Abstract
BLUF (blue light sensing using FAD) domains constitute a recently discovered class of photoreceptor proteins found in bacteria and eukaryotic algae, where they control a range of physiological responses including photosynthesis gene expression, photophobia, and negative phototaxis. Other than in well known photoreceptors such as the rhodopsins and phytochromes, BLUF domains are sensitive to light through an oxidized flavin rather than an isomerizable cofactor. To understand the physicochemical basis of BLUF domain photoactivation, we have applied femtosecond transient absorption spectroscopy to the Slr1694 BLUF domain of Synechocystis PCC6803. We show that photoactivation of BLUF domains proceeds by means of a radical-pair mechanism, driven by electron and proton transfer from the protein to the flavin, resulting in the transient formation of anionic and neutral flavin radical species that finally result in the long-lived signaling state on a 100-ps timescale. A pronounced deuteration effect is observed on the lifetimes of the intermediate radical species, indicating that proton movements underlie their molecular transformations. We propose a photoactivation mechanism that involves a successive rupture of hydrogen bonds between a conserved tyrosine and glutamine by light-induced electron transfer from tyrosine to flavin and between the glutamine and flavin by subsequent protonation at flavin N5. These events allow a reorientation of the conserved glutamine, resulting in a switching of the hydrogen-bond network connecting the chromophore to the protein, followed by radical-pair recombination, which locks the glutamine in place. It is suggested that the redox potential of flavin generally defines the light sensitivity of flavin-binding photoreceptors.
Keywords: flavoprotein, light sensing, photochemistry, reaction mechanism, spectroscopy
Organisms have evolved extensive sensory mechanisms to perceive information carried by light. Their responses are mediated by photoreceptor proteins, which are sensitive to light through prosthetic chromophore molecules. The past decade has witnessed the discovery of a large number of novel flavin-binding photoreceptors, notably the phototropins, the cryptochromes, and BLUF (blue-light sensing using FAD) domains (1). Phototropins are primarily found in plants and control several physiological responses such as phototropism, chloroplast movement, and stomatal opening (2), whereas cryptochromes are known to regulate growth and development in plants and circadian rhythms in plants and insects (3, 4). BLUF domains are a distinct family of flavin-binding photoreceptors that show no significant relationship to other sensor proteins in sequence or structure. The BLUF domain was first discovered as the N-terminal part of the flavoprotein AppA from the purple photosynthetic bacterium Rhodobacter sphaeroides and was shown to control photosynthesis gene expression in response to high-intensity blue-light irradiation and variation of the oxygen tension (5, 6). In later work, the BLUF-containing photoactivated adenylyl cyclase (PAC) photoreceptor was demonstrated to mediate photophobic responses in the green alga Euglena gracilis (7).
The generation of a light signal by photoreceptor proteins relies on the formation of a long-lived signaling state, where initial local structural changes resulting from light absorption by the chromophore are relayed to the protein surface, which undergoes specific alterations that may be sensed by signaling partner proteins. In “traditional” photoreceptors such as the rhodopsins, phytochromes and the xanthopsins, the initial local change is achieved by rapid E/Z isomerization photochemistry of their chromophore, which in turn initiates larger conformational changes by creating steric conflicts, or by charge movements among chromophore and apoprotein (1). In the absence of isomerizable groups, flavin-binding photoreceptors rely on different modes of light activation and thus provide an alternative window on how photon absorption may be coupled to biological sensory function. In the phototropins, for instance, signaling occurs through the light-induced formation of a covalent adduct between the flavin chromophore and a conserved cysteine in light, oxygen, or voltage (LOV) domains, which eventually leads to autophosphorylation of a C-terminal Ser/Thr kinase (8, 9). In analogy with the related photolyases, cryptochromes are thought to function by means of light-induced electron transfer (ET) reactions (10), but at present their mode of action remains largely obscure.
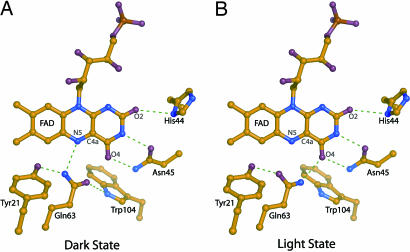
Recently, crystal and solution structures of several BLUF domains were obtained (11–14). The BLUF domain shows a ferredoxin-like fold consisting of a five-stranded β-sheet with two α-helices packed on one side of the sheet, with the noncovalently bound isoalloxazine ring of flavin adenine dinucleotide (FAD) positioned between the two α-helices. Fig. 1 shows a close-up of the FAD-binding pocket of the AppA BLUF domain (11). FAD is involved in an extensive hydrogen-bond network with residues lining the FAD binding pocket, including a highly conserved tyrosine (Tyr) and glutamine (Gln). Upon illumination with blue light, BLUF domains show a characteristic spectral red-shift of their absorption spectrum by ≈10 nm, which is a unique feature of this photoreceptor family and is thought to correspond to the signaling state (5).
Fig. 1.
X-ray structure of the R. sphaeroides AppA BLUF domain. (A and B) Close-ups of the vicinity of the FAD chromophore in two proposed orientations of the conserved Gln in dark state (A) and light state (B). The coordinates were taken from Protein Data Bank ID code 1YRX (11).
The photochemical mechanism by which BLUF domains are activated by blue light is under considerable debate. Early studies suggested that an increased π–π stacking between Tyr and FAD, or deprotonation of FAD, would induce signaling-state formation (15, 16). Jung et al. (13) proposed that in the BlrB BLUF domain, proton transfer (PT) takes place from an arginine (Arg) residue to the O2 atom of the isoalloxazine ring upon blue-light absorption, triggered by the increased basicity of O2 upon promotion of FAD to the singlet excited state. Masuda et al. (17) interpreted their results from Fourier transform infrared (FTIR) spectroscopy by a hydrogen-bond rearrangement around the FAD chromophore upon signaling-state formation. Anderson et al. (11) have proposed a photoactivation model that involves such a hydrogen-bond rearrangement accompanying a 180° rotation of the conserved Gln, as shown in Fig. 1. In this view, photon absorption leads to the breaking of hydrogen bonds from the Gln amino group to the N5 of flavin and to the Tyr and formation of a hydrogen bond to the O4 of the flavin. Ultrafast spectroscopy on the AppA BLUF domain showed that the red-shifted product state is formed in <1 ns, directly from the FAD singlet excited state and without any apparent reaction intermediate (18), yielding no clues on the validity of any of these scenarios.
Here, we use femtosecond transient absorption spectroscopy to investigate the photochemistry in the Slr1694 BLUF domain of the cyanobacterium Synechocystis PCC6803. We show that formation of the red-shifted signaling state proceeds by means of a radical-pair mechanism, driven by ET and PT processes to FAD from its protein environment and resulting in the transient formation of anionic and neutral FAD radical species. We propose a photoactivation mechanism that involves a successive rupture of hydrogen bonds between the conserved Tyr and Gln by ET and between the Gln and FAD by light-induced protonation at N5, which allows a reorientation of the conserved Gln and subsequent switch of hydrogen-bond network connecting the chromophore to the protein.
Results
Transient Absorption Spectroscopy of Slr1694.
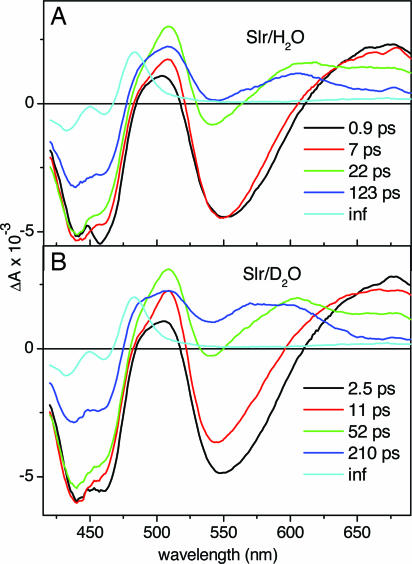
The Slr1694 protein was excited at 400 nm, and the time-resolved absorbance changes were monitored over a wavelength range from 420 to 720 nm. The data were globally analyzed in terms of a kinetic scheme with sequentially interconverting species, where each species is characterized by an evolution-associated difference spectrum (EADS). Five components were required for an adequate description of the time-resolved data, with lifetimes of 0.9, 7, 22, and 123 ps and a long-lived component. The resulting EADS are presented in Fig. 2A, and selected kinetic traces at 483, 550, 610, and 710 nm are shown in Fig. 3 as the open circles. The first EADS (black curve) in Fig. 2A can be assigned to the initially created “hot” S1 singlet excited state of the FAD chromophore, which shows a ground state bleach at wavelengths 425–475 nm, a stimulated emission band near 550 nm, and excited-state absorption at wavelengths >600 nm. This EADS evolves in 0.9 ps to the next EADS (red line), which has a lifetime of 7 ps. The 0.9-ps evolution is attributed to a vibrational cooling process of the FAD excited state (18) and involves a small blue-shift of the stimulated emission band and an increase of excited-state absorption of ≈510 nm, whereas the ground-state bleach remains the same. The EADS with a 7-ps lifetime thus corresponds to the vibrationally relaxed singlet excited state of FAD (hereafter denoted FAD*) and evolves to the next EADS (green line), which has a lifetime of 22 ps. This component shows a decrease of the stimulated emission band at 550 nm and the absorption at wavelengths >600 nm, indicating decay of FAD*. Additionally, an increase of absorption of ≈510 nm and the appearance of a distinct absorption near 600 nm is observed, whereas the ground state bleach remains the same. Thus, the 22-ps EADS can be assigned to a remaining fraction of FAD* but with a contribution of another molecular species to be determined below. The evolution in 22 ps to the next EADS (blue line), which has a lifetime of 123 ps, corresponds to a further loss of the stimulated emission band and partial loss of the ground-state bleach. The induced absorption bands at 510 and 600 nm are still present, and, additionally, a shoulder near 490 nm is formed.
Fig. 2.
EADS and their corresponding lifetimes resulting from global analysis of ultrafast transient absorption experiments on the Synechocystis Slr1694 BLUF domain upon excitation at 400 nm in H2O buffer (A) and in D2O buffer (B).
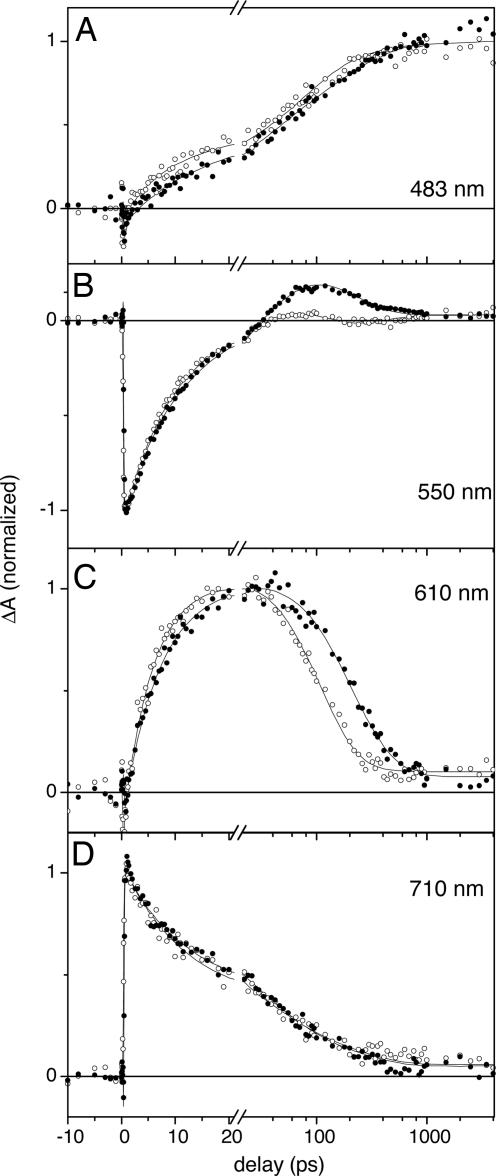
Fig. 3.
Kinetic traces of the Synechocystis Slr1694 BLUF domain in H2O (open circles) and D2O (filled circles) upon excitation at 400 nm, with detection at 483 nm (A), 550 nm (B), 610 nm (C), and 710 nm (D). The solid lines denote the result of the target analysis. Note that the time axis is linear from −10 to 20 ps and logarithmic thereafter.
In the fifth, nondecaying EADS (cyan line), the ground-state bleach has largely disappeared, and a narrow absorption feature near 483 nm has appeared. The nondecaying EADS strongly resembles the absorption of the long-lived signaling state in Slr1694 (17). We conclude that in Slr1694, the red-shifted signaling state is formed in 123 ps, consistent with previous results on the AppA BLUF domain where the long-lived product is formed on the sub-nanosecond timescale (18). Similar to AppA, FAD* in Slr1694 decays multiexponentially, albeit with substantially shorter lifetimes as judged by the decay of the stimulated emission band and the kinetic trace taken at 550 nm. Most significantly, the appearance of photocycle intermediates is observed in Slr1694, which contrasts with the situation in the AppA BLUF domain (18).
Effects of H/D Exchange.
To investigate whether the observed reaction intermediates are kinetically coupled by means of PT, ultrafast experiments were performed with deuterated Slr1694. The EADS that resulted from the sequential analysis were similar to those obtained with Slr1694 in H2O and are presented in Fig. 2B. Kinetic traces are shown in Fig. 3 as the filled circles. The lifetimes of 2.5, 11, 52, and 210 ps were consistently longer than for Slr1694 in H2O, indicating a pronounced kinetic isotope effect (KIE) upon H/D exchange of Slr1694. The 210-ps EADS (blue line) is somewhat different from the corresponding 123-ps EADS of Slr1694 in H2O because the contribution by FAD* seems to be smaller in the former. Interestingly, the shape of the 210-ps EADS in the spectral region from 500 to 700 nm strongly resembles that of a neutral flavin semiquinone FADH• radical (19), indicating the involvement of flavin radical species in the Slr1694 photoreaction, a point that will be substantiated below. The yields of signaling-state formation for Slr1694 in H2O and in D2O are identical, as evident from the amplitude of the 483-nm absorption (cyan line) relative to that of the initial ground-state bleach signal of the first EADS (black line) in each preparation. The relative amplitude of the long-lived product absorption is ≈1.6 times higher in Slr1694 as compared with AppA (18). For AppA, an absolute quantum yield of 24% was determined, implying that the quantum yield of signaling-state formation in Slr1694 amounts to ≈40%.
Fig. 3 represents selected kinetic traces together with the fitting result of the target analysis described below (solid line), taken at 483, 550, 610, and 710 nm, respectively. At 550 nm, where stimulated emission from FAD* has a main contribution, the initial decay phases of Slr1694 in H2O and D2O are almost overlaid, suggesting that no or little KIE applies to the FAD* lifetime (Fig. 3B). This phenomenon is demonstrated even more clearly in the trace at 710 nm, where FAD* has a pronounced absorption but where most other flavin species like semiquinone radicals do not absorb: the traces measured in H2O and D2O are completely overlaid (Fig. 3D). These observations are consistent with previous results on AppA, which showed no H/D exchange effect on the excited-state lifetimes (20). FAD* has an isosbestic point at 610 nm, giving a clean view on the kinetic behavior of the intermediate species. The trace shows a slower rise and slower decay in D2O, revealing the existence of two pronounced KIEs (Fig. 3C). In particular, in Slr1694 in D2O, the intermediate decay takes approximately twice as long as in H2O and occurs in ≈200 ps. The KIE can also be observed in the 483-nm trace where the rise in D2O is slower than in H2O (Fig. 3A). Thus, the decay of the intermediate state is kinetically coupled to the rise of the long-lived product, demonstrating that the intermediate is on the reaction pathway of product formation.
Target Analysis: Identification of Intermediate Reaction Dynamics.
The femtosecond transient absorption data on Slr1694 show that essentially at all delay times, a mixture of FAD*, reaction intermediate(s), or long-lived product states make up the transient spectra. To disentangle the contributions by the various molecular species, we performed a target analysis of time-resolved data of hydrated and deuterated Slr1694 samples, wherein the data are described in terms of a kinetic scheme, thereby identifying their spectral signatures and estimating their lifetimes and their connectivity. A similar approach was taken recently to dissect the PT dynamics in GFP (21). The kinetic scheme used for the target analysis, the transient concentrations of the molecular species involved, and the residuals of the fitting procedure are provided in Figs. 6 and 7 and Supporting Text, which are published as supporting information on the PNAS web site, along with a detailed description of the applied assumptions and premises. In short, four lifetimes are assumed for the decay of FAD*, which stem from inhomogeneity in the ground-state population as shown for the AppA BLUF domain (14, 18). FAD* evolves to a first intermediate Q1, which in turn evolves to a second intermediate Q2. From Q2 the long-lived photoproduct SlrRED is formed. To account for the quantum yield for SlrRED formation of ≈40%, a loss process must be included in the kinetic scheme. Because the ground-state bleach in the sequential analysis does not diminish after the first decay phase of FAD* (see Fig. 2), the loss process is introduced on the transition from Q1 to Q2. The rate constants, fractions, and quantum yields are summarized in Table 1, which is published as supporting information on the PNAS web site.
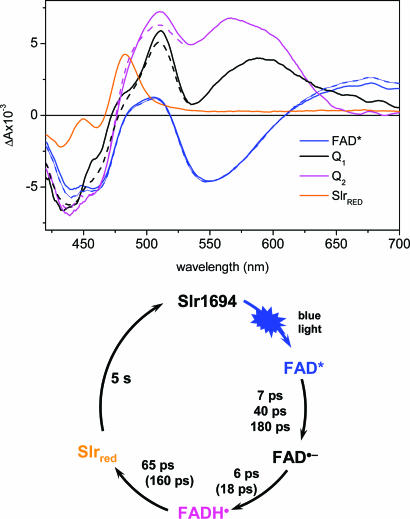
Fig. 4 shows the species-associated difference spectra (SADS) of FAD*, Q1, Q2, and SlrRED. As expected, the SADS of FAD* has the typical shape of the singlet-excited state as determined for FAD in solution (22) and in AppA (18) and as they follow from the sequential analysis in Fig. 2. The SADS of SlrRED is virtually identical to the red-shifted intermediate observed in steady state, with a main absorption peak at 483 nm (17). The SADS of Q2 has a broad absorption in the 480- to 650-nm region with peaks at 510, 566, and 610 nm. This SADS strongly resembles the spectrum of a neutral semiquinone flavin radical, FADH• (19). It is this species that is mainly responsible for the pronounced absorption around ≈510 and ≈600 nm in the 123-ps (H2O) and 210-ps (D2O) EADS in the sequential analysis, shown in Fig. 2.
Fig. 4.
Target analysis and photocycle scheme of the Synechocystis Slr1694 BLUF domain. (Upper) SADS of the various molecular species in the kinetic scheme that result from the application of a target analysis to the ultrafast time-resolved data of the Synechocystis Slr1694 BLUF domain in H2O (solid lines) and D2O (dotted lines), with FAD* (blue), Q1 (black), Q2 (magenta), and the long-lived signaling state SlrRED (orange). (Lower) The photocycle scheme of Slr1694, with time constants as they follow from the target analysis and assignments of the intermediate states as described in the text.
The SADS of Q1 shows an absorption at ≈510 nm and a broad symmetric band at ≈590 nm. The symmetric structure of the latter suggests that it corresponds to a flavin charge-transfer (CT) band rather than of a neutral semiquinone (23). The shape and amplitude of the Q1 SADS is generally similar to that of anionic semiquinone FAD•− in a CT interaction as observed in d-amino acid oxidase. The latter has a narrow absorption peak with a maximum at ≈500 nm and a broad CT band with a maximum at ≈700 nm (23, 24). Given that the absorption maximum of flavin CT bands may vary widely from one flavoprotein to another and depend on specific interactions between flavin and protein, we identify the intermediate species Q1 as a CT/anionic semiquinone FAD•− species.
Fig. 4 Lower shows a photocycle scheme for the photoactivation of Slr1694, based on the results from the target analysis. The initial light-driven reaction from FAD* involves ET from the protein environment to result in FAD•−. This event takes place multiexponentially and is insensitive to H/D exchange, with a dominant time constant of 7 ps. The resulting anionic flavin radical FAD•− is rapidly protonated in 6 ps to result in the neutral semiquinone radical FADH•. This reaction slows down 3-fold in D2O, which is consistent with a putative PT event from the protein environment to FAD. Note that as a result of rate-limiting formation times, the FAD•− radical only accumulates transiently between 5 and 15 ps. This effect can be seen from the transient concentration profiles of FAD*, Q1, Q2, and SlrRED, shown in Fig. 6. Finally, the signaling state SlrRED is formed in 65 ps from the FADH• neutral radical. This reaction is 2.4 times slower in D2O, which is reasonable because SlrRED corresponds to an oxidized FAD. The deuteration effect on the anionic and neutral radical lifetimes, and on the rise time of SlrRED, proves that the radicals are in fact intermediates on the pathway to signaling-state formation. Formation of FAD•− occurs stoichiometrically from FAD*, but ≈50% recombines to the ground state before its protonation results in formation of FADH•.
Discussion
Photoactivation Mechanism in BLUF Domains.
The observation of anionic and neutral semiquinones in the Slr1694 photocycle implies that light-driven ET is followed by PT between FAD and aromatic residues in the protein. The picosecond time scale of these events indicates that the residue(s) involved in the transfer must be in close proximity to the isoalloxazine ring of the flavin. Previous studies on flavo-enzymes have revealed that rapid light-driven ET may occur from aromatic residues to flavin (25, 26). Tyr-8 (Tyr-21 in AppA) is a good candidate because it is the closest aromatic residue to the flavin and has been shown to be critical for photocycling activity in BLUF domains (12, 15, 16). The redox properties of flavin and Tyr gives a favorable driving force for the ET reaction: the midpoint potential for flavin/flavin•− is approximately −0.8 V and for Tyr/Tyr•+ is ≈0.93 V (25, 27), providing a driving force for ET of ΔG = −0.62 eV given that the energy of the 0–0 transition of S0 → S1 is ≈ 2.35 eV (1 eV = 1.602 × 10−19 J) (27).
In the various BLUF crystal structures, the dark-state orientation of the conserved Gln remained ambiguous: according to Anderson et al. (11) the amino group of the conserved Gln would donate hydrogen bonds to N5 and the conserved Tyr, whereas Jung et al. (13) and Kita et al. (12) favored an orientation where the Gln’s amino group donates a hydrogen bond to O4 and receives a hydrogen bond from the conserved Tyr. Fourier transform infrared results by Masuda et al. (17) indicated formation of hydrogen bonds to O4 and loss of a hydrogen bond to N5 of FAD upon illumination. Recent work by Unno et al. (28) showed that mutation of the conserved Gln in the AppA BLUF domain did not alter hydrogen bonding of O4. The latter observations are consistent with a dark orientation as shown in Fig. 1A. Thus, the accumulated evidence points favorably in the direction of a hydrogen-bond switching by means of a Gln flip, as proposed by Anderson et al. (11). This mechanism also can explain why the photoactivated state is so stable and lives from seconds in Slr1694 to a half-hour in AppA. In support of this hypothesis, mutation of the conserved Tyr or the conserved Gln results in a complete loss of photocycling activity (12, 15, 16, 28, 29). With the R. sphaeroides BlrB structure at hand, Jung et al. (13) presented an alternative that would imply a light-induced PT from Arg-32 to the O2 of the flavin from FAD*. This mechanism does not involve radical flavin intermediates and is clearly at odds with the present results.
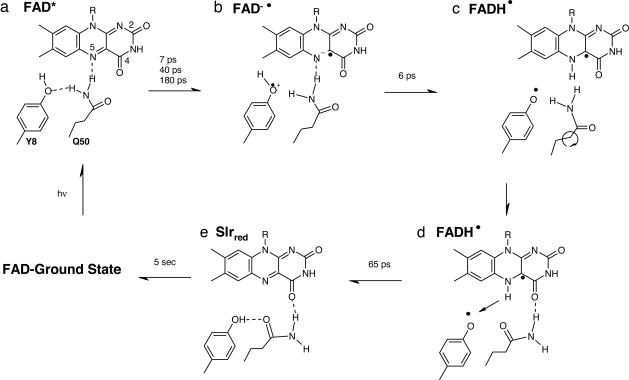
We now are in a position to propose a photochemical reaction mechanism for the photoactivation of BLUF domains, which is schematically indicated in Fig. 5. In the dark state, the mutual orientation of Gln and FAD is arranged as indicated in Fig. 5A, with hydrogen bonds from the amino group of Gln-50 to Tyr-8 and the N5 atom of FAD. Blue-light excitation induces a change in the redox potential of FAD, which causes it to accept an electron, resulting in formation of the FAD anionic semiquinone FAD•−, as indicated in Fig. 5B. In view of the close proximity between FAD and the conserved Tyr, we propose that Tyr-8 acts as the electron donor as suggested for AppA (30, 31). This event breaks the hydrogen bond between Tyr-8 and Gln-50 as a result of increased electrostatic repulsion by Tyr-8. The spectroscopic properties of FAD•− as determined from the target analysis indicate that it forms a CT complex, possibly with the oxidized Tyr-8 radical or perhaps with another positively charged residue in the vicinity of FAD.
Fig. 5.
The proposed reaction mechanism for photoinduced hydrogen-bond switching in the Synechocystis Slr1694 BLUF domain. See text for details.
Upon its reduction, the flavin becomes a strong base that attracts a proton. The closest proton source is the amino group of Gln-50, from which most likely a proton is abstracted. In turn, Gln-50 abstracts a proton from Tyr-8, which at the time is oxidized and highly reactive, resulting in the radical pair FADH•–Tyr• (Fig. 5C). The PT event takes 6 ps and slows down 3-fold upon deuteration. It is well established that protonation of the neutral flavin semiquinone radical FADH• takes place at N5 (32), which inevitably breaks the hydrogen bond between N5 and Gln-50, leaving Gln-50 unhinged and free to rotate. The hydrogen-bond rupture constitutes the triggering event that allows an ≈180° flip of Gln-50, which results in a new hydrogen bond from the amino group of Gln-50 to the O4 of the flavin (Fig. 5D). Possibly, the carbonyl of Gln-50 transiently accepts a hydrogen bond from the newly protonated N5 (configuration not shown). After 65 ps, radical-pair recombination takes place between FADH• and Tyr-8•, whereby the hydrogen atom located on N5 of the flavin returns to Tyr-8, possibly using the carbonyl of Gln-50 as a stepping stone (Fig. 5E). The reprotonated Tyr then donates a hydrogen bond to the carbonyl group of Gln-50, locking it in place. This final step is subject to a KIE of 2.4 upon H/D exchange and is attributed to formation of the long-lived signaling state. In this conformation, the amino moiety of Gln-50 donates a hydrogen bond to O4 of the flavin, and the carbonyl moiety of Gln-50 accepts a hydrogen bond from Tyr-8. Possibly, the amino moiety of Gln-50 hydrogen bonds to N5 as well (configuration not shown). The protein remains in this conformation for a long time because of high energetic barriers.
The potential involvement of the conserved Trp (Trp-91 in Slr1694, Trp-104 in AppA) is not taken into account because earlier work has shown that Trp-104 is not required for photocycling in AppA (20). The driving force is larger for Tyr than for Trp (net ΔG = −0.62 eV for Tyr and −0.4 eV for Trp), and (from the AppA crystal structure) the distances from Tyr-21 and Trp-104 to N5 of FAD are 4.5 and 5.7 Å, respectively (11), indicating that Tyr has a better spatial overlap with the flavin than Trp. The crystal structures of the R. sphaeroides BlrB and Thermosynechococcus Tll0078 BLUF domains indicated a different orientation for the conserved Trp, with a significantly increased distance to FAD (12, 13).
With the present reaction model, predictions can be made with regard to the photochemistry of the nonphotocycling Gln-50 and Tyr-8 mutants. With Gln-50 deleted, ET and possibly PT may still occur from Tyr-8 upon excitation of the flavin. In the absence of the switchable Gln-50 residue, the resulting radical pair will very likely recombine to the original ground state without the production of long-lived photoproducts. Upon deletion of Tyr-8, the light-driven ET process that underlies the photoactivation reaction is shut off, which readily explains the inability of mutants to photocycle. However, the conserved Trp (Trp-91 in Slr1694, Trp-104 in AppA) may complement the ET process to FAD without initiating the photocycle. Indeed, recent experiments on the Tyr-21 mutant of AppA showed light-driven ET from Trp-104 on a picosecond timescale, followed by radical-pair recombination on the (sub)nanosecond timescale to the original ground state (ref. 31 and M.G., W. Laan, I.H.M.v.S., R.v.G., K. J. Hellingwerf, and J.T.M.K., unpublished data).
Comparison with Other BLUF Domains.
Given the similarities in structure and sequence between all BLUF domains studied so far, a similar reaction mechanism very likely holds. In AppA, the FAD* lifetime is >1 order of magnitude longer than in Slr1694, with time constants of 90 and 590 ps. The red-shifted signaling state is apparently formed directly from FAD* (18). The reason that no transient reaction intermediates are observed probably stems from a rate-limiting initial ET rate from FAD*. Such differences in ET rates likely arise from different redox potentials of FAD, because these are sensitive to its hydrogen-bonding pattern and the vicinity of charged residues. It is interesting to note that in AppA, a positively charged His-44 hydrogen bonds to the O2 in lieu of a neutral Asn-31 in Slr1694. In the context of the present results, the formation of the FAD•− anionic radical in AppA would occur with time constants of 90 and 590 ps, which rapidly evolves into the FADH• intermediate and long-lived product AppARED on much faster timescales. In this way, the FAD•− and FADH• intermediates do not transiently accumulate during the reaction and therefore escape detection. A rate-limiting light-driven ET rather than a hydrogen transfer in the AppA BLUF domain is consistent with the absence of a KIE on the FAD excited-state lifetime upon H/D exchange (20).
Light-Induced ET: A General Theme in Flavin-Binding Photoreceptors?
Three families of flavin-binding photoreceptors have thus far been identified: the phototropins [with light, oxygen, or voltage (LOV) domains as flavin-binding domains], the cryptochromes, and the BLUF domains. Although their modes of action appear very distinct, there are interesting parallels. The present results show that the hydrogen-bond switch in BLUF domains is driven by initial ET from aromatic residues to the oxidized flavin. In the cryptochromes, similar ET processes appear to underlie their activity, with Trps as likely electron donors (10, 33). The question remains whether in LOV domains, ET or PT from a conserved Cys to the FMN chromophore constitutes the primary photochemical event (9, 34–36). If the former situation applies, we may indeed conclude that it is the redox potential of the flavin chromophore that defines the light sensitivity of flavin-binding photoreceptors, very much so as it defines the catalytic activity in most flavoenzymes (37).
Materials and Methods
Sample Preparation.
Slr1694-DNA encoding the full-length SLR protein (amino acids 1–160; GenBank accession no. D90913, NP_441709), was amplified by PCR from Synechocystis sp. PCC6803 DNA (kindly provided by I. Maldener, University of Regensburg, Regensburg, Germany) and cloned between EcoRI and HindIII sites of the pet28a(+) vector (Invitrogen). Protein was expressed in Escherichia coli strain BL21(DE3) at 18°C in 0.7 mM IPTG overnight. Slr1964 was purified on Ni-nitrilotriacetic acid resin (Qiagen, Hilden, Germany) according to the supplier’s instruction. The eluate was dialyzed 2× again 200 volumes of 10 mM NaPi/10 mM NaCl (pH 8.0) and concentrated by ultrafiltration (PM30; Millipore). For isotopic replacement, the protein was concentrated and subsequently diluted with buffered D2O at least three times.
Spectroscopy.
Femtosecond transient absorption spectroscopy was carried out with a Ti:sapphire-based regenerative amplification system as described (38). A 400-nm pump beam was obtained by frequency-doubling the output from the amplifier and attenuated to 300 nJ. Femtosecond time delays up to 5 ns between pump and probe were controlled by a delay line, and time-gated spectra at 109 delay times were recorded. The polarizations of pump and probe beams were set at the magic angle (54.7°). The instrument response function was fit to a Gaussian of 120 fs full-width at half-maximum (FWHM). The samples were loaded in a flow system of 4-ml volume, including a flow cuvette of 1-mm path length, and flowed by a peristaltic pump.
Data Analysis.
The transient absorption spectra were globally analyzed in terms of a kinetic model with sequentially interconverting EADS, i.e., 1 → 2 → 3 → …. The arrows indicate successive monoexponential decays with increasing time constants, which can be regarded as the lifetime of each species. The first EADS corresponds to the time 0 difference spectrum. The EADS are not necessarily associated with pure molecular species and often reflect mixtures of molecular species. To further unravel the pathways for product-state formation, a target analysis was applied, in which a specific kinetic scheme was tested (see Supporting Text and Figs. 6 and 7). With this procedure, the SADS of the pure molecular states are estimated. The global and target analysis procedures have been extensively reviewed in ref. 39.
Supplementary Material
Acknowledgments
M.G. was supported by the Netherlands Organization for Scientific Research (NWO) Chemical Sciences Council (NWO-CW). J.M.K. was supported by the NWO Earth and Life Sciences Council (NWO-ALW) “Molecule to Cell” program. J.T.M.K. was supported by a NWO-ALW VIDI fellowship. The work was supported by the Deutsche Forschungsgemeinschaft (to P.H.).
Abbreviations
- BLUF
blue-light sensing using FAD
- SADS
species-associated difference spectrum
- EADS
evolution-associated difference spectrum
- KIE
kinetic isotope effect
- CT
charge transfer
- ET
electron transfer
- PT
proton transfer.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.van der Horst M. A., Hellingwerf K. J. Acc. Chem. Res. 2004;37:13–20. doi: 10.1021/ar020219d. [DOI] [PubMed] [Google Scholar]
- 2.Christie J. M., Briggs W. R. In: Handbook of Photosensory Receptors. Briggs W. R., Spudich J. L., editors. Weinheim, Germany: Wiley; 2005. pp. 277–304. [Google Scholar]
- 3.Sancar A. Chem. Rev. 2003;103:2203–2237. doi: 10.1021/cr0204348. [DOI] [PubMed] [Google Scholar]
- 4.Batschauer A. In: Handbook of Photosensory Receptors. Briggs W. R., Spudich J. L., editors. Weinheim, Germany: Wiley; 2005. pp. 211–236. [Google Scholar]
- 5.Masuda S., Bauer C. E. Cell. 2002;110:613–623. doi: 10.1016/s0092-8674(02)00876-0. [DOI] [PubMed] [Google Scholar]
- 6.Gomelsky M., Klug G. Trends Biochem. Sci. 2002;27:497–500. doi: 10.1016/s0968-0004(02)02181-3. [DOI] [PubMed] [Google Scholar]
- 7.Iseki M., Matsunaga S., Murakami A., Ohno K., Shiga K., Yoshida K., Sugai M., Takahashi T., Hori T., Watanabe M. Nature. 2002;415:1047–1051. doi: 10.1038/4151047a. [DOI] [PubMed] [Google Scholar]
- 8.Crosson S., Rajagopal S., Moffat K. Biochemistry. 2003;42:2–10. doi: 10.1021/bi026978l. [DOI] [PubMed] [Google Scholar]
- 9.Swartz T. E., Bogomolni R. A. In: Handbook of Photosensory Receptors. Briggs W. R., Spudich J. L., editors. Weinheim, Germany: Wiley; 2005. pp. 305–323. [Google Scholar]
- 10.Giovani B., Byrdin M., Ahmad M., Brettel K. Nat. Struct. Biol. 2003;10:489–490. doi: 10.1038/nsb933. [DOI] [PubMed] [Google Scholar]
- 11.Anderson S., Dragnea V., Masuda S., Ybe J., Moffat K., Bauer C. Biochemistry. 2005;44:7998–8005. doi: 10.1021/bi0502691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kita A., Okajima K., Morimoto Y., Ikeuchi M., Miki K. J. Mol. Biol. 2005;349:1–9. doi: 10.1016/j.jmb.2005.03.067. [DOI] [PubMed] [Google Scholar]
- 13.Jung A., Domratcheva T., Tarutina M., Wu Q., Ko W. H., Shoeman R. L., Gomelsky M., Gardner K. H., Schlichting I. Proc. Natl. Acad. Sci. USA. 2005;102:12350–12355. doi: 10.1073/pnas.0500722102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grinstead J. S., Hsu S. T. D., Laan W., Bonvin A. M. J. J., Hellingwerf K. J., Boelens R., Kaptein R. ChemBioChem. 2006;7:187–193. doi: 10.1002/cbic.200500270. [DOI] [PubMed] [Google Scholar]
- 15.Kraft B. J., Masuda S., Kikuchi J., Dragnea V., Tollin G., Zaleski J. M., Bauer C. E. Biochemistry. 2003;42:6726–6734. doi: 10.1021/bi030055o. [DOI] [PubMed] [Google Scholar]
- 16.Laan W., van der Horst M. A., van Stokkum I. H. M., Hellingwerf K. J. Photochem. Photobiol. 2003;78:290–297. doi: 10.1562/0031-8655(2003)078<0290:icotpp>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 17.Masuda S., Hasegawa K., Ishii A., Ono T. Biochemistry. 2004;43:5304–5313. doi: 10.1021/bi049836v. [DOI] [PubMed] [Google Scholar]
- 18.Gauden M., Yeremenko S., Laan W., van Stokkum I. H. M., Ihalainen J. A., van Grondelle R., Hellingwerf K. J., Kennis J. T. M. Biochemistry. 2005;44:3653–3662. doi: 10.1021/bi047359a. [DOI] [PubMed] [Google Scholar]
- 19.Muller F., Brustlein M., Hemmerich P., Massey V., Walker W. H. Eur. J. Biochem. 1972;25:573–580. doi: 10.1111/j.1432-1033.1972.tb01730.x. [DOI] [PubMed] [Google Scholar]
- 20.Laan W., Gauden M., Yeremenko S., van Grondelle R., Kennis J. T. M., Hellingwerf K. J. Biochemistry. 2006;45:51–60. doi: 10.1021/bi051367p. [DOI] [PubMed] [Google Scholar]
- 21.Kennis J. T. M., Larsen D. S., van Stokkum I. H. M., Vengris M., van Thor J. J., van Grondelle R. Proc. Natl. Acad. Sci. USA. 2004;101:17988–17993. doi: 10.1073/pnas.0404262102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stanley R. J. Antioxidants Redox Signaling. 2001;3:847–866. doi: 10.1089/15230860152665028. [DOI] [PubMed] [Google Scholar]
- 23.Miura R. Chem. Rec. 2001;1:183–194. doi: 10.1002/tcr.1007. [DOI] [PubMed] [Google Scholar]
- 24.Nishina Y., Sato K., Shi R. W., Setoyama C., Miura R., Shiga S. J. Biochem. 2001;130:637–647. doi: 10.1093/oxfordjournals.jbchem.a003029. [DOI] [PubMed] [Google Scholar]
- 25.Zhong D. P., Zewail A. H. Proc. Natl. Acad. Sci. USA. 2001;98:11867–11872. doi: 10.1073/pnas.211440398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mataga N., Chosrowjan H., Taniguchi S., Tanaka F., Kido N., Kitamura M. J. Phys. Chem. B. 2002;106:8917–8920. [Google Scholar]
- 27.Harriman A. J. Phys. Chem. 1987;91:6102–6104. [Google Scholar]
- 28.Unno M., Masuda S., Ono T., Yamauchi S. J. Am. Chem. Soc. 2006;128:5638–5639. doi: 10.1021/ja060633z. [DOI] [PubMed] [Google Scholar]
- 29.Hasegawa K., Masuda S., Ono T. A. Plant Cell Physiol. 2005;46:136–146. doi: 10.1093/pcp/pci003. [DOI] [PubMed] [Google Scholar]
- 30.Zirak P., Penzkofer A., Schiereis T., Hegemann P.,A., J., Schlichting I. Chem. Phys. 2005;315:142–154. [Google Scholar]
- 31.Dragnea V., Waegele M., Balascuta S., Bauer C. E., Dragnea B. Biochemistry. 2005;44:15978–15985. doi: 10.1021/bi050839x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muller F., Hemmerich P., Ehrenberg A., Palmer G., Massey V. Eur. J. Biochem. 1970;14:185–196. doi: 10.1111/j.1432-1033.1970.tb00277.x. [DOI] [PubMed] [Google Scholar]
- 33.Zeugner A., Byrdin M., Bouly J. P., Bakrim N., Giovani B., Brettel K., Ahmad M. J. Biol. Chem. 2005;280:19437–19440. doi: 10.1074/jbc.C500077200. [DOI] [PubMed] [Google Scholar]
- 34.Schleicher E., Kowalczyk R. M., Kay C. W. M., Hegemann P., Bacher A., Fischer M., Bittl R., Richter G., Weber S. J. Am. Chem. Soc. 2004;126:11067–11076. doi: 10.1021/ja049553q. [DOI] [PubMed] [Google Scholar]
- 35.Kennis J. T. M., Crosson S., Gauden M., van Stokkum I. H. M., Moffat K., van Grondelle R. Biochemistry. 2003;42:3385–3392. doi: 10.1021/bi034022k. [DOI] [PubMed] [Google Scholar]
- 36.Sato Y., Iwata T., Tokutomi S., Kandori H. J. Am. Chem. Soc. 2005;127:1088–1089. doi: 10.1021/ja0436897. [DOI] [PubMed] [Google Scholar]
- 37.Ghisla S., Massey V. Eur. J. Biochem. 1989;181:1–17. doi: 10.1111/j.1432-1033.1989.tb14688.x. [DOI] [PubMed] [Google Scholar]
- 38.Gradinaru C. C., Kennis J. T. M., Papagiannakis E., van Stokkum I. H. M., Cogdell R. J., Fleming G. R., Niederman R. A., van Grondelle R. Proc. Natl. Acad. Sci. USA. 2001;98:2364–2369. doi: 10.1073/pnas.051501298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Stokkum I. H. M., Larsen D. S., van Grondelle R. Biochim. Biophys. Acta. 2004;1657:82–104. doi: 10.1016/j.bbabio.2004.04.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.