Abstract
Complex social behavior in Microtus voles and other mammals has been postulated to be under the direct genetic control of a single locus: the arginine vasopressin 1a receptor (avpr1a) gene. Using a phylogenetic approach, we show that a repetitive element in the promoter region of avpr1a, which reportedly causes social monogamy, is actually widespread in nonmonogamous Microtus and other rodents. There was no evidence for intraspecific polymorphism in regard to the presence or absence of the repetitive element. Among 25 rodent species studied, the element was absent in only two closely related nonmonogamous species, indicating that this absence is certainly the result of an evolutionarily recent loss. Our analyses further demonstrate that the repetitive structures upstream of the avpr1a gene in humans and primates, which have been associated with social bonding, are evolutionarily distinct from those in rodents. Our evolutionary approach reveals that monogamy in rodents is not controlled by a single polymorphism in the promoter region of the avpr1a gene. We thus resolve the contradiction between the claims for an evolutionarily conserved genetic programming of social behavior in mammals and the vast evidence for highly complex and flexible mating systems.
Keywords: avpr1a gene, mammals, mating system, Microtus, voles
The extent to which behavior is encoded in genes is highly debated (1–3). Although some candidate genes affecting behavior have been identified in invertebrates [e.g., in the fruit fly (as reviewed in ref. 4) and the honey bee (5)], the genetic bases of vertebrate behavior are far from being elucidated. Links between genetic polymorphisms and behavior have been reported for very few species (6, 7), but high behavioral plasticity and modifiability by environmental parameters generally suggest the involvement of several genes and pathways (8).
A prime example for the genetic control of complex social behavior in vertebrates by a polymorphism at a single locus is the arginine vasopressin 1a receptor (avpr1a) gene region characterized in Microtus voles (9–11). The alteration of gene expression and receptor distribution in the brain by the presence of a highly repetitive array of short tandem repeat (STR) sequences in the 5′ regulatory region of the avpr1a gene has been linked to social behavior in Microtus voles and other mammals (12–14). These avpr1a STRs were detected in two socially monogamous vole species (Microtus ochrogaster and Microtus pinetorum) (13–15), whereas they were absent from the regulatory region of two socially nonmonogamous species (Microtus pennsylvanicus and Microtus montanus) (13). The transfer of the avpr1a gene region that included the STRs from socially monogamous M. ochrogaster to Mus musculus led to modified avpr1a gene expression in the brain and more affiliative behavior in male mice (13). Moreover, transgenic M. pennsylvanicus expressing both the species-specific and additionally M. ochrogaster receptors in the brain showed increased pair-bonding behavior, which was taken as evidence that avpr1a STRs induce monogamy (12, 14–17). Repetitive genetic structures upstream of the avpr1a gene in humans and primates were then also hypothesized to be involved in social bonding (15).
If the presence of STRs upstream of the mammalian avpr1a gene was important for the initiation of pair bonding as a prerequisite for monogamous behavior in mammals (see ref. 18), and especially in Microtus rodents, one would predict that avpr1aSTRs should be found in only a few species because <5% of all mammals are monogamous (19). Furthermore, if avpr1a STRs in different taxa were phylogenetically nonindependent, patterns in regard to presence/absence and structure of these genetic elements should be similar in closely related species. We tested these predictions by examining the presence or absence of avpr1a STRs in Microtus species from three continents and in rodents from other genera along a molecular phylogeny. Moreover, we compared the structure and location of the different avpr1a STRs present in mammals to elucidate the general evolutionary context of these repetitive elements.
Results
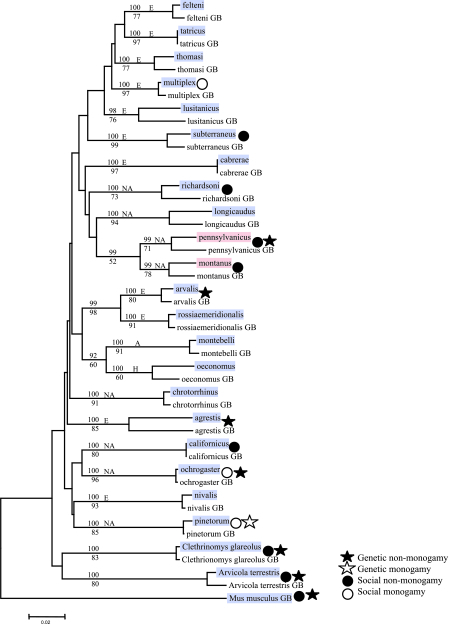
Our analysis revealed the presence of avpr1a STRs in 19 of the 21 examined Microtus species from three different continents (Europe, North America, and Asia), as well as in three other rodent genera and families (the bank vole, Clethrionomys glareolus; the water vole, Arvicola terrestris; and the house mouse, Mus musculus). We found that avpr1a STRs were absent only in the two North American species M. pennsylvanicus and M. montanus (Fig. 1). Furthermore, we found no intraspecific polymorphism for the presence or absence of the avpr1a STRs in the 16 species for which we analyzed several individuals from different populations (total N = 116) (Table 1). Consistent with the initial report linking the avpr1a STRs with monogamous behavior (13), the STRs were present in the North American species M. ochrogaster (N = 10) and M. pinetorum (N = 1) and were absent in all individuals investigated for M. pennsylvanicus (N = 11) and M. montanus (N = 14). Several populations were screened for each species.
Fig. 1.
Segregation pattern of avpr1a STRs and mating system parameters along a molecular phylogeny (cytb) of Microtus and other rodents. Species in which avpr1a STRs are present are highlighted in blue, and species in which they are absent are highlighted in red. Symbols refer to genetic and social monogamy or nonmonogamy. The phylogeny was based on cytb sequences of the analyzed species in comparison with published molecular data [sequences from GenBank (GB)]. Bootstrap values >50 for the neighbor-joining and maximum-likelihood methods are shown above and below the branches, respectively. Species from all continents inhabited by the Microtus genus are included: E, Europe; NA, North America; A, Asia; H, Holarctic.
Table 1.
Number of individuals analyzed for the presence/absence of STRs in the 5′ regulatory region of the avpr1a gene, and accession numbers of cytb sequences
| Species | No. ofindividuals analyzedfor avpr1a STRs | Accession number |
|
|---|---|---|---|
| New cytb sequence | Previously published cytb sequence | ||
| Microtus ochrogaster | 10 | DQ663671 | AF163901 |
| Microtus pennsylvanicus | 11 | DQ663649 | AF119279 |
| Microtus montanus | 14 | DQ663650 | AF119280 |
| Microtus californicus | 6 | DQ663651 | AF163891 |
| Microtus chrotorrhinus | 2 | DQ663652 | AF163893 |
| Microtus montebelli | 2 | DQ663653 | AF163900 |
| Microtus pinetorum | 1 | DQ663654 | AF163904 |
| Microtus richardsoni | 2 | DQ663655 | AF163905 |
| Microtus longicaudus | 4 | DQ663656 | AF187230 |
| Microtus oeconomus | 3 | DQ663657 | AY220028 |
| Microtus agrestis | 11 | DQ663658 | AY167210 |
| Microtus arvalis | 29 | DQ663659 | AY220770 |
| Microtus cabrerae | 1 | DQ663660 | AY513788 |
| Microtus felteni | 1 | DQ663661 | AY513798 |
| Microtus lusitanicus | 1 | DQ663662 | AY513812 |
| Microtus multiplex | 1 | DQ663663 | AY513816 |
| Microtus rossiaemeridionalis | 2 | DQ663664 | AY513819 |
| Microtus subterraneus | 3 | DQ663665 | AY513833 |
| Microtus tatricus | 4 | DQ663666 | AY513837 |
| Microtus thomasi | 1 | DQ663667 | AY513840 |
| Microtus nivalis | 1 | DQ663668 | AY513845 |
| Arvicola terrestris | 1 | DQ663669 | AF159400 |
| Clethrionomys glareolus | 3 | DQ663670 | AY309421 |
| Mus musculus | 2 | — | AB205312 |
All our cytochrome b (cytb) sequences clustered in phylogenetic trees together with the GenBank entries for the same species with a high bootstrap support (see Fig. 1). The topologies of the phylogenies were consistent when we used the maximum-likelihood and neighbor-joining methods, and they were consistent with previous phylogenetic analyses of cytb covering additional species (20). All phylogenetic trees grouped M. pennsylvanicus and M. montanus as sister taxa.
Available behavioral and genetic data on the mating systems of the investigated species do not support a strict connection between the presence of avpr1a STRs and monogamy (see Fig. 1; also see Table 2, which is published as supporting information on the PNAS web site). For example, social monogamy is actually only partly supported by behavioral data for M. ochrogaster because it is not observed in all populations (21). Moreover, at least three other Microtus species (Microtus californicus, Microtus richardsoni, and Microtus subterraneus), plus the rodent species from other genera and families used as outgroups in our analyses, are all socially nonmonogamous although they feature avpr1a STRs (see Table 2 for references).
Genotyping of Microtus arvalis mothers and their offspring provided unambiguous evidence for genetic nonmonogamy in the presence of avpr1a STRs (Table 3, which is published as supporting information on the PNAS web site). More than two paternal alleles were present in all eight litters from different populations of M. arvalis, which means that all examined females had mated with several males, even though these females possessed avpr1a STRs (see Supporting Text, which is published as supporting information on the PNAS web site). Additionally, multiple paternity has been demonstrated independently for three further Microtus species for which we detected avpr1a STRs, including M. ochrogaster (Table 2). The non-Microtine rodents investigated here all feature avpr1a STRs and also are not genetically monogamous (Fig. 1 and see Table 2 for references).
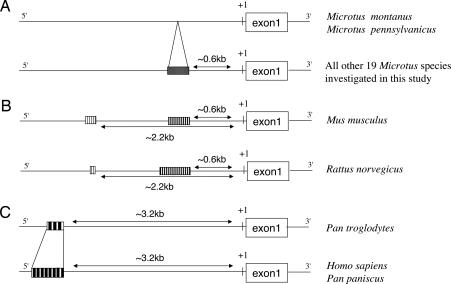
Our comparative analyses of a 5-kb DNA sequence upstream of the avpr1a gene revealed that house mice (Mus musculus) and rats (Rattus norvegicus) have an array of STRs at the same distance (≈600 bp) from the avpr1a gene, as in Microtus, but their repeat motifs differ from all sequenced Microtus species (Fig. 2). Humans, bonobos (Pan paniscus), and chimpanzees (Pan troglodytes) feature long stretches of STRs upstream of the avpr1a gene, but these STRs are further upstream (3.2 kb) and show other repeat motifs than all rodents examined (Fig. 2).
Fig. 2.
Schematic view of STRs in the 5′ region of the avpr1a gene of various mammals. (A) All Microtus species investigated in this study show an array of STRs ≈0.6 kb upstream of exon1, except for two species, M. montanus and M. pennsylvanicus, which lack this element. (B) STRs are present in Mus musculus and R. norvegicus at the same position as in Microtus but have a different repeat motif. An additional short array of STRs is located 2.2 kb upstream of exon1. (C) Homo sapiens, P. paniscus, and P. troglodytes exhibit STR arrays of different lengths located ≈3.2 kb upstream of exon1 of the avpr1a gene (see ref. 15). The repeat motifs in these primates differ from those in rodents.
Discussion
Our results on the segregation of avpr1a STRs in the evolution of the Microtus genus and other mammals refute the general validity of a potential link between the presence of these genetic elements and monogamy in rodents and other taxa (12–14). The prevalence of the avpr1a STRs among rodents is in clear contrast to the general rarity of social monogamy in mammals (19). If the presence of STRs upstream of the avpr1a gene were, by itself, indicative of a tendency toward monogamous behavior, our results would imply that all investigated species other than M. pennsylvanicus and M. montanus should be monogamous. Interestingly, our phylogenetic analyses of the mitochondrial cytb gene show that these two species are very closely related (Fig. 1 and ref. 20), suggesting that the absence of the avpr1a STRs is an evolutionarily novel trait in the Microtus genus and that the presence of the STRs is the ancestral state in rodents.
In contrast to previous suggestions (10, 11, 13–15, 17), our results demonstrate that the presence of STRs upstream of the avpr1a gene is not directly linked to the mating system or basic social organization of these mammals (Fig. 1). Therefore, the alteration of sociobehavioral traits such as pair bonding or more affiliative behavior observed after the transfer of avpr1a gene regions between species (13–15) may not necessarily indicate a transition to monogamy but rather could result from disturbed epistatic interactions. Even though length variation within the avpr1a STRs has been reported to be associated with behavioral differences within particular populations of M. ochrogaster (15) and could thus be involved in the modulation of some social behaviors, the potential link of the presence of avpr1a STRs with monogamy is not as direct and general as previously suggested. Moreover, other neuropeptides such as oxytocin and dopamine have also been demonstrated to modify social and reproductive behavior in mammals (22), and especially in Microtus rodents (11, 23–26). Receptors for these neuropeptides also show different distribution patterns in the brain between socially more affiliative and less affiliative vole species (27). For example, the dopamine system can modulate partner preference displayed by avpr1a- transgenic M. pennsylvanicus (28), which indicates that these complex behavioral patterns are regulated by several neural circuits (29). Given the multifactorial nature of these systems, it seems important that future studies explicitly consider the large variation in both behavioral and neuroanatomical patterns that exists between individuals or populations (see ref. 29).
It is important to note that STRs located upstream of the avpr1a gene in various mammalian taxa may have different evolutionary backgrounds and may not be functionally equivalent (see ref. 10). There is no indication for the evolution of a common genetic mechanism modulating behavior in different mammalian species, although the patterns observed might be largely comparable and involve similar peptides (29). Given the different structure and location of the repeats, the primate avpr1a STRs do not seem evolutionarily closely related to the rodent avpr1a STRs. Polymorphism within STRs upstream of the avpr1a gene has been associated with psychosocial variation in humans (30), but a functional connection between the STRs upstream of the avpr1a gene in humans or primates and mating behavior remains to be shown. In light of our results, we postulate that such variation in STR length in higher primates is not linked to basic differences in the social or mating systems of these species.
In conclusion, our analyses show that monogamy evolved independently from the presence of a putative sole genetic switch in mammals. The absence of a simple, evolutionarily conserved genetic programming of monogamous behavior in mammals is consistent with the large body of literature on intraspecific variation in social organization and mating systems (18, 31). A rigid genetic coding of such a complex and important behavior is unlikely (8) and would certainly be detrimental for species such as small mammals that have highly fluctuating densities and must adapt their reproductive strategy to changing environments.
Materials and Methods
avpr1a STRs Analyses.
We analyzed presence/absence data and the structural patterns of avpr1a STRs in the genomes of 21 Microtus species chosen from the entire distributional range of the genus (11 European, 1 Asian, 8 North American, and 1 Holarctic species) and for the outgroup taxa A. terrestris, C. glareolus, and Mus musculus (Fig. 1 and Table 1). DNA was extracted from tissue samples by using magnetic beads (MagneSil BLUE; Promega) and a standard phenol/chloroform protocol (see ref. 32).
For PCR amplification of the region containing the STRs, we used PCR primers from ref. 13, as well as newly developed primers (forward 5′-AAAAACAGCTCCCCCTGCT-3′; reverse 5′-GGGGCGCGTATAATCTACCT-3′). PCR amplification was performed in a reaction volume of 25 μl on a GeneAmP PCR system (9700; Applied Biosystems). The reaction volume for one sample consisted of 2 μl of DNA, 12 μl of H2O, 1.5 μl of MgCl2, 4.8 μl of 2.5 mM dNTPs, 2.5 μl of buffer including MgCl2, 0.2 μl of Taq polymerase (Qiagen), and 1 μl of each primer (10 μM).
Cycling conditions included an initial denaturation step at 95°C for 2 min, followed by 40 cycles of denaturation at 95°C for 1 min, primer annealing at 50°C for 1 min, sequence extension at 72°C for 1 min, and a final extension step at 72°C for 30 min. PCR products were separated on a 1.5% agarose gel, and length differences were scored by comparison with a 100-bp ladder (Invitrogen). For confirmation of locus specificity, cleaned PCR products (GenElute PCR clean-up kit; Sigma) were sequenced by using amplification primers or an internal sequencing primer (5′-TGGTGTTGTCTATCCGTGTGTG-3′). For the sequencing reaction, Big Dye Terminator version 3.1 ready reaction mix (Applied Biosystems) was used in a reaction volume of 10 μl. Cycle sequencing PCR conditions consisted of an initial denaturation step for 50 sec at 96°C, followed by 30 cycles of denaturation at 96°C for 10 sec, annealing at 55°C for 10 sec, and extension at 72°C for 4 min 30 sec. The products were cleaned by using the DyeEx 96 spin kit (Qiagen) and were separated and detected on an ABI Prism 3100 genetic analyzer (Applied Biosystems).
Amplification of the avpr1a STR region results in short sequences of 200–300 bp for species without STRs and longer sequences of 600–800 bp if STRs are present (13). The DNA sequences of the STRs themselves are highly variable in the length of the different motifs and are difficult to align; however, regions up- and downstream of the insertion point of the STRs are conserved across species and can, therefore, be used to verify the locus-specificity of the sequences (13, 15).
We examined published sequence information on the avpr1a gene, and on the corresponding noncoding sequence 5 kb upstream, for the presence and structure of STRs. We retrieved available sequences for human, chimpanzee (P. troglodytes), mouse (Mus musculus), and rat (R. norvegicus) from GenBank (www.nbci.nlm.nih.gov) and Ensembl (www.ensembl.org). Data on bonobo (P. paniscus) were obtained from ref. 15. Alignment of the sequences was done with the Clustal W algorithm (33), implemented in bioedit 5.0.9 (34), and revealed the location, length, and repeat motif of STRs in the different species.
Survey of Mating Systems.
We performed literature searches for behavioral and genetic data on mating systems of the different Microtus species and other rodents analyzed here. Detailed information on monogamous or nonmonogamous behavior is unfortunately lacking for most species; however, multiple paternity within litters may serve as a very conservative proxy for genetic nonmonogamy. To differentiate between social and genetic monogamy, we followed ref. 18 (p. 267), in which “social monogamy” is described as a close relationship with one partner at a time, whereas “genetic monogamy” refers to reproduction with a single partner confirmed by molecular analyses. The term “nonmonogamy” is used to summarize all types of mating systems that involve more than two partners (e.g., polygyny and promiscuity).
To further examine the potential association of the presence of avpr1a STRs and monogamy, we analyzed paternity in litters of European M. arvalis. We analyzed eight females from different populations for the presence of avpr1a STRs and analyzed their 45 offspring at seven microsatellite loci for multiple paternity. Multiple paternity can be reliably identified if more than two paternal alleles at a microsatellite locus are present in littermates with the same mother. This procedure provides a conservative measure of multiple paternities because only fathers that passed on different alleles at a locus to the offspring can be identified. To assess the number of different paternal alleles in the offspring, the genotype of each offspring was compared with the genotype of its mother (see Supporting Text and Table 3 for details).
Cytochrome b Analyses.
Phylogenetic reconstructions and verification of species identity for all analyzed taxa were based on 930-bp DNA sequence data from the mitochondrial cytb. Molecular analyses of cytb followed the procedure described by Fink et al. (35), with the addition of an amplification primer pair (forward 5′-CATCAGACACAGCAACAGCA-3′; reverse 5′-TGAATGGGTATTCGACTGGTT-3′) and two internal sequencing primers (5′-CCGTNATAGCAACAGCATT-3′ and 5′-TTGGATCCTGTTTCGTGTAAGAA-3′). Additional cytb sequences were retrieved from GenBank (www.nbci.nlm.nih.gov). Accession numbers are given in Table 1. Sequences were aligned by using the Clustal W algorithm (33) implemented in bioedit 5.0.9 (34), and the alignment was revised manually. Phylogenetic trees were obtained by using neighbor-joining (36) methods implemented in mega 2.1 (37) and maximum-likelihood algorithms in paup 4.0b (38), for which the most suitable model of DNA substitution was estimated with modeltest 3.06 (37). For the maximum-likelihood method, the most suitable model was the general time-reversible model (39) with six substitution types: A ↔ C, 0.9169; A ↔ G, 11.8880; A ↔ T, 1.6388; C ↔ G, 0.6181; C ↔ T, 11.7057; and G ↔ T, 1.0000. The gamma shape parameter was estimated as 0.9418, and 55.6% invariable sites were detected. The nucleotide frequencies were as follows: A, 0.3440; C, 0.3421; G, 0.0862; and T, 0.2278.
Supplementary Material
Acknowledgments
We thank the editor and two reviewers for constructive comments on the manuscript, as well as G. Bertorelle, M. Jaarola, I. Keller, and A. Tzika for critical reading of an earlier version. We are grateful to the following individuals and institutions for providing access to samples: A. Bannikova, S. Braaker, R. Burri, Bündner Naturmuseum, F. Catzeflis, C. Conroy, B. Cushing, T. Derting, M. Jaarola, T. Maddalena, N. Martinkova, J.-P. Müller, Museum of Vertebrate Zoology of the University of California, M. Pfunder, R. Pita, J. Suchomel, J. Robovsky, J. Runge, and P. Vogel.
Abbreviations
- avpr1a
arginine vasopressin 1a receptor
- STR
short tandem repeat
- cytb
cytochrome b.
Footnotes
References
- 1.Robinson G. E., Grozinger C. M., Whitfield C. W. Nat. Rev. Genet. 2005;6:257–270. doi: 10.1038/nrg1575. [DOI] [PubMed] [Google Scholar]
- 2.Robinson G. E. Trends Ecol. Evol. 1999;14:202–205. doi: 10.1016/s0169-5347(98)01536-5. [DOI] [PubMed] [Google Scholar]
- 3.Baker B. S., Taylor B. J., Hall J. C. Cell. 2001;105:13–24. doi: 10.1016/s0092-8674(01)00293-8. [DOI] [PubMed] [Google Scholar]
- 4.Sokolowski M. B. Nat. Rev. Genet. 2001;2:879–890. doi: 10.1038/35098592. [DOI] [PubMed] [Google Scholar]
- 5.Ben-Shahar Y., Robichon A., Sokolowski M. B., Robinson G. E. Science. 2002;296:741–744. doi: 10.1126/science.1069911. [DOI] [PubMed] [Google Scholar]
- 6.Lank D. B., Smith C. M., Hanotte O., Burke T., Cooke F. Nature. 1995;378:59–62. [Google Scholar]
- 7.Mandiyan V. S., Coats J. K., Shah N. M. Nat. Neurosci. 2005;8:1660–1662. doi: 10.1038/nn1589. [DOI] [PubMed] [Google Scholar]
- 8.Bućan M., Abel T. Nat. Rev. Genet. 2002;3:114–123. doi: 10.1038/nrg728. [DOI] [PubMed] [Google Scholar]
- 9.Fitzpatrick M., Ben-Shahar Y., Smid H., Vet L., Robinson G., Sokolowski M. Trends Ecol. Evol. 2005;20:96–104. doi: 10.1016/j.tree.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 10.Young L. J., Wang Z. Nat. Neurosci. 2004;7:1048–1054. doi: 10.1038/nn1327. [DOI] [PubMed] [Google Scholar]
- 11.Keverne E. B., Curley J. P. Curr. Opin. Neurobiol. 2004;14:777–783. doi: 10.1016/j.conb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 12.Hammock E. A. D., Young L. J. Mol. Biol. Evol. 2004;21:1057–1063. doi: 10.1093/molbev/msh104. [DOI] [PubMed] [Google Scholar]
- 13.Young L. J., Nilsen R., Waymire K. G., MacGregor G. R., Insel T. R. Nature. 1999;400:766–768. doi: 10.1038/23475. [DOI] [PubMed] [Google Scholar]
- 14.Lim M. M., Wang Z., Olazabal D. E., Ren X., Terwilliger E. F., Young L. J. Nature. 2004;429:754–757. doi: 10.1038/nature02539. [DOI] [PubMed] [Google Scholar]
- 15.Hammock E. A. D., Young L. J. Science. 2005;308:1630–1634. doi: 10.1126/science.1111427. [DOI] [PubMed] [Google Scholar]
- 16.Pitkow L. J., Sharer C. A., Ren X., Insel T. R., Terwilliger E. F., Young L. J. J. Neurosci. 2001;21:7392–7396. doi: 10.1523/JNEUROSCI.21-18-07392.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lim M. M., Hammock E. A. D., Young L. J. J. Neuroendocrinol. 2004;16:325–332. doi: 10.1111/j.0953-8194.2004.01162.x. [DOI] [PubMed] [Google Scholar]
- 18.Reichard U. H., Boesch C. Monogamy: Mating Strategies and Partnerships in Birds, Humans and Other Mammals. Cambridge, U.K.: Cambridge Univ. Press; 2003. [Google Scholar]
- 19.Kleiman D. G. Q. Rev. Biol. 1977;52:39–69. doi: 10.1086/409721. [DOI] [PubMed] [Google Scholar]
- 20.Jaarola M., Martinkova N., Gunduz I., Brunhoff C., Zima J., Nadachowski A., Amori G., Bulatova N. S., Chondropoulos B., Fraguedakis-Tsolis S., et al. Mol. Phylogenet. Evol. 2004;33:647–663. doi: 10.1016/j.ympev.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 21.Cushing B. S., Martin J. O., Young L. J., Carter C. S. Horm. Behav. 2001;39:48–58. doi: 10.1006/hbeh.2000.1633. [DOI] [PubMed] [Google Scholar]
- 22.Curley J. P., Keverne E. B. Trends Ecol. Evol. 2005;20:561–567. doi: 10.1016/j.tree.2005.05.018. [DOI] [PubMed] [Google Scholar]
- 23.Young L. J. Horm. Behav. 1999;36:212–221. doi: 10.1006/hbeh.1999.1548. [DOI] [PubMed] [Google Scholar]
- 24.Insel T. R., Young L. J. Curr. Opin. Neurobiol. 2000;10:784–789. doi: 10.1016/s0959-4388(00)00146-x. [DOI] [PubMed] [Google Scholar]
- 25.Gingrich B., Liu Y., Cascio C., Wang Z., Insel T. R. Behav. Neurosci. 2000;114:173–183. doi: 10.1037//0735-7044.114.1.173. [DOI] [PubMed] [Google Scholar]
- 26.Wang Z., Yu G., Cascio C., Liu Y., Gingrich B., Insel T. R. Behav. Neurosci. 1999;113:602–611. doi: 10.1037//0735-7044.113.3.602. [DOI] [PubMed] [Google Scholar]
- 27.Lim M. M., Nair H. P., Young L. J. J. Comp. Neurol. 2005;487:75–92. doi: 10.1002/cne.20532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aragona B. J., Liu Y., Yu Y. J., Curtis T. J., Detwiler J. M., Insel T. R., Wang Z. Nat. Neurosci. 2006;9:133–139. doi: 10.1038/nn1613. [DOI] [PubMed] [Google Scholar]
- 29.Nair H. P., Young L. J. Physiology. 2006;21:146–152. doi: 10.1152/physiol.00049.2005. [DOI] [PubMed] [Google Scholar]
- 30.Bachner-Melman R., Dina C., Zohar A. H., Constantini N., Lerer E., Hoch S., Sella S., Nemanov L., Gritsenko I., Lichtenberg P., et al. PloS Genet. 2005;1:394–403. doi: 10.1371/journal.pgen.0010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Müller A. E., Thalmann U. Biol. Rev. 2000;75:405–435. doi: 10.1017/s0006323100005533. [DOI] [PubMed] [Google Scholar]
- 32.Heckel G., Burri R., Fink S., Desmet J.-F., Excoffier L. Evolution. 2005;59:2231–2242. [PubMed] [Google Scholar]
- 33.Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hall T. A. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 35.Fink S., Excoffier L., Heckel G. Mol. Ecol. 2004;13:3501–3514. doi: 10.1111/j.1365-294X.2004.02351.x. [DOI] [PubMed] [Google Scholar]
- 36.Saitou N., Nei M. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 37.Kumar S., Tamura K., Jakobsen I. B., Nei M. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- 38.Swofford D. L. paup*, Phylogenetic Analysis Using Parsimony (*and Other Methods) Sunderland, MA: Sinauer; 1999. Version 4.0B. [Google Scholar]
- 39.Tavaré S., Balding D. J., Griffiths R. C., Donnelly P. Genetics. 1997;145:505–518. doi: 10.1093/genetics/145.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.