Abstract
The transcription factor E2F1 is known to regulate cell proliferation and has been thought to modulate tumorigenesis via this mechanism alone. Here we show that mice deficient in E2F1 exhibit enhanced angiogenesis. The proangiogenic phenotype in E2F1 deficiency is the result of overproduction of vascular endothelial growth factor (VEGF) and is prevented by VEGF blockade. Under hypoxic conditions, E2F1 down-regulates the expression of VEGF promoter activity by associating with p53 and specifically down-regulating expression of VEGF but not other hypoxia-inducible genes, suggesting a promoter structure context-dependent regulation mechanism. We found that the minimum VEGF promoter mediating transcriptional repression by E2F1 features an E2F1- binding site with four Sp-1 sites in close proximity. These data disclose an unexpected function of endogenous E2F1: regulation of angiogenic activity via p53-dependent transcriptional control of VEGF expression.
Keywords: E2F, endothelial cell
The coordinated regulation of cell cycle progression and apoptosis is critical for the maintenance of homeostasis in living organisms. In mammals, the intimate relationship between cell cycle and programmed cell death is well illustrated by the evolution of our understanding of the role of the transcription factor E2F1. Initially described as an inducer of cell cycle progression (1–4), the generation of an E2F1 deficient mouse was anticipated to result in a hypoplastic, potentially lethal phenotype. In contrast, these mice exhibit a striking combination of tumors and tissue atrophy (5–7).
Vascular homeostasis is also dependent on appropriate regulation of vascular cell cycle and apoptosis. Vascular injury, induced by tissue ischemia or vascular trauma, for example after balloon angioplasty, induces the local proliferation and migration of endothelial cells required to restore micro- and macrovascular integrity. Dysregulation of these processes can have severe consequences. For example, excessive proliferation of endothelial cells is associated with tumor growth and metastasis (8), whereas a deficient endothelial proliferative response to ischemia can result in tissue necrosis. The role of E2F1 in regulating vascular proliferation remains controversial (9–13).
We have previously shown that tumor necrosis factor α (TNF-α), expressed at sites of vascular injury, suppresses endothelial cell (EC) proliferation by repressing E2F1 activity (11). More recently, we demonstrated that ectopic E2F1 overexpression at sites of angioplasty-induced injury promotes EC growth and suppresses apoptosis, thereby promoting functional endothelial recovery (12). In contrast, others (9) have shown that inhibition of E2F signaling in venous bypass grafts prevents the development of occlusive thickening of the vessel wall highlighting the complex role of E2F signaling in vascular biology.
To further define the role of E2F1 in EC proliferation and differentiation, we initiated a series of experiments in mice deficient in E2F1 (E2F1−/−) to evaluate angiogenesis, a process critically dependent on endothelial cell cycle control and apoptosis. Based on our prior data, we hypothesized that the absence of E2F1 would impair EC proliferation and result in an impaired angiogenic response to injury.
Results and Discussion
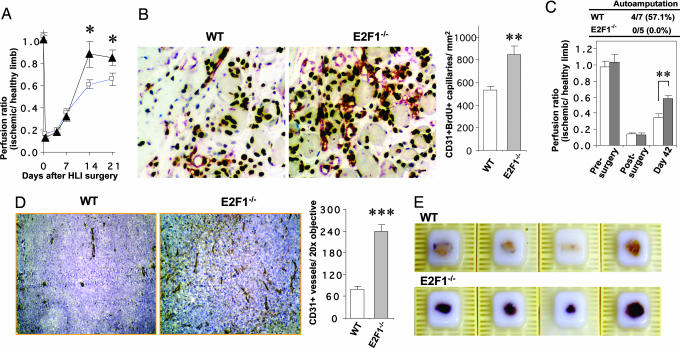
We induced hind limb ischemia (HLI) in E2F1 deficient (E2F1−/−) and control wild-type (WT) mice. The E2F1−/− mice exhibited significantly faster recovery of blood perfusion after surgery than WT mice (P < 0.05 at day 14 and day 21; Fig. 1A). Using CD31 and BrdU double labeling, we confirmed a significantly higher density of newly formed capillaries in the ischemic limbs of E2F1−/− mice than in control mice (P < 0.01 at day 14, n = 5; Fig. 1B). After HLI in older (20 month old) animals, in which the challenge of ischemic recovery is greater, the ischemic limbs of E2F1−/− mice exhibited more rapid blood flow recovery and were markedly less likely to experience autoamputation than age-matched WT controls (Fig. 1C), providing further evidence for a significant role for E2F1 in ischemia-induced angiogenesis.
Fig. 1.
E2F1−/− mice exhibit enhanced angiogenic activity. (A) Serial measurement of blood flow recovery after the induction of surgical HLI in WT (□) and E2F1−/− (▴) mice, using laser Doppler perfusion imaging (LDPI) (n = 35 per group; ∗, P < 0.05). (B) (Left and Center) Representative histology of newly formed capillaries in ischemic hind limbs [identified by CD31 (red), BrdU (brown), and hematoxylin/eosin triple staining]. (Magnification: ×400.) (Right) Quantification of CD31+ BrdU+ double-positive capillary density (six randomly chosen fields per ischemic limb were averaged; n = 5 ischemic limbs per group) 14 days after HLI. (C) HLI in old (20 month) mice. (Upper) Autoamputation rate at day 42 after HLI. (Lower) Laser Doppler blood flow recovery (data at day 42 were collected from the nonautoamputated mice) (Open bar, WT; gray bar, E2F1−/−; ∗∗, P < 0.01). (D) (Left) Representative histology of vasculature at day 14 of LLC-1 tumor challenge in WT and E2F1−/− mice (CD31, brown and hematoxylin/eosin staining). (Original magnification: ×200.) (Right) Quantification of CD31+ capillary density of tumor tissue at day 14 of tumor challenge in WT or E2F1−/− mice. (Six randomly chosen fields per tumor were averaged; n = 5 tumors per group; ∗∗∗, P < 0.001.) (E) Photograph showing hemorrhagic tumors in E2F1−/− mice.
Because these findings were unexpected, we evaluated neovascularization in a second model to determine whether increased angiogenesis in the absence of E2F1 was a generalized finding or specific to ischemic injury.
We performed a tumor challenge by inoculating the LLC-1 cells into WT and E2F1−/− mice. One strength of this model for our purposes is that it allowed us to study the contribution of angiogenesis by the host ECs to tumor growth (14, 15). We injected Lewis lung carcinoma cells (2 × 106) s.c. in E2F1−/− and WT mice (n = 5 mice per group) and found that tumors grown in E2F1−/− mice were more highly vascularized (P < 0.001 at day 14; Fig. 1D), had higher permeability, and were more likely to demonstrate intratumor hemorrhage (Fig. 1E). These findings provided evidence that increased angiogenesis in the absence of E2F1 is a phenomenon that occurs during both recovery from ischemic injury and during tumor growth, and that E2F1 activity in the vasculature was responsible for at least a portion of the angiogenic potential of tumors.
Given the role of E2F1 as a cell cycle regulator, we considered the possibility that the increased angiogenesis we observed in E2F1−/− mice could be caused by enhanced proliferative activity of endothelial cells. We therefore assessed the activity of primary mouse aortic endothelial cells (MAECs) from E2F1−/− and WT controls and found no difference in proliferation, apoptosis, migration, and tube formation under normoxic conditions (Fig. 5 A–D, which is published as supporting information on the PNAS web site).
To investigate the in vivo response of ECs to an angiogenic stimulus, a cornea micropocket neovascularization assay was performed by using implanted vascular endothelial growth factor (VEGF) pellets. Again, we found no significant difference between E2F1−/− and WT mice in the number or length of newly formed vessels in the cornea (Fig. 5E). Taken together, these data provided evidence that the in vitro phenotype and in vivo response of E2F1 null mice to angiogenic stimuli were similar to those of WT mice and that there were no E2F1-mediated changes in EC phenotype to explain the difference in angiogenesis seen in ischemia and tumor models.
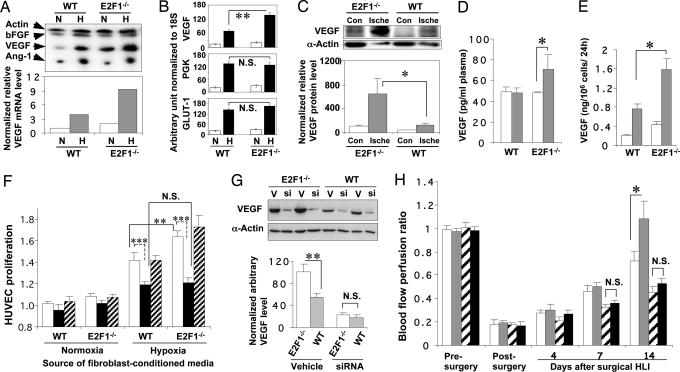
We then considered the possibility that the E2F1 transcription factor might be regulating angiogenesis by modulating the expression of genes that direct new vessel formation. Accordingly, we used a ribonuclease protection assay to screen mRNA expression of various angiogenic factors in cultured primary fibroblasts from E2F1−/− and WT mice. When exposed to hypoxic conditions for 24 h, cells from the WT mice showed a significant increase in VEGF and angiopoietin-1 (Ang-1) but not in basic fibroblast growth factor (bFGF) (or α-actin as a control) (Fig. 2A). E2F1−/− cells have a higher basal level of VEGF transcripts than cells from WT mice and, when exposed to hypoxic conditions for 24 h, demonstrated a higher hypoxic induction of VEGF. The expression of Ang-1, bFGF, and α-actin in E2F1−/− cells was similar to WT cells (Fig. 2A). We confirmed elevated VEGF mRNA expression in E2F1−/− cells with quantitative real-time RT-PCR (Fig. 2B). mRNA expression of phosphoglyceratekinase (PGK) and glucose transporter 1 (GLUT-1), two other hypoxia-inducible genes, was similar in E2F1−/− and WT cells (Fig. 2B).
Fig. 2.
Loss of E2F1 increases VEGF production and VEGF-dependent EC proliferation and angiogenesis. (A) Representative ribonuclease protection assay photograph (Upper) or quantification (Lower). (B) Quantitative real-time RT-PCR (n = 3 per group; ∗∗, P < 0.01) for mRNA expression of hypoxia-inducible genes in primary lung fibroblasts (WT or E2F1−/−) under conditions of normoxia (“N”) or hypoxia (“H”) for 24 h. (C) Representative immunoblotting for VEGF (Upper) and densitometric quantification of relative VEGF levels (Lower) in the ischemic limbs of mice 14 days after HLI (n = 5 per group; ∗, P < 0.05). (D) ELISA for plasma VEGF levels of mice 14 days after tumor challenge (open bar, vehicle; gray bar, LLC-1 tumor; n = 6 per group; *, P < 0.05). (E) ELISA for hypoxia-induced VEGF secretion in E2F1−/− and WT primary fibroblasts (open bar, normoxia; gray bar, hypoxia; n = 4 per group; ∗, P < 0.05). (F) 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay for proliferation of human umbilical vein endothelial cells exposed to 10% FBS/M199 medium supplemented with 20% conditioned medium collected from WT fibroblasts or from E2F1−/− fibroblasts that were cultured under either normoxic or hypoxic conditions for 24 h (open bar, no antibody; gray bar, anti-VEGF antibody at 10 μg/ml; striped bar, control IgG at 10 μg/ml; the two bars indicated by the two ends of each bracket are compared; ∗∗, P < 0.01; ∗∗∗, P < 0.001). This figure is representative of three MTT assays. The contribution of VEGF overexpression to enhanced blood flow recovery in HLI E2F1−/− mice was evaluated by siRNA-mediated VEGF gene knockdown in the ischemic limb. (G) Representative example (Upper) and quantification (Lower) of tissue VEGF levels (Western blotting) at day 7 after HLI surgery and repeated (days 1 and 3) multisite siRNA i.m. injection. (H) Serial laser Doppler perfusion imaging measurement of blood flow recovery after HLI surgery plus in vivo VEGF knockdown with siRNA (open bar, WT with control GFP siRNA; gray bar, E2F1−/− with control GFP siRNA; striped bar, WT with VEGF siRNA; filled bar, E2F1−/− with VEGF siRNA; n = 12 per group; ∗, P < 0.05).
Postnatal angiogenesis comprises an integrated orchestration of physiological processes, including dilation of existing vessels, increased vascular permeability, extracellular matrix degradation, EC proliferation, migration, differentiation and assembly, cord and lumen formation, recruitment of periendothelial cells, and remodeling of the preliminary vessels (16, 17). Such processes require the coordination of a cohort of angiogenic factors (16, 18), among which angiopoietins play a critical role. In this regard, it is interesting to note that, although VEGF stimulates EC proliferation and by itself may be capable of forming fragile and permeable capillaries, Ang-1 stabilizes the vessel walls (19), resulting in long-lasting functional blood vessels (17, 18). Our data indicate that E2F1 specifically inhibits the expression of VEGF but not Ang-1 or bFGF. These data may explain our observation that cells from E2F1-deficient mice showed a rapid increase in highly permeable vessels that ultimately resulted in hemorrhage, possibly because of a relative deficiency of Ang-1.
Next, we assessed in vivo VEGF expression in E2F1−/− vs. WT mice after HLI. At 14 days after surgery, we found significantly higher VEGF protein levels in the ischemic hind limbs of E2F1−/− mice compared with WT mice (P < 0.05, n = 6 per group; Fig. 2C). VEGF levels in the nonischemic limb of E2F1−/− mice were slightly higher but not significantly different from controls (Fig. 2C). The fold induction of VEGF protein in the ischemic limb of E2F1−/− mice was significantly greater than in the ischemic limb of WT mice (5.18 ± 1.52 vs. 2.41 ± 0.33, P < 0.05). In addition, ELISA analysis revealed that E2F1−/− mice had significantly higher plasma VEGF levels than WT mice 14 days after tumor challenge (P < 0.05; Fig. 2D). These data suggested that increased VEGF expression is responsible, at least in part, for the increased angiogenesis seen in E2F1−/− mice.
We also found that cultured cells from E2F1−/− mice produced and secreted more functional VEGF in vitro after exposure to hypoxia than cells from WT controls (P<0.05, n = 3 per group; Fig. 2E). Conditioned medium from the hypoxic E2F1−/− fibroblasts stimulated a higher rate of human umbilical vein endothelial cell (HUVEC) proliferation than medium from hypoxic WT cells (Fig. 2F), whereas pretreatment with VEGF neutralizing antibody completely blocked the increase in HUVEC proliferation induced by conditioned medium from E2F1−/− cells (Fig. 2F). Finally, when we injected small interfering RNA (siRNA) against VEGF locally in the ischemic limbs of E2F1 null and WT mice, there was no significant difference between E2F1−/− and WT mice in the rate of blood perfusion recovery (Fig. 2 G and H). These experiments confirmed that increased VEGF production was responsible for the enhanced angiogenesis observed in E2F1−/− mice.
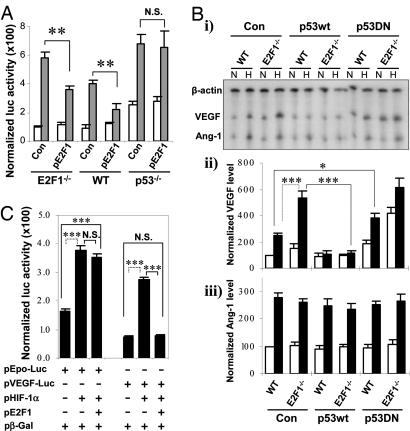
VEGF promoter-luciferase reporter assays confirmed exaggerated hypoxic induction of VEGF promoter activity in E2F1−/− cells, which was normalized when the expression of E2F1 was restored (Fig. 3A), indicating that loss of E2F1 was responsible for the increase in VEGF mRNA expression and suggesting that E2F1 may serve to keep hypoxia-induced VEGF transcription in check.
Fig. 3.
E2F1 represses p53-dependent VEGF mRNA expression. (A) Cotransfection of VEGF promoter (2.6 kb)-luciferase plasmid and E2F1 expression plasmid vs. control plasmid into WT, E2F1−/−, or p53−/− lung fibroblasts, which were subsequently exposed to normoxia (open bar) or hypoxia (gray bar) for 24 h and assayed for luciferase activity (n = 3 per group; ∗∗, P < 0.01). As shown, E2F1 overexpression repressed VEGF promoter activity in E2F1-null cells and WT cells but not in p53-null cells. (B) Ribonuclease protection assay was performed on primary fibroblasts (WT or E2F1−/−) under conditions of normoxia (“N”) or hypoxia (“H”) for 24 h after transfection with a control empty plasmid, p53WT, or p53DN plasmid. The β-actin-normalized value of VEGF or Ang-1 mRNA in WT cells under normoxia conditions was arbitrarily designated as 100, based on which the relative values of other treatment groups were then extrapolated. Representative examples (i) and quantitative analyses for VEGF (ii) and Ang-1 (iii) from three independent experiments are shown. As shown in ii, the increased expression of VEGF in E2F-deficient fibroblasts was nullified by overexpression of WT p53. In contrast, expression of Ang-1 (iii) was not p53-dependent. (C) Differential effects of E2F1 and HIF-1α on VEGF and erythropoietin promoter activity. WT fibroblasts were cotransfected with indicated plasmids and exposed to hypoxia for 24 h before the luciferase and β-galactosidase assays (n = 3 per group; ∗∗∗, P < 0.001).
The p53 tumor suppressor is a transcription factor that regulates the downstream effects of E2F1 for a variety of biological activities (3, 20, 21). p53 activation has also been shown to suppress tumor angiogenesis and VEGF transcription (22). Prior reports have shown that hypoxia induces both E2F1 and p53 with similar kinetics (23). Based on these observations, we wondered whether p53 could be involved in the E2F1-mediated suppression of VEGF. We performed a series of transfections with either a WT p53 plasmid, pCMV-p53 (p53WT), or a dominant-negative mutant p53 plasmid, pCMV-p53mt135 (p53DN) (24). As shown in Fig. 3B (i and ii, right), transfection with p53WT in E2F1−/− cells restored the repression of VEGF mRNA expression under hypoxic conditions. In contrast, transfection of p53DN in WT cells de-repressed VEGF expression (Fig. 3B i and ii), whereas expression of Ang-1 and bFGF were not altered (Fig. 3B i and iii and data not shown). Most notably, hypoxia-induced VEGF promoter activity was significantly repressed by over-expression of E2F1 in E2F1−/− and WT but not p53-deficient (p53−/−) cells (Fig. 3A). These results are consistent with findings reported from other laboratories (25) regarding VEGF expression in p53−/− cells, suggesting that the suppression of VEGF mRNA expression by E2F1 requires intact p53 function.
p53 has been shown to down-regulate VEGF directly through modulating VEGF promoter activity (22, 26) and indirectly through inducing hypoxia-inducible factor (HIF)-1α proteosomal degradation (25, 27, 28). In addition, investigators have mapped a minimal VEGF promoter with Sp1 sites that appear to be essential for p53-mediated repression (22). Our mRNA analysis demonstrated that expression of other classic hypoxia-inducible genes, including Ang-1, PGK, and GLUT-1, was unaltered in E2F1−/− vs. WT cells (Fig. 2 A and B and data not shown), providing evidence that p53-mediated HIF-1α degradation was not responsible for altered VEGF expression.
To further confirm these observations, we performed cotransfection experiments by using plasmids expressing E2F1 and HIF1α and reporter constructs for the VEGF and erythropoietin promoters, both of which are known targets of HIF1α. Under hypoxic conditions, the activity of both promoters was significantly increased by over-expression of HIF1α.
These cotransfection studies revealed that VEGF, but not Epo promoter activity, was significantly down-regulated by cotransfection of pE2F1 (Fig. 3C), providing confirmatory evidence that E2F1 specifically modulates VEGF transcription, but not HIF-1α transactivity in general.
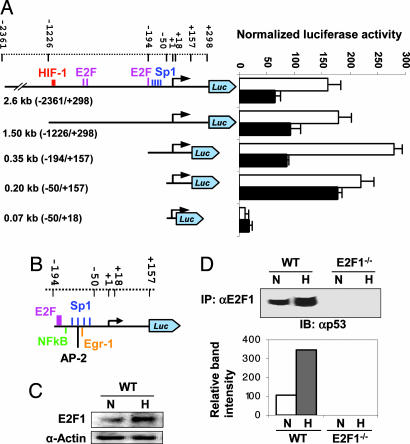
To define the minimum VEGF promoter that mediates transcriptional repression by E2F1, we performed serial cotransfection experiments by using various VEGF promoter truncation mutant-Luc reporter constructs with pE2F1 in E2F1−/− cells under hypoxic conditions. We were able to map the minimal promoter to −194 from the transcriptional start site (Fig. 4A). This minimum VEGF promoter features an E2F1-binding site with four Sp-1 sites in close proximity (Fig. 4B). This finding was of interest because the same Sp-1 sites within the VEGF promoter are targets of p53-mediated VEGF transcriptional repression (22, 26). Therefore, it is likely that the E2F1-induced, p53-dependent, VEGF-specific transcriptional repression responsible for enhanced angiogenesis in the absence of functional E2F1 involves an interaction between E2F1 and p53 on the VEGF promoter, probably in a promoter structure context-dependent manner (29). Indeed, hypoxia induced not only an increase in E2F1 and p53 protein levels (Fig. 4C and data not shown), but also a significant increase in E2F1:p53 physical association, as demonstrated by in vivo coimmunoprecipitation (Fig. 4D). These findings corroborate our hypothesis that repression of VEGF transcription is mediated by a E2F1 and p53 protein:protein interaction.
Fig. 4.
E2F1:P53 interaction down-regulates VEGF promoter activity. (A) Schema of cotransfection of VEGF promoter truncation mutant-Luc reporter constructs with pE2F1 in E2F1−/− fibroblasts to determine the minimal VEGF promoter that mediates VEGF suppression by E2F1. (B) Schematic representation of cis-elements in the minimal VEGF promoter. The E2F1-binding element at approximately −134 to −127 is labeled. Other known elements in this region include an NF-κB site and four Sp1 sites. (C) Western blotting of E2F1 protein in the WT fibroblasts exposed to normoxia (“N”) or hypoxia (“H”). (D) Representative (Upper) and quantification of (Lower) coimmunoprecipitation. Anti-murine E2F1 antibody immunoprecipitates were immunoblotted with an anti-murine p53 antibody.
Our data demonstrate that a significant portion of p53 activity is associated with E2F1. This p53 activity appears to be distinct from the role of p53 in HIF-1α inhibition that was previously documented (25, 27, 28) and was also reflected in our study (Fig. 3B). A physical interaction between E2F1 and p53 has been reported and was linked to regulation of cell cycle and apoptosis of transformed cells in the setting of normoxia (30, 31). We were surprised to find that under hypoxic conditions the E2F1:p53 interaction appeared to affect angiogenesis predominantly through direct regulation of VEGF transcription. Further study is needed to confirm the in vivo E2F1:p53 complex assembly on the VEGF promoter. It is possible that this interaction may also influence other downstream effectors of angiogenesis. p53 has also been previously reported to inhibit tumor angiogenesis through other mechanisms, such as inhibition of human FGF-2 activity by suppressing translation and modification of its conformation (32), down-regulating bFGF-binding protein (33), and up-regulating thrombospondin-1 (34). Our data do not exclude the possibility that these mechanisms may contribute to the enhanced angiogenic phenotype of E2F1−/− mice.
The up-regulation of p53 expression and induction of apoptosis in response to hypoxia represents a major selective advantage for those tumor cells that have diminished apoptotic potential because of a defect of the p53 pathway (35). Also, such defects confer a second advantage, because p53 also inhibits tumor angiogenesis by regulating the production and activity of angiogenic and anti-angiogenic factors (22, 26, 32, 36, 37).
Hypoxia may be one of the most important physiological inducers of p53, but the upstream mechanisms of p53 induction, activity modification, and accumulation are not well understood. Unlike DNA damage or oncogenic activation, G1 phase arrest by hypoxia is not strictly dependent on p53 function, although it is associated with an increase in nuclear p53 protein (38). This action suggests that hypoxia induces p53 activity by a mechanism other than DNA damage and activates a different spectrum of downstream genes (39).
Several mechanisms have been proposed for the induction of p53 by hypoxia. Although p53 has been shown to promote HIF-1α degradation by the ubiquitination pathway, HIF-1α can stabilize p53 (27, 40). In addition, hypoxia-induced nuclear accumulation of p53 is consistently associated with down-regulation of Mdm2 or Hdm2 in human cells (40–42). The E2F1/ARF/Mdm2/p53 pathway is a well accepted mechanism for E2F1-induced p53 activation in which transcription of ARF is up-regulated by E2F1, counteracting Mdm2 neutralization of p53 through ubiquitination-mediated proteasomal degradation. In an independent experiment, we found that under hypoxic conditions E2F1 down-regulates ARF promoter activity (G.Q. and D.W.L., unpublished work). It is therefore less likely that the E2F1/ARF/Mdm2 axis is playing a role in p53 activation (43). Rather, we observed a direct physical association of E2F1 and p53, showing that E2F1 induces p53 accumulation through a protein:protein interaction (30, 44). Interestingly, Rb protein is reversibly de-phosphorylated and bound in the nucleus during hypoxia (45). Dephosphorylated Rb has been shown to stabilize and protect E2F1 from degradation, and the protein:protein interaction in the Rb/E2F1/p53 axis thereby provides a potential mechanism for hypoxia-induced p53 accumulation. Further study is warranted to characterize the functional domains that mediate the E2F1:p53 interaction and identify other downstream effectors of angiogenesis.
Conclusions
The tissue and cell type-specific effects of the E2F family of transcription factors have been documented in E2F1-deficient mice in a variety of biological processes, such as embryonic development, cell differentiation, proliferation, and apoptosis (5, 46–49). Our data add an additional layer of complexity to the role of E2F1 in cell cycle control and apoptosis and raise important questions about the specific effects of different E2Fs in the cardiovascular system. E2F1-deficient mice have impaired apoptotic function, which was previously presumed to explain the increased incidence of cancer in these mice (5, 7, 48). However, our studies suggest that the increase in tumor growth in E2F1-deficient mice might also result from an increase in the vascular supply of tumors secondary to dysregulated VEGF expression.
We therefore propose a model of promoter structure context-dependent transcriptional regulation of VEGF based on overexpression of E2F1, p53 protein:protein interaction, p53-dependent regulation, and minimal VEGF promoter structure. One of the best illustrated examples of such regulation is the E2F1:Sp1 interaction and synergy on the murine tk gene promoter (29). In our current investigation, we used site-directed mutagenesis and chromatin immunoprecipitation (ChIP) techniques to determine the precise cis-elements mediating transcriptional regulation.
Our findings suggest that, in addition to its seemingly contradictory roles as oncogene and tumor suppressor in tumorigenesis, E2F1 may suppress tumor growth by inhibiting tumor angiogenesis. A physiological role for this pathway could be the regulation of angiogenesis, including that observed after ischemia. As such, E2F1 provides a potential therapeutic target for angiogenesis-dependent disease processes.
Methods
Cells.
Primary human umbilical vein endothelial cells (50), mouse aortic endothelial cells (51), and lung fibroblasts (52) were harvested, cultured, characterized as previously described, and used at passage 4–8.
Cells were incubated at 37°C in a chamber containing 5% CO2 and 1% O2 to induce hypoxic conditions. Gases were balanced in N2, unless otherwise specified.
Plasmid Transient Transfection and Reporter Assays.
Please refer to Supporting Methods, which is published as supporting information on the PNAS web site, for detailed methods.
3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyl Tetrazolium Bromide (MTT) Proliferation Assay.
Please refer to Supporting Methods for detailed methods.
ELISA.
VEGF in mouse plasma or cell culture media was quantified with a mouse VEGF ELISA kit (Quantikine; R & D Systems).
Immunoprecipitation and Immunoblotting.
Protein extraction from mouse limb tissue or from cultured cells was performed following standard techniques (53). The lysate of cultured cells was immunoprecipitated with rabbit anti-murine E2F1 polyclonal antibody (Santa Cruz Biotechnology). After electrophoresis and membrane transfer, the lysate was probed with a goat anti-murine p53 polyclonal antibody (Santa Cruz Biotechnology) by using Western blot techniques (54). Densitometry was performed by using National Institutes of Health image software.
Ribonuclease Protection Assay (RPA).
Please refer to Supporting Methods for detailed methods.
Real-Time RT-PCR.
Please refer to Supporting Methods for detailed methods.
In Vivo Studies.
Age- and sex-matched E2F1-deficient (E2F1−/−) and littermate WT control mice were bred from E2F1+/− mice that had been generated by crossing E2F1−/− with WT (both on the background of F2 of the C57BL/6 × 129; The Jackson Laboratory). Mice were bred, maintained, and operated following protocols approved by St. Elizabeth’s Medical Center of Boston Institutional Animal Care and Use Committee.
HLI Model.
Please refer to Supporting Methods for detailed methods.
In Vivo siRNA Transfection.
Murine VEGF siRNA and control GFP siRNA were designed, synthesized, and purified by Dharmacon Research (Lafayette, CO). The VEGF siRNA has been shown to specifically suppress VEGF expression both in vitro and in vivo (55). To block the effect of VEGF on angiogenesis in vivo, a 500 pM concentration of VEGF siRNA conjugated with TransIT reagent (Mirus; Madison, WI) was injected intramuscularly in the ischemic hind limb at 1, 4, 7, and 10 days after HLI. The blood flow recoveries were monitored by using laser Doppler perfusing imaging, and VEGF levels in ischemic limbs were quantified using Western blot analysis.
LLC-1 Tumor Models.
Please refer to Supporting Methods for detailed methods.
Immunohistochemistry.
Please refer to Supporting Methods for detailed methods.
Statistics.
Data were presented as average ± SEM. Comparison between two means was performed with an unpaired Student’s t test, whereas ANOVA with Fisher’s protected least significant differences and Bonferroni–Dunn post hoc analysis were used for comparisons of more than two means. Statistical significance was assigned if P < 0.05.
Supplementary Material
Acknowledgments
We thank Ms. Mickey Neely and Ms. Deirdre Couchon for secretarial assistance. Ad-E2F1 was a kind gift of Dr. J. Nevins (Duke University, Durham, NC). pVEGF promoter-luciferase constructs were generously provided by Dr. Debabrata Mukhopadhyay (Beth Israel Deaconess Medical Center, Boston). Epo-Luc construct was generously provided by Dr. H. Franklin Bunn (Brigham and Women’s Hospital, Harvard Medical School, Boston). This work was supported by National Institutes of Health Grants HL-53354, HL-57516, HL-80137, HL-63414, HL-77428, and HL-66957 (to D.W.L.) and American Heart Association Grant 0430135N (to G.Q.).
Abbreviations
- Ang-1
angiopoietin-1
- bFGF
basic fibroblast growth factor
- EC
endothelial cell
- HLI
hind limb ischemia
- siRNA
small interfering RNA.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Johnson D. G., Schwarz J. K., Cress W. D., Nevins J. R. Nature. 1993;365:349–352. doi: 10.1038/365349a0. [DOI] [PubMed] [Google Scholar]
- 2.DeGregori J., Leone G., Ohtani K., Miron A., Nevins J. R. Genes Dev. 1995;9:2873–2887. doi: 10.1101/gad.9.23.2873. [DOI] [PubMed] [Google Scholar]
- 3.Kowalik T. F., DeGregori J., Schwarz J. K., Nevins J. R. J. Virol. 1995;69:2491–2500. doi: 10.1128/jvi.69.4.2491-2500.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwarz J. K., Bassing C. H., Kovesdi I., Datto M. B., Blazing M., George S., Wang X. F., Nevins J. R. Proc. Natl. Acad. Sci. USA. 1995;92:483–487. doi: 10.1073/pnas.92.2.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamasaki L., Jacks T., Bronson R., Goillot E., Harlow E., Dyson N. J. Cell. 1996;85:537–548. doi: 10.1016/s0092-8674(00)81254-4. [DOI] [PubMed] [Google Scholar]
- 6.Weinberg R. A. Cell. 1996;85:457–459. doi: 10.1016/s0092-8674(00)81244-1. [DOI] [PubMed] [Google Scholar]
- 7.DeGregori J., Leone G., Miron A., Jakoi L., Nevins J. R. Proc. Natl. Acad. Sci. USA. 1997;94:7245–7250. doi: 10.1073/pnas.94.14.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Folkman J. N. Engl. J. Med. 1971;285:1182–1186. doi: 10.1056/NEJM197111182852108. [DOI] [PubMed] [Google Scholar]
- 9.Morishita R., Gibbons G. H., Horiuchi M., Ellison K. E., Nakama M., Zhang L., Kaneda Y., Ogihara T., Dzau V. J. Proc. Natl. Acad. Sci. USA. 1995;92:5855–5859. doi: 10.1073/pnas.92.13.5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mann M. J., Whittemore A. D., Donaldson M. C., Belkin M., Conte M. S., Polak J. F., Orav E. J., Ehsan A., Dell’Acqua G., Dzau V. J. Lancet. 1999;354:1493–1498. doi: 10.1016/S0140-6736(99)09405-2. [DOI] [PubMed] [Google Scholar]
- 11.Spyridopoulos I., Principe N., Krasinski K. L., Xu S., Kearney M., Magner M., Isner J. M., Losordo D. W. Circulation. 1998;98:2883–2890. doi: 10.1161/01.cir.98.25.2883. [DOI] [PubMed] [Google Scholar]
- 12.Goukassian D. A., Kishore R., Krasinski K., Dolan C., Luedemann C., Yoon Y. S., Kearney M., Hanley A., Ma H., Asahara T., et al. Circ. Res. 2003;93:162–169. doi: 10.1161/01.RES.0000082980.94211.3A. [DOI] [PubMed] [Google Scholar]
- 13.Kishore R., Luedemann C., Bord E., Goukassian D., Losordo D. W. Circ. Res. 2003;93:932–940. doi: 10.1161/01.RES.0000102400.22370.20. [DOI] [PubMed] [Google Scholar]
- 14.Adams J. M., Cory S. Science. 1991;254:1161–1167. doi: 10.1126/science.1957168. [DOI] [PubMed] [Google Scholar]
- 15.Hursting S. D. Curr. Opin. Oncol. 1997;9:487–491. doi: 10.1097/00001622-199709050-00015. [DOI] [PubMed] [Google Scholar]
- 16.Carmeliet P., Conway E. M. Nat. Biotechnol. 2001;19:1019–1020. doi: 10.1038/nbt1101-1019. [DOI] [PubMed] [Google Scholar]
- 17.Conway E. M., Collen D., Carmeliet P. Cardiovasc. Res. 2001;49:507–521. doi: 10.1016/s0008-6363(00)00281-9. [DOI] [PubMed] [Google Scholar]
- 18.Yancopoulos G. D., Davis S., Gale N. W., Rudge J. S., Wiegand S. J., Holash J. Nature. 2000;407:242–248. doi: 10.1038/35025215. [DOI] [PubMed] [Google Scholar]
- 19.Thurston G., Rudge J. S., Ioff E., Zhou H., Ross L., Croll D. D., Glazer M., Holash J., McDonald D. M., Yancopoulos G. D. Nat. Med. 2000;6:460–463. doi: 10.1038/74725. [DOI] [PubMed] [Google Scholar]
- 20.Qin X. Q., Livingston D. M., Kaelin W. G., Jr., Adams P. D. Proc. Natl. Acad. Sci. USA. 1994;91:10918–10922. doi: 10.1073/pnas.91.23.10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pan H., Yin C., Dyson N. J., Harlow E., Yamasaki L., Van Dyke T. Mol. Cell. 1998;2:283–292. doi: 10.1016/s1097-2765(00)80273-7. [DOI] [PubMed] [Google Scholar]
- 22.Zhang L., Yu D., Hu M., Xiong S., Lang A., Ellis L. M., Pollock R. E. Cancer Res. 2000;60:3655–3661. [PubMed] [Google Scholar]
- 23.O’Connor D. J., Lu X. Oncogene. 2000;19:2369–2376. doi: 10.1038/sj.onc.1203540. [DOI] [PubMed] [Google Scholar]
- 24.Vogelstein B., Kinzler K. W. Cell. 1992;70:523–526. doi: 10.1016/0092-8674(92)90421-8. [DOI] [PubMed] [Google Scholar]
- 25.Ravi R., Mookerjee B., Bhujwalla Z. M., Sutter C. H., Artemov D., Zeng Q., Dillehay L. E., Madan A., Semenza G. L., Bedi A. Genes Dev. 2000;14:34–44. [PMC free article] [PubMed] [Google Scholar]
- 26.Pal S., Datta K., Mukhopadhyay D. Cancer Res. 2001;61:6952–6957. [PubMed] [Google Scholar]
- 27.An W. G., Kanekal M., Simon M. C., Maltepe E., Blagosklonny M. V., Neckers L. M. Nature. 1998;392:405–408. doi: 10.1038/32925. [DOI] [PubMed] [Google Scholar]
- 28.Blagosklonny M. V., Giannakakou P., Wojtowicz M., Romanova L. Y., Ain K. B., Bates S. E., Fojo T. J. Clin. Endocrinol. Metab. 1998;83:2516–2522. doi: 10.1210/jcem.83.7.4984. [DOI] [PubMed] [Google Scholar]
- 29.Fry C. J., Farnham P. J. J. Biol. Chem. 1999;274:29583–29586. doi: 10.1074/jbc.274.42.29583. [DOI] [PubMed] [Google Scholar]
- 30.O’Connor D. J., Lam E. W., Griffin S., Zhong S., Leighton L. C., Burbidge S. A., Lu X. EMBO J. 1995;14:6184–6192. doi: 10.1002/j.1460-2075.1995.tb00309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsieh J. K., Yap D., O’Connor D. J., Fogal V., Fallis L., Chan F., Zhong S., Lu X. Mol. Cell. Biol. 2002;22:78–93. doi: 10.1128/MCB.22.1.78-93.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galy B., Creancier L., Prado-Lourenco L., Prats A. C., Prats H. Oncogene. 2001;20:4613–4620. doi: 10.1038/sj.onc.1204630. [DOI] [PubMed] [Google Scholar]
- 33.Sherif Z. A., Nakai S., Pirollo K. F., Rait A., Chang E. H. Cancer Gene Ther. 2001;8:771–782. doi: 10.1038/sj.cgt.7700361. [DOI] [PubMed] [Google Scholar]
- 34.Dameron K. M., Volpert O. V., Tainsky M. A., Bouck N. Science. 1994;265:1582–1584. doi: 10.1126/science.7521539. [DOI] [PubMed] [Google Scholar]
- 35.Graeber T. G., Osmanian C., Jacks T., Housman D. E., Koch C. J., Lowe S. W., Giaccia A. J. Nature. 1996;379:88–91. doi: 10.1038/379088a0. [DOI] [PubMed] [Google Scholar]
- 36.Kim M. S., Kwon H. J., Lee Y. M., Baek J. H., Jang J. E., Lee S. W., Moon E. J., Kim H. S., Lee S. K., Chung H. Y., et al. Nat. Med. 2001;7:437–443. doi: 10.1038/86507. [DOI] [PubMed] [Google Scholar]
- 37.Volpert O. V., Dameron K. M., Bouck N. Oncogene. 1997;14:1492–1502. doi: 10.1038/sj.onc.1200977. [DOI] [PubMed] [Google Scholar]
- 38.Graeber T. G., Peterson J. F., Tsai M., Monica K., Fornace A. J., Jr., Giaccia A. J. Mol. Cell. Biol. 1994;14:6264–6277. doi: 10.1128/mcb.14.9.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koumenis C., Alarcon R., Hammond E., Sutphin P., Hoffman W., Murphy M., Derr J., Taya Y., Lowe S. W., Kastan M., Giaccia A. Mol. Cell. Biol. 2001;21:1297–1310. doi: 10.1128/MCB.21.4.1297-1310.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzuki H., Tomida A., Tsuruo T. Oncogene. 2001;20:5779–5788. doi: 10.1038/sj.onc.1204742. [DOI] [PubMed] [Google Scholar]
- 41.Alarcon R., Koumenis C., Geyer R. K., Maki C. G., Giaccia A. J. Cancer Res. 1999;59:6046–6051. [PubMed] [Google Scholar]
- 42.Zhu Y., Mao X. O., Sun Y., Xia Z., Greenberg D. A. J. Biol. Chem. 2002;277:22909–22914. doi: 10.1074/jbc.M200042200. [DOI] [PubMed] [Google Scholar]
- 43.Rogoff H. A., Pickering M. T., Debatis M. E., Jones S., Kowalik T. F. Mol. Cell. Biol. 2002;22:5308–5318. doi: 10.1128/MCB.22.15.5308-5318.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nip J., Strom D. K., Eischen C. M., Cleveland J. L., Zambetti G. P., Hiebert S. W. Oncogene. 2001;20:910–920. doi: 10.1038/sj.onc.1204171. [DOI] [PubMed] [Google Scholar]
- 45.Amellem O., Stokke T., Sandvik J. A., Pettersen E. O. Exp. Cell Res. 1996;227:106–115. doi: 10.1006/excr.1996.0255. [DOI] [PubMed] [Google Scholar]
- 46.Wu L., Timmers C., Maiti B., Saavedra H. I., Sang L., Chong G. T., Nuckolls F., Giangrande P., Wright F. A., Field S. J., et al. Nature. 2001;414:457–462. doi: 10.1038/35106593. [DOI] [PubMed] [Google Scholar]
- 47.Saavedra H. I., Wu L., de Bruin A., Timmers C., Rosol T. J., Weinstein M., Robinson M. L., Leone G. Cell Growth Differ. 2002;13:215–225. [PubMed] [Google Scholar]
- 48.Field S. J., Tsai F. Y., Kuo F., Livingston A. M., Orkin S. H., Greenberg M. E. Cell. 1996;85:549–561. doi: 10.1016/s0092-8674(00)81255-6. [DOI] [PubMed] [Google Scholar]
- 49.DeGregori J. Biochim. Biophys. Acta. 2002;1602:131–150. doi: 10.1016/s0304-419x(02)00051-3. [DOI] [PubMed] [Google Scholar]
- 50.Murohara T., Witzenbichler B., Spyridopoulos I., Asahara T., Ding B., Sullivan A., Losordo D. W., Isner J. M. Arterioscler. Thromb. Vasc. Biol. 1999;19:1156–1161. doi: 10.1161/01.atv.19.5.1156. [DOI] [PubMed] [Google Scholar]
- 51.Kondo S., Yin D., Aoki T., Takahashi J. A., Morimura T., Takeuchi J. Exp. Cell Res. 1994;213:428–432. doi: 10.1006/excr.1994.1219. [DOI] [PubMed] [Google Scholar]
- 52.Yarovinsky T. O., Hunninghake G. W. Am. J. Physiol. 2001;281:L1248–L1256. doi: 10.1152/ajplung.2001.281.5.L1248. [DOI] [PubMed] [Google Scholar]
- 53.Couffinhal T., Silver M., Zheng L. P., Kearney M., Witzenbichler B., Isner J. M. Am. J. Pathol. 1998;152:1667–1679. [PMC free article] [PubMed] [Google Scholar]
- 54.Kishore R., Spyridopoulos I., Luedemann C., Losordo D. W. Circ. Res. 2002;91:307–314. doi: 10.1161/01.res.0000031744.06353.d3. [DOI] [PubMed] [Google Scholar]
- 55.Reich S. J., Fosnot J., Kuroki A., Tang W., Yang X., Maguire A. M., Bennett J., Tolentino M. J. Mol. Vis. 2003;9:210–216. [PubMed] [Google Scholar]
- 56.Asahara T., Masuda H., Takahashi T., Kalka C., Pastore C., Silver M., Kearne M., Magner M., Isner J. M. Circ. Res. 1999;85:221–228. doi: 10.1161/01.res.85.3.221. [DOI] [PubMed] [Google Scholar]
- 57.Murasawa S., Llevadot J., Silver M., Isner J. M., Losordo D. W., Asahara T. Circulation. 2002;106:1133–1139. doi: 10.1161/01.cir.0000027584.85865.b4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.