Abstract
Nogo isoforms (Nogo-A and -B) have been implicated in regulating neural and cardiovascular functions, such as cell spreading and chemotaxis. Unlike the loop domain (Nogo-66) found in all Nogo isoforms that can interact with a neural-specific Nogo-66 receptor, the receptor for the amino terminus of Nogo-B that mediates vascular function is unknown. Here, we identify a previously uncharacterized Nogo-B receptor specific for the amino terminus of Nogo-B and show that Nogo-B receptor localizes with the ligand Nogo-B during VEGF and wound healing angiogenesis in vivo, mediates chemotaxis in a heterologous expression system and chemotaxis, and 3D tube formation in native endothelial cells. Thus, identification of this receptor may lead to the discovery of agonists or antagonists of this pathway to regulate vascular remodeling and angiogenesis.
Keywords: migration, angiogenesis, reticulon, vascular remodeling, expression cloning
Nogo isoforms are part of a superfamily of proteins called reticulons (1). Nogo-A is a myelin-associated inhibitor of axonal sprouting, Nogo-B is expressed in many cells in culture and is the primary Nogo isoform expressed in blood vessels, and Nogo-C is expressed in the CNS and skeletal muscle (2). Nogo-A and -B isoforms have a common amino terminus for the first 184 aa, and all three isoforms contain a conserved reticulon homology domain (RHD). A 66-aa loop domain termed Nogo-66 in the RHD can interact with a glycosylphosphatidylinositol-linked cell-surface Nogo-66 receptor (NgR) (3), and this receptor mediates, in part, the inhibitory function of Nogo-A on neuronal outgrowth. In contrast to Nogo-A, the amino terminus of Nogo-B (AmNogo-B) promotes the adhesion and chemotaxis of endothelial cells and negatively regulates platelet-derived growth factor-induced chemotaxis in smooth muscle cells (4). The receptor(s) mediating the actions of Nogo-B are unknown but are necessary to elucidate the potential functions of this ligand.
Here, we show that AmNogo-B binds to a previously uncharacterized Nogo-B receptor (NgBR). Structure–function analysis of the amino terminus of Nogo-A (AmNogo-A) and AmNogo-B defines the domains of Nogo-B necessary for binding to NgBR. NgBR colocalizes with the ligand Nogo-B during angiogenesis in vivo and mediates chemotaxis and 3D tube formation of endothelial cells in vitro. Thus, identification of this previously undescribed receptor may lead to the discovery of agonists or antagonists of this pathway to regulate vascular remodeling and angiogenesis.
Results and Discussion
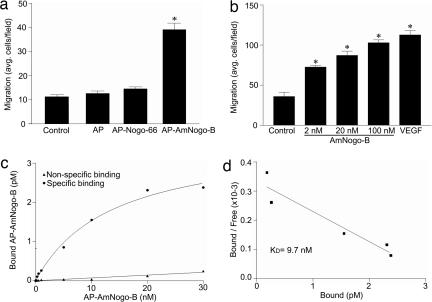
Initially, we examined the migratory response of human umbilical vein endothelial cells (HUVEC) in response to a gradient of recombinant, purified alkaline phosphatase (AP) fusion protein expressing AmNogo-B (Fig. 1a, amino acids 1–200; AP–AmNogo-B) or a recombinant purified AmNogo-B (Fig. 1b). AP alone or an AP fusion of Nogo-66 (AP–Nogo-66) does not promote HUVEC migration, but AmNogo-B dose-dependently increases migration of endothelial cells, similarly in magnitude to VEGF. Next, we examined the binding of AP–AmNogo-B to the surface of endothelial cells. As seen in Fig. 1 c and d, AP–AmNogo-B binds to a specific, saturable, high-affinity binding site on endothelial cells with an estimated Kd of 9.7 nM. We have shown that the expression of NgR is below the limits of detection in vascular cells (4). Thus, the ability of soluble AmNogo-B, but not Nogo-66, to induce chemotaxis and the specific binding of AP–AmNogo-B to endothelial cells suggests that AmNogo-B may interact with a unique receptor.
Fig. 1.
Interaction of AmNogo-B with cell-surface receptor in HUVEC. (a) Comparison of Nogo-B domains on HUVEC migration. Cell migration was examined in modified Boyden chambers by using purified recombinant AP, AP fusion Nogo-B loop domain (AP–Nogo-66), and AP fusion AmNogo-B domain (AP–AmNogo-B, 10 nM each). (b) Dose-dependent migration in response to purified recombinant AmNogo-B or VEGF (1.1 nM). (c) Surface binding of AP–AmNogo-B to native HUVEC. Nonspecific binding was determined by measuring binding in the presence of a 100-fold molar excess of recombinant AmNogo-B. (d) The Scatchard plot of data from c with a calculated Kd = 9.7 nM.
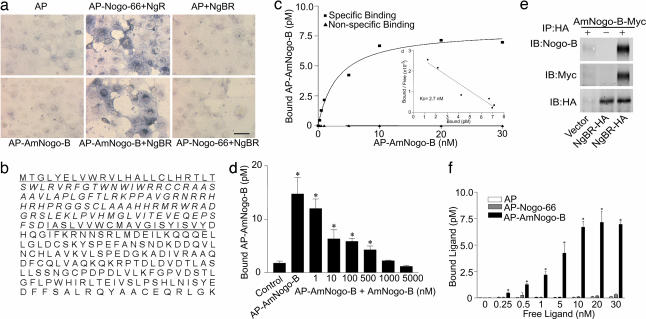
To identify a specific receptor for AmNogo-B, we used a recombinant AP–AmNogo-B fusion protein to screen a cDNA expression library from human heart (0.5 × 106 independent clones) transfected into COS or CHO cells. Cells were transfected with pools (5,000 clones) of cDNA and screened by using AP versus AP–AmNogo-B binding and detection of the AP product by near-infrared cell imaging (Li-Cor, Lincoln, NE) or AP activity assays. As seen in Fig. 2a (first column of wells, Upper and Lower), COS cells do not bind AP or AP–AmNogo-B but selectively bind AP–Nogo-66 when transfected with the cDNA encoding NgR (Fig. 2a Upper, second column). After several rounds of screening, amplification, and sib selection, a single cDNA was isolated and, when transfected into COS cells, afforded the binding of AmNogo-B (Fig. 2a Lower, second column) but not AP (Fig. 2a Upper, third column) or AP–Nogo-66 (Fig. 2a Lower, third column). The isolated cDNA (2,636 bp) encodes an ORF of 293 aa that we define as a NgBR (Fig. 2b). The deduced amino acid sequence reveals a signal peptide sequence (23 aa) (Fig. 2b, underlined), a putative ectodomain of 93 aa, and a Type 1A transmembrane domain of 19 aa (Fig 2b, doubly underlined), followed by a cytoplasmic domain of 158 aa. Analysis of the NgBR cDNA sequence and translation product revealed 100% identity to a full-length sequence listed as a hypothetical protein in humans (BC013026.2 or NM138459.2) found on human chromosome 6q 22.31, with orthologs in mouse, chicken, and zebrafish. Comparing NgBR with other sequences in the National Center for Biotechnology Information database revealed that regions of the cytoplasmic domain have a high degree of similarity (49%) to the cis-prenyltransferase family of lipid-modifying enzymes, such as human cis-isoprenyltransferase and bacterial undecaprenyl phosphate synthase. However, direct assays for lipid transferase activity were negative (Fig. 6, which is published as supporting information on the PNAS web site, with mouse liver as a positive control), suggesting that the NgBR may act as a scaffold for the binding of isoprenyl lipids and/or prenylated proteins. Mechanistically, NgBR may signal via the recruitment/sequestration of prenylated proteins, such as Ras, joining the growing list of other proteins that use prenyl groups in proteins to facilitate binding to partners, such as prenylated Ras binding to galectin-1 (5, 6) or cGMP phosphodiesterase δ (7) and prenylated Cdc42 (8)or Rac2 (9)interacting with RhoGDI. However, additional studies are necessary to test this assertion.
Fig. 2.
Identification and characterization of AmNogo-B receptor. (a) COS-7 cells were transfected with an expression vector encoding human NgBR or NgR, followed by binding of AP, AP–AmNogo-B, or AP–Nogo-66. (Scale bar, 100 μm.) (b) The deduced amino acid sequence of NgBR. (c) Surface binding of AP–AmNogo-B to CHO cells expressing NgBR. Nonspecific binding was determined as above. (c Inset) Scatchard plot of data from c with a calculated Kd of 2.74 nM. (d) Binding of 10 nM AP–AmNogo-B to CHO cells expressing NgBR and competition by increasing concentration of purified AmNogo-B. Values are represented as mean ± SEM (∗, P < 0.01; n = 4). (e) Extracts of CHO cells expressing control vector or NgBR-HA were incubated with or without purified AmNogo-B (+ or −) and then immunoprecipitated with Anti-HA matrix. Bound proteins were detected by immunoblotting. (f) Preferential binding of AP–AmNogo-B in CHO cells expressing NgBR. Increasing concentrations of recombinant AP, AP–Nogo-66, or AP–AmNogo-B were incubated with CHO cells expressing NgBR for 2 h at 4°C. Each value represents mean ± SEM (∗, P < 0.05; n = 4).
Transient transfection of the NgBR cDNA into CHO cells allows the specific, saturable high-affinity binding similar to that seen in endothelial cells, with an estimated Kd of 2.7 nM (Fig. 2c and Inset). Moreover, purified AmNogo-B, dose-dependently displaces the binding of AP–AmNogo-B, consistent with these ligands competing for the same binding site (Fig. 2d). Next, we examined the ability of the transfected receptor to bind AmNogo-B in vitro. Lysates prepared from vector or NgBR-hemagglutinin (HA)-transfected CHO cells were mixed with purified AmNogo-B-myc and HA-tagged receptor immunopurified. As seen in Fig. 2e, AmNogo-B (detected with Nogo-B and anti-myc antisera) interacted with lysates only when the receptor was expressed (compare lane 1 with lane 3). Stable expression of the receptor in CHO cells permits the binding of AP–AmNogo-B but not AP or AP–Nogo-66 (Fig. 2f).
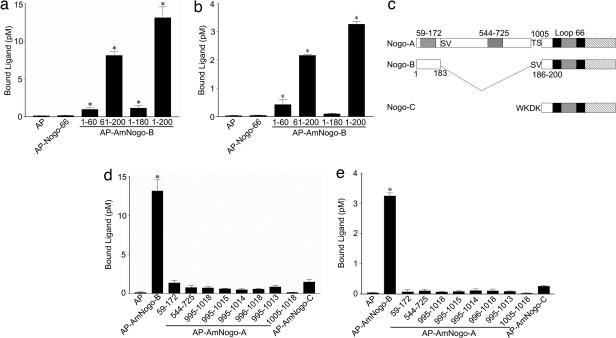
Next, we characterized the regions of AmNogo-B responsible for binding to NgBR in cells stably expressing the receptor and HUVEC. AP, AP–Nogo-66 or AP–AmNogo-B (amino acids 1–180) did not bind to the receptor, whereas constructs expressing AP–Nogo-B (61–200) or full-length AP–Am Nogo-B did, suggesting that amino acids between 180 and 200 were critical for binding in NgBR-expressing cells (Fig. 3a) and in HUVEC (Fig. 3b), but the stretch of aspartates and glutamates from amino acids 32–51 found in AmNogo-A and -B were not critical. Interestingly, amino acids 1–183 are identical between Nogo-A and -B (Fig. 3c). In Nogo-A, there is a unique exon encoding amino acids 184–1,004, thereby generating a much longer amino terminus. Moreover, amino acids 1,005–1,019 in Nogo-A are identical to amino acids 186–200 in Nogo-B and to amino acids 12–26 in Nogo-C. Previous studies with AmNogo-A have identified two biologically active domains, amino acids 59–172, a region that reduces spreading of multiple cell types, and amino acids 544–725, a region that is neural-specific in reducing axon spreading. As seen in Fig. 3 d and e (in NgBR-expressing cells and HUVEC, respectively) AP–Nogo-A (59–172) and AP–Nogo-A (544–725) do not bind to NgBR, suggesting that these ligands likely bind to a unique, unidentified receptor. In addition, constructs expressing several regions of Nogo-A overlapping with regions of identify in Nogo-B, as well as the amino terminus of Nogo-C (amino acids 1–26), do not bind to NgBR strongly, suggesting that the cloned receptor that expressed in HUVEC is specific for AmNogo-B as a ligand. However, we cannot rule out the possibility that NgBR may serve as a coreceptor for native full-length Nogo-A.
Fig. 3.
Comparison of Nogo domains and specificity of NgBR receptor. (a) Surface binding of AP–AmNogo-B domains to CHO cells expressing NgBR. Ligands (10 nM) were incubated with CHO cells expressing NgBR for 2 h at 4°C. Each value represents mean ± SEM (n = 4). (b) Surface binding of AP–AmNogo-B domains to HUVEC. Ligands (10 nM) were incubated with HUVEC for 2 h at 4°C. Each value represents mean ± SEM (n = 4). (c) Alignments of three Nogo isoforms. (d) Surface binding of AP–AmNogo-A domains or AP–AmNogo-C to CHO cells expressing NgBR. Ligands (10 nM) were incubated with CHO cells expressing NgBR for 2 h at 4°C. Each value represents mean ± SEM (n = 4). (e) Surface binding of AP–AmNogo-A domains or AP–AmNogo-C to HUVEC. Ligands (10 nM) were incubated with HUVEC for 2 h at 4°C. Each value represents mean ± SEM (n = 4). ∗, P < 0.05.
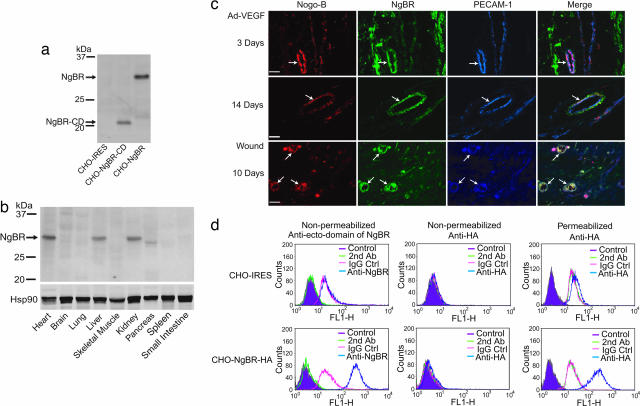
Next, we developed a polyclonal Ab directed at the putative ectodomain of the receptor to examine the expression of NgBR in cells and tissue. CHO cells transfected with vector alone do not exhibit immunoreactivity, whereas CHO cells expressing a truncated form of NgBR lacking the cytoplasmic domain (missing amino acids 181–293; NgBR-CD) and WT NgBR yield the predicted molecular masses of 21 and 30 kDa, respectively (Fig. 4a). NgBR protein is highly expressed in mouse heart, liver, kidney, and pancreas (Fig. 4b). Next, because Nogo-B regulates vascular remodeling in vivo and promotes endothelial cell adhesion and migration in vitro, we examined the patterns of Nogo-B and NgBR in two models of angiogenesis: intradermal injection of adenoviral VEGF into the ear (Ad-VEGF) and healing of full-thickness wounds (Fig. 4c). Nogo-B (Fig. 4c, column 1, red) and NgBR (Fig. 4c, column 2, green) are present in both endothelial cells and pericytes in a subset of growing (day 3, Top) and more mature (day 14, Middle) angiogenic vessels. Immunoreactive Nogo-B and NgBR are found in a subset of PECAM-1-positive endothelial cells at both time points (Fig. 4c, see pink areas of colabeling in merge). In a different model of angiogenesis, Nogo-B and NgBR colocalize with PECAM-1-positive endothelial cells after 10 days of wound healing. Finally, to examine the topography of NgBR, we performed FACs analysis of CHO cells expressing vector alone [CHO-internal ribosome entry site (IRES)] or CHO cells stably expressing full-length NgBR with a C- terminal HA tag (CHO-NgBR-HA) (Fig. 4d). As seen in Fig. 4d Upper, there is nonspecific labeling with all antibodies tested in both nonpermeabilized and permeabilized CHO-IRES cells. In contrast, as seen in Fig. 4d Lower, in nonpermeabilized cells expressing NgBR-HA, anti-NgBR detects a surface epitope (Fig. 4d Lower Left), and labeling with anti-HA is identical to nonimmune IgG control antisera, defining the N-terminal epitope on the cell surface. Permeabilization of the cells permitted detection of the C-terminal HA epitope, consistent with the predicted topography of the cloned cDNA with the N terminus extracellular and the C terminus intracellular.
Fig. 4.
Distribution of the NgBR protein in mouse tissues and presence during tissue angiogenesis. (a) Characterization of polyclonal Ab against the putative ectodomain of NgBR. The Ab shows only the immunoreactivity to the truncated form of NgBR lacking the cytoplasmic domain (missing amino acids 181–293; NgBR-CD) and wild-type NgBR at the predicated molecular masses of 21 and 30 kDa, respectively. (b) Distribution of the NgBR protein in mouse tissues. The positions of protein markers (kDa) are at the left. (c) Localization of Nogo-B, NgBR, and PECAM-1 in angiogenic blood vessels (arrows). Frozen sections of ear angiogenesis (7 μm thick) or paraffin sections (6 μm thick) of wound-healing-associated angiogenesis were stained with anti-Nogo-B Ab and Alexa Fluor 568-conjugated secondary Ab (red), anti-NgBR Ab and Alexa Fluor 488-conjugated secondary Ab (green), and anti-PECAM-1 Ab and Alexa Fluor 647-conjugated secondary Ab (blue). Magnification, ×400. (Scale bar, 30 μm.) (d) Surface immunostaining of the NgBR receptor by flow cytometry. Vector-transduced cells (CHO-IRES, Upper) or cells expressing NgBR-HA (CHO-NgBR-HA, Lower) were either unmanipulated (magenta), incubated with secondary Ab alone (orange), incubated with IgG isotype control Ab (pink) or anti-NgBR or anti-HA (blue), and sorted by FACS. The receptor was detected by anti-NgBR (Lower Left) as determined by a rightward shift in the histogram. In nonpermeabilized cells, the anti-HA did not detect the receptor.
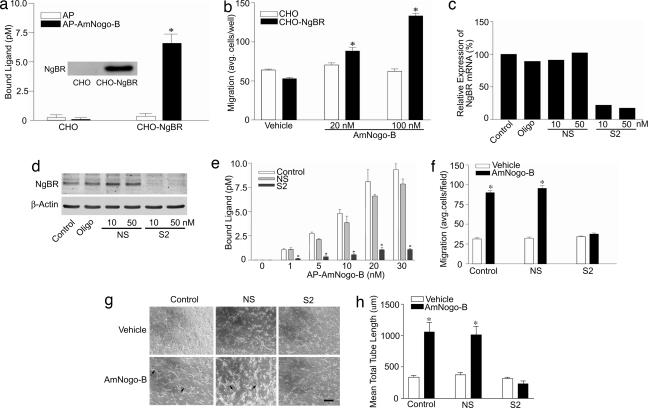
To examine the function of NgBR, we examined AmNogo-B-mediated chemotaxis in CHO cells stably transfected with the cDNA for the NgBR. As seen in Fig. 5a, there is minimal binding of AP and AP–AmNogo-B to CHO cells, whereas stable transfection of CHO cells with the cDNA for NgBR permits binding of AP–AmNogo-B (see Fig. 5a Inset for levels of NgBR expressed). Next, CHO cells or CHO cells expressing NgBR were placed into a Boyden chamber and the chemotactic response to soluble AmNogo-B examined. As seen in Fig. 5b, transfection of NgBR is required for AmNogo-B-mediated chemotaxis, suggesting that the cloned receptor is essential for ligand binding and signal transduction. To examine the role of the endogenous receptor, we developed an RNA interference strategy [short interfering RNA (siRNA)] to reduce the expression of NgBR in cells. Sequence 1 (S1) targets the coding region of the mRNA and sequence 2 (S2) targets the 3′ untranslated region (see Fig. 7, which is published as supporting information on the PNAS web site, for characterization of siRNAs). Treatment of HUVEC with S2 siRNA but not nonsilencing (NS) RNA, reduces the levels of NgBR mRNA (determined by quantitative PCR; Fig. 5c), NgBR protein (Fig. 5d), the binding of AP–AmNogo-B (Fig. 5e), and abolishes AmNogo-B-mediated chemotaxis of HUVEC (Fig. 5f). We also examined the effects of AmNogo-B in an in vitro model of tubulogenesis by placing HUVEC into 3D culture. AmNogo-B stimulated an increase in tube formation (phase-contrast images in Fig. 5g and quantified in Fig. 5h), an effect attenuated by a reduction in NgBR by siRNA S2. These data document that endogenous NgBR is required for the in vitro angiogenic actions of AmNogo-B in endothelial cells.
Fig. 5.
NgBR is required for the chemotactic actions of AmNogo-B. (a) Establishment of stable CHO cell lines expressing NgBR. (a Inset) The levels of NgBR protein by Western blotting. (b) NgBR is required for AmNogo-B-induced chemotaxis in a Boyden chamber assay (n = 4). (c and d) S2 siRNA transfection down-regulates NgBR mRNA and protein levels in HUVEC cells compared with no-treatment control, oligofectamine control (Oligo), and NS control. (e and f) S2 siRNA transfection abolishes AP–AmNogo-B binding and chemotactic response of HUVEC to soluble AmNogo-B. (g and h) NgBR is necessary for AmNogo-B-induced tube formation. After treatment with either NS or S2 siRNA, HUVEC were suspended in type I collagen gels and treated with vehicle or 80 nM AmNogo-B. After 24 h of incubation, cells were photographed by using openlab (g), and total network length was quantified (h). Representative images (g) are shown for two independent experiments (n = 8 in h). (Scale bar, 20 μm.) ∗, P < 0.05.
These data define a signaling receptor for AmNogo-B that is expressed in a variety of tissues and in endothelial cells. Experiments showing that NgBR is necessary and sufficient for binding and migration in a heterologous expression system in CHO cells, as well as in native endothelial cells, demonstrate that the cloned cDNA is functionally competent to signal upon binding Nogo-B. We have demonstrated that Nogo-B is highly expressed in intact blood vessels, present in caveolae/lipid rafts isolated from cultured endothelial cells, and exerts a role in vascular remodeling as an endogenous regulator of vascular cell functions after injury (4). Precisely how Nogo-A or -B ligands are released from cells is unknown, and this concept is complicated by many predicted and experimentally tested topographies of Nogo isoforms (4, 10–12) but may occur because of low level expression on the cell surface or be released after injury or damage. Identification of how AmNogo-B couples to NgBR, and the development of agonists and/or antagonists to block AmNogo-B–NgBR interaction will facilitate elucidating the contribution of Nogo-B/NgBR to other cardiovascular functions.
Materials and Methods
Cell Culture.
HUVEC were cultured in M199 with 20%FBS and endothelial cell growth supplement (ECGS). HEK293T and COS-7 cells were cultured in high-glucose DMEM with 10% FBS. CHO cells were cultured in MEM-α with 5% FBS.
Real-Time RT-PCR.
Total RNA from cells was isolated by using the RNeasy kit (Qiagen, Valencia, CA). Reverse transcription was then performed by using 100 ng of RNA and the Superscript First-Strand Synthesis System kit (Invitrogen). Real-time PCR analysis was done with the iCycler iQ detection system using the iQ SYBR green Supermix kit (Bio-Rad). The NgBR mRNA level was normalized by house keeping gene 18S. We used the following primers: for NgBR, forward (5′-TGCCAGTTAGTAGCCCAGAAGCAA-3′) and reverse (5′-TGATGTGCCAGGGAAGAAAGCCTA-3′); for 18S, forward (5′-CGGCGAC GACCCATTCGAAC-3′) and reverse (5′-GAATCGAACCCTGATTCCCC GTC-3′).
AmNogo-B Recombinant Proteins.
To express AmNogo-B, the human Nogo-B cDNA of residues 1–200 was ligated into pcSecTag2-HygroC (Invitrogen) by using the Ig κ-chain signal peptide of pSecTag2 with an in-frame Myc-His tag or pcAP-5 in frame with the signal sequence, His tag, and placental AP coding region. The resultant plasmid DNA was transfected into HEK293T cells, and secreted AmNogo-B or AP–AmNogo-B was purified with Ni-affinity chromatography.
AmNogo-B Receptor-Binding Assays and Expression Cloning.
To detect AP–AmNogo-B binding, cultures were washed with Hanks’ balanced salt solution (HBSS) containing 20 mM Hepes, pH 7.5, and 1 mg/ml BSA (HBH). The plates were then incubated with AP–AmNogo-B in DMEM containing 20 mM Hepes, pH 7.5, and 1 mg/ml BSA for 2 h at 4°C. The bound AP-AmNogo-B was detected by using the Blue Substrate kit (Vector Laboratories). The blue staining was examined by using the ODYSSEY Infrared Imaging System (Li-Cor, Lincoln, NE) and confirmed by microscopy. Alternatively, the bound AP–AmNogo-B was extracted with Triton X-100, and AP activity was colorimetrically quantified by using p-nitro-phenyl phosphate (Sigma) as substrate after heat inactivation of endogenous AP as described (13, 14). Nonspecific binding was determined by measuring binding in the presence of a 100-fold molar excess of recombinant AmNogo-B lacking the AP fusion protein. Specific binding was determined by subtracting the nonspecific binding from total binding. For determination of apparent Kd, the binding of AP–AmNogo-B was measured in triplicate as described above and the Kd value quantified by using the Scatchard plot program of GraphPad prism (one-site binding, linear regression) using the ratio of bound ligand to free ligand as the y axis and bound ligand (pM) as the x axis.
For expression cloning of AmNogo-B receptor, pools of 5,000 arrayed clones from a human heart cDNA library (OriGene Technologies, Rockville, MD) were transfected into COS-7 cells, and AP–AmNogo-B binding was assessed. We isolated single NgBR cDNA clones by sib selection and sequenced them. A NgBR-HA was created in pIRESneo vector (BD, Palo Alto, CA) with HA tag at the carboxyl-terminal. To access the physical interaction of NgBR with AmNogo-B, we incubated 50 μg of solubilized extracts of CHO cells expressing control vector (pIRES-neo) or NgBR-HA with 25 μg of purified AmNogo-B or buffer for 2 h at room temperature. The HA-tagged NgBR was immunoisolated with anti-HA immunobeads (Roche, Indianapolis, IN) and associated proteins analyzed by Western blotting.
Ear Angiogenesis and Wounding Model.
Adenovirus encoding murine VEGF-A 164 (109 viral particles) were injected intradermally into the right ears of CD1 mice. The left ears were injected with the same amount of control virus encoding β-gal. At the different time points, animals were killed and the ears removed and embedded in optimal cutting temperature (OCT) compound (Tissue-Tek; Sakura, Torrance, CA). A full-thickness wound (≈5-mm diameter) was created by excising the skin and the underlying panniculus carnosus in C57Bl6 mice. At 10 days after wounding, skin biopsy specimens from six mice were collected for immunohistochemistry analysis. Frozen sections (7 μm) were immunostained with goat polyclonal anti-Nogo-B (Santa Cruz Biotechnology), rabbit polyclonal anti-NgBR (Imgenex, San Diego, CA), and rat monoclonal anti-mouse PECAM-1 (BD Biosciences–Pharmingen) primary antibodies and Alexa Fluor 568 donkey anti-goat-, Alexa Fluor 488 donkey anti rabbit-, and Alexa Fluor 647 chicken anti-rat-conjugated secondary antibodies (Invitrogen).
Migration Experiments.
A modified Boyden chamber was used (Costar transwell inserts; Corning). The transwell inserts were coated with a solution of 0.1% gelatin (Sigma) in PBS at 4°C overnight and then air-dried. VEGF at 50 ng/ml (1.1 nM) or recombinant AmNogo-B at various concentrations dissolved in medium 199 containing 0.1% BSA was added in the bottom chamber of Boyden apparatus. HUVEC (2 × 105 cells) suspended in a 100-μl aliquot of medium 199 containing 0.1% BSA was added to the upper chamber. After 5 h incubation, cells on both sides of the membrane were fixed and stained with a Diff-Quik staining kit (Baxter Healthcare, Miami, FL). The average number of cells from five randomly chosen high-power (×400) fields on the lower side of the membrane was counted.
NgBR Ab Production.
The peptide [CRNRRHHRHPRG, residues from 64–74 (carboxy terminal from the signal peptide)] was used to immunize rabbits (Imgenex). The antiserum was purified by using the same peptide-conjugated SulfoLink Coupling Gel (Pierce) and diluted (1:500) for immunoblots.
Western Blot Analysis.
COS-7 or CHO cells were transfected with NgBR-HA plasmid DNA by using Lipofectamine 2000 (Invitrogen). Expression of NgBR-HA was detected by using anti-HA (Roche) and NgBR (Imgenex), respectively, β-actin (Sigma) or Hsp90 (BD Biosciences–Pharmingen) was used to control for loading.
siRNA Transfection.
NgBR siRNA1(S1 forward: CCAGAAUUUGCAAAUAGUA, S1 reverse: UACUAUUUGCAAAUUCUGG) and NgBR siRNA2 (S2 forward: GGAAAUACAUAGACCUACA, S2 reverse: UGUAGGUCUAUGUAUUUCC) siRNA oligonucleotides with 3′ dTdT overhangs were synthesized by Qiagen. Control siRNA in experiments refers to a nonsilencing siRNA (NS forward: UUCUCCGAACGUGUCACGU, NS reverse: ACGUGACACGUUCGGAGAA) designed and synthesized by Qiagen. HUVEC and CHO stable cell lines were transfected with siRNA by using Oligofectamine (Invitrogen). Quantification of NgBR mRNA and protein, ligand binding assay, and migration assay were performed at 72 h after transfection.
3D Tube-Formation Assay.
HUVEC were resuspended (final concentration 1 × 106) in a mixture containing rat tail type I collagen (1.5 mg/ml), 1/10 volume 10× M199, and 1M Hepes and neutralized with NaOH. Droplets (0.1 ml each) of the cell/collagen mixture were placed in cell culture dishes and allowed to polymerize for 15 min at 37°C. Growth medium containing either vehicle or agonist was then added to each well. HUVEC were allowed to form tube-like structures for 1–2 days. To evaluate tube formation in 3D cultures, cells were photographed by using the program openlab (Improvision, Lexington, MA), and total-network length (defined as an elongation of cell into tube-like structures typically seen in 3D cultures) was quantified in five fields for each replicate per experiment by using the measurement tools provided with openlab.
Isoprenyl Lipid Transferase Activity Assay.
NgBR was immunoisolated from CHO cells expressing NgBR-HA by using anti-HA matrix beads. Lipid transferase activity was measured by determining the amount of [1-14C]IPP (isopentenyl pyrophosphate) incorporated into butanol-extractable polyprenyl diphosphates as described (15). The activity was assayed in a 50-μl reaction containing 50 mM Hepes, pH 7.5, 2 mM MgCl2, 5 mM KF, 1 mM DTT, 0.5% CHAPS, 50 μM [1-14C]IPP [0.15 μCi (1 Ci = 37 GBq) per reaction], and 50 μM allylic isoprenoid diphosphate (such as FPP, farnesyl diphosphate; GGPP, geranylgeranyl diphosphate). The reaction was started by the addition of 1 μg of protein and allowed to proceed for 20 min at 37°C. Mouse liver extract (1 μg) was used as a positive control for the assay. The reaction was stopped by the addition of 0.5 ml of 1-butanol saturated with water, followed by the addition of 0.5 ml of 2 M KCl. An aliquot of the butanol phase was removed for scintillation counting using SafeScint scintillation mixture (American Bioanalytic, Natick, MA).
Supplementary Material
Acknowledgments
We thank Dr. Joseph Schlessinger for comments on the manuscript, Dr. Dean Crick for helpful discussions regarding cis-isoprenyltransferases, Weihua Huang for sequence alignments, and Dr. Themis Kyriakides for tissue blocks from wound-healing experiments. This work was supported by grants from the National Institutes of Health and the Sandler Program for Asthma Research (to W.C.S.) and from Phillip Morris (to R.Q.M.).
Abbreviations
- AmNogo-A
amino terminal of Nogo-A
- AmNogo-B
amino terminal of Nogo B
- AP
alkaline phosphatase
- HA
hemagglutinin
- HUVEC
human umbilical vein endothelial cells
- NgR
Nogo receptor
- NgBR
Nogo-B receptor
- NS RNA
nonsilencing RNA
- S1
sequence 1
- S2
sequence 2
- siRNA
short interfering RNA.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Oertle T., Schwab M. E. Trends Cell Biol. 2003;13:187–194. doi: 10.1016/s0962-8924(03)00035-7. [DOI] [PubMed] [Google Scholar]
- 2.Oertle T., Huber C., van der Putten H., Schwab M. E. J. Mol. Biol. 2003;325:299–323. doi: 10.1016/s0022-2836(02)01179-8. [DOI] [PubMed] [Google Scholar]
- 3.Fournier A. E., GrandPre T., Strittmatter S. M. Nature. 2001;409:341–346. doi: 10.1038/35053072. [DOI] [PubMed] [Google Scholar]
- 4.Acevedo L., Yu J., Erdjument-Bromage H., Miao R. Q., Kim J. E., Fulton D., Tempst P., Strittmatter S. M., Sessa W. C. Nat. Med. 2004;10:382–388. doi: 10.1038/nm1020. [DOI] [PubMed] [Google Scholar]
- 5.Rotblat B., Niv H., Andre S., Kaltner H., Gabius H. J., Kloog Y. Cancer Res. 2004;64:3112–3118. doi: 10.1158/0008-5472.can-04-0026. [DOI] [PubMed] [Google Scholar]
- 6.Elad-Sfadia G., Haklai R., Ballan E., Gabius H. J., Kloog Y. J. Biol. Chem. 2002;277:37169–37175. doi: 10.1074/jbc.M205698200. [DOI] [PubMed] [Google Scholar]
- 7.Zhang H., Liu X. H., Zhang K., Chen C. K., Frederick J. M., Prestwich G. D., Baehr W. J. Biol. Chem. 2004;279:407–413. doi: 10.1074/jbc.M306559200. [DOI] [PubMed] [Google Scholar]
- 8.Hoffman G. R., Nassar N., Cerione R. A. Cell. 2000;100:345–356. doi: 10.1016/s0092-8674(00)80670-4. [DOI] [PubMed] [Google Scholar]
- 9.Scheffzek K., Stephan I., Jensen O. N., Illenberger D., Gierschik P. Nat. Struct. Biol. 2000;7:122–126. doi: 10.1038/72392. [DOI] [PubMed] [Google Scholar]
- 10.GrandPre T., Nakamura F., Vartanian T., Strittmatter S. M. Nature. 2000;403:439–444. doi: 10.1038/35000226. [DOI] [PubMed] [Google Scholar]
- 11.Dodd D. A., Niederoest B., Bloechlinger S., Dupuis L., Loeffler J. P., Schwab M. E. J. Biol. Chem. 2005;280:12494–12502. doi: 10.1074/jbc.M411827200. [DOI] [PubMed] [Google Scholar]
- 12.Voeltz G. K., Prinz W. A., Shibata Y., Rist J. M., Rapoport T. A. Cell. 2006;124:573–586. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
- 13.Flanagan J. G., Leder P. Cell. 1990;63:185–194. doi: 10.1016/0092-8674(90)90299-t. [DOI] [PubMed] [Google Scholar]
- 14.Flanagan J. G., Cheng H. J. Methods Enzymol. 2000;327:198–210. doi: 10.1016/s0076-6879(00)27277-7. [DOI] [PubMed] [Google Scholar]
- 15.Ericsson J., Greene J. M., Carter K. C., Shell B. K., Duan D. R., Florence C., Edwards P. A. J. Lipid Res. 1998;39:1731–1739. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.