzed by plotting the initial velocities as a function of the inhibitor concentration. To determine the IC50 at 30°C, the data were fit to the following equations,
or
where I is the inhibitor concentration, vi represents the initial rate at given concentrations of inhibitor, and vc represents the initial velocity of the control reaction without inhibitor.
To determine whether peptide 920 was competing with acyl-ACP, R-3-hydroxymyristoyl-ACP (Km set to 5 μM) was varied from 1 to 100 μM, whereas UDP-GlcNAc was held constant at 1 μM. Similarly, to test the competition with the other substrate, UDP-GlcNAc (Km = 0.8 mM) (23) was varied from 1 μM to 5 mM, whereas R-3-hydroxymyristoyl-ACP was held constant at 50 μM. The higher concentration of R-3-hydroxymyristoyl-ACP was needed to ensure sensitivity in product detection, because the KM for UDP-GlcNAc is relatively high (23). Competition experiments were performed by using the assay conditions described in refs. 23 and 53. The IC50 values were determined by using Eqs. 1 and 2. The apparent Ki for the inhibition of LpxA was estimated based on:
Acknowledgments
We thank Karen Soldano and Dr. James C. Phillips for technical assistance and Dr. David Six for help with the preparation of this manuscript. This research was supported by National Institutes of Health Grants GM-51310 (to C.R.H.R.) and CA-95130 (to D.T.G.). A.H.W. was supported by Duke University Cell and Molecular Biology Training Program Grant GM-07184 for part of this project.
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2AQ9).
References
- 1.Raetz C. R. H. Annu. Rev. Biochem. 1990;59:129–170. doi: 10.1146/annurev.bi.59.070190.001021. [DOI] [PubMed] [Google Scholar]
- 2.Raetz C. R. H., Whitfield C. Annu. Rev. Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brade H., Opal S. M., Vogel S. N., Morrison D. C. New York: Dekker; 1999. p. 950. [Google Scholar]
- 4.Nikaido H. Microbiol. Mol. Biol. Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poltorak A., He X., Smirnova I., Liu M. Y., Huffel C. V., Du X., Birdwell D., Alejos E., Silva M., Galanos C., et al. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 6.Hoshino K., Takeuchi O., Kawai T., Sanjo H., Ogawa T., Takeda Y., Takeda K., Akira S. J. Immunol. 1999;162:3749–3752. [PubMed] [Google Scholar]
- 7.Akira S., Takeda K. Nat. Rev. Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 8.Wyckoff T. J. O., Lin S., Cotter R. J., Dotson G. D., Raetz C. R. H. J. Biol. Chem. 1998;273:32369–32372. doi: 10.1074/jbc.273.49.32369. [DOI] [PubMed] [Google Scholar]
- 9.Brabetz W., Muller-Loennies S., Holst O., Brade H. Eur. J. Biochem. 1997;247:716–724. doi: 10.1111/j.1432-1033.1997.00716.x. [DOI] [PubMed] [Google Scholar]
- 10.Onishi H. R., Pelak B. A., Gerckens L. S., Silver L. L., Kahan F. M., Chen M. H., Patchett A. A., Galloway S. M., Hyland S. A., Anderson M. S., Raetz C. R. H. Science. 1996;274:980–982. doi: 10.1126/science.274.5289.980. [DOI] [PubMed] [Google Scholar]
- 11.Jackman J. E., Fierke C. A., Tumey L. N., Pirrung M., Uchiyama T., Tahir S. H., Hindsgaul O., Raetz C. R. H. J. Biol. Chem. 2000;275:11002–11009. doi: 10.1074/jbc.275.15.11002. [DOI] [PubMed] [Google Scholar]
- 12.Anderson M. S., Bulawa C. E., Raetz C. R. H. J. Biol. Chem. 1985;260:15536–15541. [PubMed] [Google Scholar]
- 13.Anderson M. S., Raetz C. R. H. J. Biol. Chem. 1987;262:5159–5169. [PubMed] [Google Scholar]
- 14.Anderson M. S., Bull H. S., Galloway S. M., Kelly T. M., Mohan S., Radika K., Raetz C. R. H. J. Biol. Chem. 1993;268:19858–19865. [PubMed] [Google Scholar]
- 15.Raetz C. R. H., Roderick S. L. Science. 1995;270:997–1000. doi: 10.1126/science.270.5238.997. [DOI] [PubMed] [Google Scholar]
- 16.Jain N. U., Wyckoff T. J., Raetz C. R. H., Prestegard J. H. J. Mol. Biol. 2004;343:1379–1389. doi: 10.1016/j.jmb.2004.08.103. [DOI] [PubMed] [Google Scholar]
- 17.Kisker C., Schindelin H., Alber B. E., Ferry J. G., Rees D. C. EMBO J. 1996;15:2323–2330. [PMC free article] [PubMed] [Google Scholar]
- 18.Beaman T. W., Binder D. A., Blanchard J. S., Roderick S. L. Biochemistry. 1997;36:489–494. doi: 10.1021/bi962522q. [DOI] [PubMed] [Google Scholar]
- 19.Olsen L. R., Roderick S. L. Biochemistry. 2001;40:1913–1921. doi: 10.1021/bi002503n. [DOI] [PubMed] [Google Scholar]
- 20.Wang X. G., Olsen L. R., Roderick S. L. Structure (London) 2002;10:581–588. doi: 10.1016/s0969-2126(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 21.Beaman T. W., Blanchard J. S., Roderick S. L. Biochemistry. 1998;37:10363–10369. doi: 10.1021/bi980759b. [DOI] [PubMed] [Google Scholar]
- 22.Beaman T. W., Sugantino M., Roderick S. L. Biochemistry. 1998;37:6689–6696. doi: 10.1021/bi980106v. [DOI] [PubMed] [Google Scholar]
- 23.Wyckoff T. J., Raetz C. R. H. J. Biol. Chem. 1999;274:27047–27055. doi: 10.1074/jbc.274.38.27047. [DOI] [PubMed] [Google Scholar]
- 24.Lee B. I., Suh S. W. Proteins. 2003;53:772–774. doi: 10.1002/prot.10436. [DOI] [PubMed] [Google Scholar]
- 25.Benson R. E., Gottlin E. B., Christensen D. J., Hamilton P. T. Antimicrob. Agents Chemother. 2003;47:2875–2881. doi: 10.1128/AAC.47.9.2875-2881.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davis I. W., Murray L. W., Richardson J. S., Richardson D. C. Nucleic Acids Res. 2004;32:W615–W619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neu H. C. Science. 1992;257:1064–1073. doi: 10.1126/science.257.5073.1064. [DOI] [PubMed] [Google Scholar]
- 28.Walsh C. T. Nat. Rev. Microbiol. 2003;1:65–70. doi: 10.1038/nrmicro727. [DOI] [PubMed] [Google Scholar]
- 29.Conway S. P., Brownlee K. G., Denton M., Peckham D. G. Am. J. Respir. Med. 2003;2:321–332. doi: 10.1007/BF03256660. [DOI] [PubMed] [Google Scholar]
- 30.Garau J., Gomez L. Curr. Opin. Infect. Dis. 2003;16:135–143. doi: 10.1097/00001432-200304000-00010. [DOI] [PubMed] [Google Scholar]
- 31.Clements J. M., Coignard F., Johnson I., Chandler S., Palan S., Waller A., Wijkmans J., Hunter M. G. Antimicrob. Agents Chemother. 2002;46:1793–1799. doi: 10.1128/AAC.46.6.1793-1799.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kline T., Andersen N. H., Harwood E. A., Bowman J., Malanda A., Endsley S., Erwin A. L., Doyle M., Fong S., Harris A. L., et al. J. Med. Chem. 2002;45:3112–3129. doi: 10.1021/jm010579r. [DOI] [PubMed] [Google Scholar]
- 33.Coggins B. E., Li X., McClerren A. L., Hindsgaul O., Raetz C. R. H., Zhou P. Nat. Struct. Biol. 2003;10:645–651. doi: 10.1038/nsb948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pirrung M. C., Tumey L. N., McClerren A. L., Raetz C. R. H. J. Am. Chem. Soc. 2003;125:1575–1586. doi: 10.1021/ja0209114. [DOI] [PubMed] [Google Scholar]
- 35.Whittington D. A., Rusche K. M., Shin H., Fierke C. A., Christianson D. W. Proc. Natl. Acad. Sci. USA. 2003;100:8146–8150. doi: 10.1073/pnas.1432990100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williamson J. M., Anderson M. S., Raetz C. R. H. J. Bacteriol. 1991;173:3591–3596. doi: 10.1128/jb.173.11.3591-3596.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roujeinikova A., Baldock C., Simon W. J., Gilroy J., Baker P. J., Stuitje A. R., Rice D. W., Slabas A. R., Rafferty J. B. Structure (London) 2002;10:825–835. doi: 10.1016/s0969-2126(02)00775-x. [DOI] [PubMed] [Google Scholar]
- 38.White S. W., Zheng J., Zhang Y. M., Rock C. O. Annu. Rev. Biochem. 2005;74:791–831. doi: 10.1146/annurev.biochem.74.082803.133524. [DOI] [PubMed] [Google Scholar]
- 39.Cronan J. E. Annu. Rev. Microbiol. 2003;57:203–224. doi: 10.1146/annurev.micro.57.030502.090851. [DOI] [PubMed] [Google Scholar]
- 40.Khosla C., Gokhale R. S., Jacobsen J. R., Cane D. E. Annu. Rev. Biochem. 1999;68:219–253. doi: 10.1146/annurev.biochem.68.1.219. [DOI] [PubMed] [Google Scholar]
- 41.Liou G. F., Khosla C. Curr. Opin. Chem. Biol. 2003;7:279–284. doi: 10.1016/s1367-5931(03)00016-4. [DOI] [PubMed] [Google Scholar]
- 42.Walsh C. T. Science. 2004;303:1805–1810. doi: 10.1126/science.1094318. [DOI] [PubMed] [Google Scholar]
- 43.Galloway S. M., Raetz C. R. H. J. Biol. Chem. 1990;265:6394–6402. [PubMed] [Google Scholar]
- 44.Fernandez-Lopez S., Kim H. S., Choi E. C., Delgado M., Granja J. R., Khasanov A., Kraehenbuehl K., Long G., Weinberger D. A., Wilcoxen K. M., Ghadiri M. R. Nature. 2001;412:452–455. doi: 10.1038/35086601. [DOI] [PubMed] [Google Scholar]
- 45.McClerren A. L., Zhou P., Guan Z., Raetz C. R. H., Rudolph J. Biochemistry. 2005;44:1106–1113. doi: 10.1021/bi048001h. [DOI] [PubMed] [Google Scholar]
- 46.Pfitzner U., Raetz C. R. H., Roderick S. L. Proteins Struct. Funct. Genet. 1995;22:191–192. doi: 10.1002/prot.340220212. [DOI] [PubMed] [Google Scholar]
- 47.Otwinowski Z., Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 48.Vagin A., Teplyakov A. J. Appl. Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 49.Jones T. A., Kjeldgaard M. In: From First Map to Final Model. Bailey S., Hubbard R., Waller D., editors. Warrington, U.K.: Sci. Eng. Res. Counc. Daresbury Lab; 1994. pp. 1–13. [Google Scholar]
- 50.Brunger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., et al. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 51.Word J. M., Lovell S. C., LaBean T. H., Taylor H. C., Zalis M. E., Presley B. K., Richardson J. S., Richardson D. C. J. Mol. Biol. 1999;285:1711–1733. doi: 10.1006/jmbi.1998.2400. [DOI] [PubMed] [Google Scholar]
- 52.Lovell S. C., Davis I. W., Arendall W. B., III, de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., Richardson D. C. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 53.Odegaard T. J., Kaltashov I. A., Cotter R. J., Steeghs L., van der Ley P., Khan S., Maskell D. J., Raetz C. R. H. J. Biol. Chem. 1997;272:19688–19696. doi: 10.1074/jbc.272.32.19688. [DOI] [PubMed] [Google Scholar]