Abstract
The peptidoglycan (PG) layers surrounding bacterial cells play an important role in determining cell shape. The machinery controlling when and where new PG is made is not understood, but is proposed to involve interactions between bacterial actin homologs such as Mbl, which forms helical cables within cells, and extracellular multiprotein complexes that include penicillin-binding proteins. It has been suggested that labeled antibiotics that bind to PG precursors may be useful for imaging PG to help determine the genes that control the biosynthesis of this polymer. Here, we compare the staining patterns observed in Bacillus subtilis using fluorescent derivatives of two PG-binding antibiotics, vancomycin and ramoplanin. The staining patterns for both probes exhibit a strong dependence on probe concentration, suggesting antibiotic-induced perturbations in PG synthesis. Ramoplanin probes may be better imaging agents than vancomycin probes because they yield clear staining patterns at concentrations well below their minimum inhibitory concentrations. Under some conditions, both ramoplanin and vancomycin probes produce helicoid staining patterns along the cylindrical walls of B. subtilis cells. This sidewall staining is observed in the absence of the cytoskeletal protein Mbl. Although Mbl plays an important role in cell shape determination, our data indicate that other proteins control the spatial localization of the biosynthetic complexes responsible for new PG synthesis along the walls of B. subtilis cells.
Keywords: helix, ramoplanin, vancomycin, Mbl, bacterial cytoskeleton
Bacterial cells are surrounded by layers of peptidoglycan (PG), a crosslinked carbohydrate polymer that functions as a protective exoskeleton. These PG layers enable bacteria to withstand high internal osmotic pressures and also play an important role in maintaining cell shape (1, 2). Because PG is essential for survival and is a defining feature of bacteria, considerable effort has been focused on understanding its structure and assembly. We know a great deal about how the subunits that comprise PG are synthesized inside the cell (3), but we do not yet understand how these subunits are converted into a complex, 3D polymer on the cell surface (4, 5).
Early models of bacterial cell growth held that new PG is made and incorporated at midcell around the time of cell division (6). Subsequent metabolic labeling experiments in Bacillus subtilis and Escherichia coli showed that PG is also made along the cylindrical walls of the cells (7–13). Recent experiments using fluorescent vancomycin, an antibiotic that inhibits PG biosynthesis by binding to PG intermediates (i.e., a substrate-binding antibiotic), suggest that cylindrical wall synthesis in B. subtilis occurs in a helix-like pattern (14). It is currently believed that large biosynthetic machines containing different sets of synthetic and hydrolytic enzymes are involved in wall and septal synthesis during bacterial cell growth and division (15–20). It has been proposed that the spatial localization of these machines, and hence the pattern in which new PG is produced, is determined by cytoskeletal proteins inside bacterial cells. In B. subtilis, the actin homolog Mbl, which forms a dynamic helical filament beneath the membrane, was proposed to direct the incorporation of new PG along the sidewalls of the cells. This proposal was based on experiments showing that fluorescent vancomycin stains the walls of WT cells in a helix-like pattern, but stains mbl− cells only at the septa (14).
We have been investigating the mechanism of action of another antibiotic, ramoplanin (2a; Fig. 1A), which also binds to PG intermediates (21). Ramoplanin binds only to the reducing end of the nascent glycan chain, found at the initiation sites of PG synthesis, and lipid II (Fig. 1B) (21, 22). Vancomycin, in contrast, recognizes a dipeptide unit, d-Ala-d-Ala, that is found along the growing glycan chain and in lipid II. In some organisms, d-Ala-d-Ala is also present in mature PG (Fig. 1B) (23). The specificity of ramoplanin suggested that it might have particular utility for detecting sites of nascent PG synthesis.
Fig. 1.
Structures and cellular targets of vancomycin and ramoplanin. (A) Structures of compounds discussed in the text. (B) The extracellular stage of PG biosynthesis. Vancomycin recognizes d-Ala-d-Ala (red); ramoplanin recognizes diphospho-MurNAc (blue). In B. subtilis, L-Lys is replaced with meso-diaminopimelic acid.
We report a comparison of the patterns observed in B. subtilis using fluorescent ramoplanin and vancomycin. Staining patterns for both labeled antibiotics are concentration-dependent, suggesting that both probes perturb PG synthesis when used near inhibitory concentrations. At low concentrations, ramoplanin probes stain the nascent division sites, the cell poles, and the sidewalls of cells in a helix-like pattern; vancomycin probes yield similar sidewall staining patterns but only when used at relatively high concentrations and only when unlabeled vancomycin is also present. We discuss differences in the recognition chemistry of the probes that may explain these differences in behavior. Finally, using either vancomycin or ramoplanin probes, we show that sidewall staining is observed in both mbl− and WT cells. Our data indicate that if an underlying cytoskeletal scaffold in B. subtilis directs the sidewall localization of the machinery involved in PG synthesis, Mbl is not an essential part of that scaffold.
Results
Preparation and Evaluation of Fluorescent Vancomycin and Ramoplanin Derivatives.
Substrate-binding antibiotics used to image PG synthesis must be labeled at sites that do not interfere with their ability to bind to PG intermediates. Vancomycin (1a) contains two amines that are amenable to modification. The more reactive amine is located on the disaccharide attached to the convex surface of the molecule. The disaccharide does not play any role in d-Ala-d-Ala binding (24), so we attached fluorescein to this amine to produce a fluorescent derivative of vancomycin (Van-FL) (1b). We also prepared a derivative of vancomycin, desleucyl-van-FL (1c), in which the N-methyl-leucine moiety, a critical part of the peptide binding pocket, was removed (25). 1c does not bind d-Ala-d-Ala and thus serves as a negative control to establish that patterns observed with 1b reflect binding to PG intermediates. Finally, we investigated a BodipyFl derivative of vancomycin (1d) to examine whether the structure of the fluorophore influences probe behavior.
Ramoplanin (2a) also contains two amines, Orn-4 and Orn-10. Orn-4 can be modified without altering binding to lipid II; however, even small changes to Orn-10 affect lipid II binding (26). Therefore, we attached fluorescein and BodipyFl to Orn-4 to prepare ramoplanin probes 2b and 2d. We also prepared a ramoplanin derivative containing fluorescein at Orn-10 (2c) to produce a structural isomer of 2b that cannot bind lipid II.
Biological Evaluation of Labeled Probes.
We chose to compare compounds in terms of minimum inhibitory concentrations (MICs) rather than absolute concentrations because MICs better reflect the degree to which cell physiology is perturbed. The MICs of ramoplanin (2a), vancomycin (1a), Ramo-4FL (2b), and Van-FL (1b) were measured against WT B. subtilis PY79 in CH media (see Supporting Methods, which is published as supporting information on the PNAS web site), and the values are shown in Table 1. Vancomycin has potent activity against PY79, whereas ramoplanin has moderate activity. Attaching fluorescein leads to increased MICs for both antibiotics, although the increase is greater for the vancomycin probe (>100-fold compared with a 4-fold increase for ramoplanin). It was previously reported that the MICs of vancomycin and Van-FL are comparable (14). However, the Van-FL sample used for the MIC measurements contained at least a 4-fold excess of unlabeled vancomycin; accordingly, the MIC measured was likely the MIC of vancomycin.
Table 1.
MICs (in μg/ml) of test compounds against B. subtilis
| Compound | CH media |
LB media |
|
|---|---|---|---|
| WT (PY79) | WT (PY79) | Mbl− | |
| (1a) Vancomycin | 0.13 | 0.16 | 0.16 |
| (1b) Van-F1 | 20.0 | 10.0 | 10.0 |
| (1c) Desleucyl-van-F1 | >80.0 | ND | ND |
| (1d) Van-BDP | 2.5 | 0.63 | 0.63 |
| (2a) Ramoplanin | 5.0 | 1.25 | 1.25 |
| (2b) Ramo-4F1 | 20.0 | 10.0 | 10.0 |
| (2c) Ramo-10F1 | 40.0 | 40.0 | 40.0 |
| (2d) Ramo-4BDP | 10.0 | 10.0 | 10.0 |
ND, not determined.
The reduced biological activity of 1b and 2b raises questions about whether the fluorescein labels substantively alter the affinity for the compounds or simply reduce how much material reaches lipid II and other PG precursors on the cell surface. To determine whether the fluorophore on 2b interferes with lipid II binding in vitro, we investigated the ability of 2b to inhibit Escherichia coli PBP1b, a prototypical transglycosylase. Like ramoplanin, 2b was found to inhibit transglycosylation by binding to lipid II with a stoichiometry of 2:1 antibiotic/lipid II (data not shown) (27). The Kd value estimated from the inhibition curve was comparable to that for ramoplanin itself. Thus, the increased MIC of 2b is not caused by a decrease in its intrinsic affinity for lipid II. We did not examine Van-FL for binding to d-Ala-d-Ala, but studies of related vancomycin derivatives support the hypothesis that 1b binds substrate comparably to the parent compound (24), making it unlikely that the increased MIC reflects a change in affinity for d-Ala-d-Ala.
Fluorescein is large and negatively charged, and the cell wall of B. subtilis is rich in anionic teichoic acids, which repel negatively charged molecules (28). Furthermore, the fluorescein label decreases the solubility of the antibiotic derivatives relative to the parent compounds. Thus, the increased MICs may be caused by less effective partitioning of labeled probes through the PG layers because of repulsive interactions and/or poor solubility. Consistent with this hypothesis, we have found that probes containing the smaller, neutral BodipyFl have lower MICs than the fluorescein derivatives (Table 1).
Concentration Dependence of Fluorescent Staining Patterns.
As a first step in our analysis of these antibiotic probes, we analyzed the effects of probe concentration on the staining patterns. B. subtilis cells were treated with increasing concentrations of Ramo-4FL (2b) and Van-FL (1b) for 5–10 min and examined by fluorescence microscopy. At low concentrations (×0.1–0.2 MIC) of 2b, we observed bright but narrow bands at both the old and new division sites in septate chains of vegetatively growing cells. As the probe concentration increased to the MIC, the pattern changed. The bands at the old poles (Fig. 2 B and C, arrowheads) grew faint, whereas the bands at midcell (Fig. 2B, arrows) became thick. Because these antibiotics may alter the normal pattern of PG synthesis, we believe that the patterns observed at low concentrations provide a more accurate reflection of how PG synthesis normally proceeds than the patterns observed at higher concentrations.
Fig. 2.
Staining of B. subtilis PY79 with ramoplanin analogs. (A) Examples of sidewall staining observed with 2a:2b. (B–H Left) Probe-treated cells. (B–H Center) TMA-DPH-treated cells (mb, membrane stain). (B–H Right) Overlays of probe-treated (green) and TMA-DPH-treated (red) cells. Arrowheads and arrows point to old division sites (poles) and new division sites (septa), respectively. (A) Fluorescent images of 1:1 mixture of 2a:2b at 0.5 μg/ml each, along with a schematic representation of the helix. (B) 2b at 0.1× MIC (2 μg/ml). (C) 2b at 1.0× MIC (20 μg/ml). (D) A 1:1 mixture of 2a:2c at 0.5 μg/ml each (higher concentrations look similar). (E) A 1:1 mixture of 2a:2b at 0.5 μg/ml each. (F) A 1:1 mixture of 2a:2b at 1.0 μg/ml each. (G) A 1:1 mixture of 2a:2b at 2.5 μg/ml each. (H) 2d at 0.1× MIC (1.0 μg/ml). (Scale bars, 2 μm.) See Fig. 5, which is published as supporting information on the PNAS web site, for larger fields.
We were unable to carry out a parallel set of concentration studies with 1b because this probe aggregates, producing high backgrounds. However, we achieved acceptable staining of B. subtilis cells with 1b at concentrations around 0.5× MIC (data not shown). A pattern similar to that seen with the ramoplanin probe at comparable concentrations was observed: narrow fluorescent bands at the old poles and thicker bands at the new division sites. The similarity in the staining patterns for 2b and 1b is consistent with the hypothesis that both compounds detect early PG intermediates.
The Fluorescent Patterns Correlate with Binding to PG Precursors.
Although the similar staining patterns observed for 2b and 1b were encouraging, both compounds contain fluorescein, and it was important to rule out the possibility that the observed patterns reflect interactions of the fluorophore with cell surface components. Therefore, we tested probe compounds 1c and 2c that were incapable of binding to lipid II or other PG precursors. The damaged Van-FL derivative 1c did not stain cells at any concentrations tested, whereas the Ramo-10FL compound (2c) accumulated in blobs at midcell like the membrane dye 1-(4-trimethylammoniumphenyl)-6-phenyl-1,3,5-hexatriene p-toluenesulfonate (TMA-DPH), but did not produce the banded pattern observed with 2b (Fig. 6, which is published as supporting information on the PNAS web site). We conclude that the banded pattern observed with both 1b and 2b correlates with binding to PG precursors.
Sidewall Staining with Ramoplanin Derivatives.
In the experiments described above, sidewall staining was not observed in any cells. We were puzzled by these results because studies with radioactive PG building blocks had shown that new PG is incorporated along the sidewalls of B. subtilis (2), and recently a helix-like sidewall staining pattern was observed with a mixture of vancomycin (1a) and Van-FL (1b) (14). We wondered whether the lack of wall staining could be related to less effective partitioning of the probes through sidewall PG compared with polar PG. It is speculated that there are differences in the cell wall at the poles and along the sides of rod-shaped bacteria; these differences may affect the extent of compound penetration (4, 29). We reasoned that if compound penetration was a problem, then reducing the adverse effects of the fluorophore should result in sidewall staining.
We considered two possible ways to minimize problems caused by the fluorophore in ramoplanin probes. The first was to exploit the finding that ramoplanin binds to lipid II as a dimer. All other things being equal, a heterodimer containing one problematic fluorescent label should be a better probe than a homodimer containing two such labels. When equimolar unlabeled ramoplanin (2a) was added to 2b, we observed intense sidewall staining, even at a 2b concentration 10-fold below that used to observe septal staining with the pure compound (Fig. 2A). The sidewall staining typically takes the form of peripheral dots and transverse bands, suggestive of a helix-like structure. A particularly clear example of a helical pattern can be observed in Fig. 2C Left. The helix in this image is left-handed; in other cells (see, e.g., Fig. 2C Right), the helices appear right-handed. Still to be determined is whether the helical handedness does, in fact, vary, because the implications for PG biosynthesis would be significant.
To verify that the sidewall staining reflects the location of PG precursors, we treated B. subtilis cells with a 1:1 mixture of ramoplanin (2a) and the control probe (2c), which does not bind lipid II. No sidewall staining was observed in any cells, confirming that the peripheral staining seen in the 2a:2b mixture depends on binding to PG precursors (Fig. 2D).
We next investigated the concentration dependence of the staining pattern observed with 2a:2b mixtures. As the concentration of the probe mixture increases, staining along the sidewall decreases first, followed by staining at the old poles (Fig. 2 E-G, blue arrowhead). At concentrations near the MIC, fluorescent bands are observed at midcell (Fig. 2G, pink arrow) but staining elsewhere is mostly gone. Thus, the patterns obtained with the probe mixture change with concentration, as was seen with the pure probe.
We also explored a second approach to minimize adverse effects of the fluorophore. This approach involved preparing a different probe, Ramo-4BDP (2d), which contains BodipyFl. Used by itself 2d was able to stain B. subtilis cells not only at the poles but also along the sidewalls (Fig. 2H). The sidewall staining was similar to that obtained with the 2a:2b mixture. These results support the hypothesis that probe structure can affect probe access to PG intermediates on the membrane surface.
Sidewall Staining with Vancomycin Derivatives.
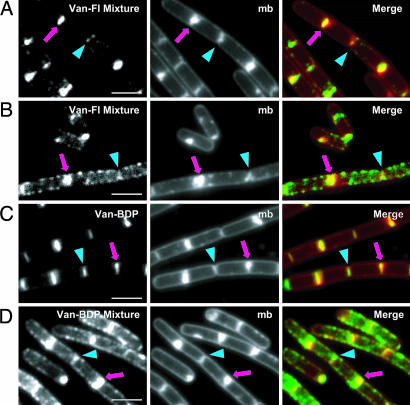
Like 2b, 1b does not stain the sidewalls of B. subtilis when used by itself. It was previously reported, however, that mixtures of Van-FL and unlabeled vancomycin resulted in helix-like staining of the sidewalls (14). Although we do not understand why a vancomycin/Van-FL mixture should work better than the pure probe, we investigated the use of the mixture. At low to moderate vancomycin concentrations (sub-MIC levels), we observed septal staining but no sidewall staining (Fig. 3A). Because vancomycin is a bacteriostatic antibiotic, and because incubation times before imaging are short, it is possible to use vancomycin probe concentrations above the MIC without causing cell lysis. When we increased the concentration of probe mixture above the MIC, staining appeared along the sidewalls of the cells and the new division sites (Fig. 3B) but decreased at the old division sites. The observed wall staining pattern (Fig. 3B) was similar to the helical pattern previously reported for vancomycin/Van-FL (35), with peripheral dots and transverse bands. In some respects, this pattern resembles that observed at low concentrations of 2a:2b (Fig. 2E), but the decreased staining at the old poles in the 1a:1b-treated B. subtilis cells (Fig. 3B) suggests a disruption in normal biosynthetic processes.
Fig. 3.
Staining of B. subtilis PY79 with vancomycin analogs. (Left) Probe-treated cells. (Center) TMA-DPH-treated cells. mb, membrane stain. (Right) Overlays of probe-treated (green) and TMA-DPH-stained cells (red). Arrowheads and arrows point to old division sites (poles) and new division sites (septa), respectively. (A) A 1:1 mixture of 1a:1b at 0.13 μg/ml each. (B) A 1:1 mixture of 1a:1b at 0.4 μg/ml. (C) 1d at 0.4× MIC (1.0 μg/ml). (D) A 1:1 mixture of 1a:1d at 0.4 μg/ml. (Scale bars, 2 μm.) See Fig. 7, which is published as supporting information on the PNAS web site, for larger fields.
We next investigated whether BodipyFL vancomycin (Van-BDP) (1d) alone would stain the sidewalls like its ramoplanin counterpart 2d. Van-BDP has been used to visualize PG synthesis, but there is no information on how it compares with Van-Fl (23). The MIC of 1d is lower than that of 1b (Table 1), but like 1b, 1d only stains B. subtilis cells at the nascent and old division sites when used by itself (Fig. 3C). Sidewall staining is not observed unless unlabeled vancomycin is also added (Fig. 3D). There was less background because of aggregation with the 1a/1d mixtures than with the 1a/1b mixtures.
Effect of Mbl on PG Staining.
One motivation for investigating fluorescent substrate-binding antibiotics is that such compounds may be useful for probing the phenotypes of bacterial strains having mutations in genes proposed to be involved in PG synthesis. Based on experiments in which Van-FL staining patterns for WT and mbl− cells were compared, it was proposed that Mbl acts as an intracellular scaffold that directs wall PG synthesis in B. subtilis (14). Mbl is an actin homolog that plays an important role in shape determination in B. subtilis (30), and mbl null mutants are short and show morphological aberrations compared with WT cells. As a control for our ramoplanin probes we decided to examine PG staining in an mbl mutant. We constructed the mbl null mutant and compared PG staining patterns for the WT and mutant strain by using Ramo-4BDP (2d) alone or the 2a:2d mixture (Fig. 4A and data not shown). Although lower concentrations of 2d were needed to detect staining of the mbl− cells, we observed staining patterns similar to those observed for the WT PY79 strain. Fluorescence was observed at both the old and new division sites and along the sidewalls of the cells. The sidewall staining shows the characteristic peripheral dots and transverse bands, but the pattern is less regular compared with the WT strain. This finding may reflect the decreased length and distortions of the cells or more disorder in the sidewall biosynthetic machinery.
Fig. 4.
Comparison of mblΔ and WT strains stained with 2d (Upper) or a 1a:1d mixture (Lower). (A Left) mblΔ stained with 2d (0.2 μg/ml). (A Right) WT stained with 2d (1.0 μg/ml). (B Left) mblΔ stained with 1a:1d (1:1 ratio at 0.08 μg/ml). (B Right) WT stained with 1:1 mixture of 1a:1d (1:1 ratio at 0.5 μg/ml). (Scale bars, 2 μm.)
Prompted by the unexpected results obtained with Ramo-4BDP (2d), we examined the staining of mbl− cells with Van-BDP. The staining pattern obtained with a 1:1 mixture of 1a:1d (with 1a at ×0.5 MIC) was similar to that for Ramo-4BDP. Staining was evident at both the division sites and along the sidewalls. We cannot explain the difference in our results to those reported previously (14). Because sidewall staining of mbl− cells is observed with both ramoplanin and vancomycin probes, we conclude that Mbl is probably not essential for the sidewall localization of PG synthesis machinery in B. subtilis.
Discussion
PG biosynthesis has been the subject of extensive study because of its importance in cell survival and its role in maintaining cell shape. Rod-shaped bacteria such as B. subtilis maintain their morphology by directing synthesis of new PG along the walls of the cells during elongation. Although this much is accepted, there still remains the question of what determines the localization of the biosynthetic machinery involved in wall synthesis. In the late 1990s, it was discovered that bacteria contain actin homologs, including Mbl and MreB in B. subtilis (31). GFP-fusion studies have shown that these actin homologs form dynamic helical cables just beneath the cytoplasmic membrane (30, 31). It was speculated, therefore, that the external assembly of PG is directed by interactions between these internal scaffolds and the machinery for PG synthesis. Demonstrating a correlation between these or other internal scaffolds and PG synthesis requires having methods to detect where new PG is made on the bacterial cell surface.
Efforts to monitor PG synthesis go back decades. Most previous attempts to study the synthesis of new PG have involved studies in which radiolabeled precursors are incorporated into PG and detected only after considerable sample processing (31). These studies have revealed that PG is made at the nascent septum and along the walls of rod-shaped bacteria, but they do not provide adequate spatial resolution to determine the patterns of incorporation. Thus, for many years there has been a debate about whether wall PG synthesis is diffuse or zonal (1, 13). Metabolic labeling experiments also suffer from poor temporal resolution, and the timing of different modes of PG synthesis continues to be debated. Methods that enable direct visualization of PG synthesis in live B. subtilis cells could provide answers to longstanding questions about the process.
Daniel and Errington (14) recently introduced a novel method for detecting sites of PG synthesis in Gram-positive organisms that involves the use of fluorescent vancomycin to detect d-Ala-d-Ala found in PG precursors. Using fluorescent vancomycin, they suggested that PG is synthesized in a helical pattern along the walls of B. subtilis cells. We were intrigued by the possibility that a substrate-binding antibiotic could serve as a probe to detect new PG; however, vancomycin can recognize growing PG chains as well as initiation sites, and its affinity for d-Ala-d-Ala is only in the low micromolar range. Anticipating that a higher-affinity substrate binder with greater specificity for PG initiation sites would provide better resolution, we began to investigate derivatives of the antibiotic ramoplanin. This molecule recognizes structural elements found only in lipid II and at the reducing end of the growing PG polymer (21). Assuming that lipid II is translocated where it is used, and that glycan chain polymerization proceeds by the addition of disaccharide units to the reducing end of the growing polymer, as proposed (32, 33), ramoplanin should bind only at the initiation sites of PG synthesis. We compared fluorescent ramoplanin derivatives to fluorescent vancomycin to ascertain whether there are any differences in the behavior of these probes.
We have found that the patterns observed with both vancomycin and ramoplanin probes depend on concentration. Because both compounds are antibiotics that act by inhibiting PG synthesis, we hypothesize that the patterns observed at lower concentrations are more representative of normal biosynthetic processes than the patterns observed at higher concentrations. At low concentrations (relative to MIC values), we observed helix-like staining patterns along the sidewalls of B. subtilis by using 2d alone or a mixture of 2a and 2b. We also observed intense bands at the newly forming septa as well as weaker bands at the poles of cells that have divided. Control experiments established that these staining patterns correlate with the ability of the probes to bind to lipid II. These results support the idea that PG is synthesized in a helicoid pattern along the walls of the cells and at the new division sites and suggest that it continues for some period at the poles after septation. Although it is widely believed that the poles become metabolically inert immediately after cell division, our data suggest that PG synthesis persists at these sites. This observation is consistent with studies by Mobley et al. (10).
The staining patterns observed with vancomycin analogs are similar but not identical to those obtained with ramoplanin analogs. Importantly, helical wall staining cannot be observed with any vancomycin derivatives unless unlabeled vancomycin is also added at relatively high concentrations. Under these conditions, staining at the old division sites has decreased. By contrast, mixtures of ramoplanin (2a)/Ramo-4FL (2b) and pure Ramo-4BDP (2d) stain both the cylindrical cell walls and the old and new division sites of B. subtilis cells.
We do not understand why adding unlabeled vancomycin should facilitate wall staining in B. subtilis cells. Unlike ramoplanin, which forms 2:1 complexes with PG precursors, vancomycin forms only 1:1 complexes. One possibility is that unlabeled vancomycin helps create new sites for the labeled vancomycin to bind. It has been shown that vancomycin preferentially inhibits the transpeptidation step of PG synthesis in E. coli compared with the transglycosylation step (34). Therefore, it binds to d-Ala-d-Ala moieties found in the growing glycan strand in preference to the initiation sites of PG synthesis. If vancomycin behaves similarly in B. subtilis, the addition of vancomycin may block transpeptidation without completely blocking addition of new monomer units to the growing glycan strands. New sites for binding near the initiation sites of PG synthesis may be created by the addition of vancomycin, and the labeled probe could accumulate at these sites. Such an explanation would be consistent with the similarities in helical wall staining between vancomycin probes and ramoplanin probes, while also providing a rationale for why it is absolutely necessary to add unlabeled vancomycin to visualize sidewall staining with labeled vancomycin.
Surprisingly, we discovered that both vancomycin and ramoplanin probes stain the sidewalls of mbl− cells in a pattern that is qualitatively similar to the pattern observed in WT cells, albeit more compressed. Because the mbl null mutants are short and sometimes deformed, it is clear that Mbl plays an important role in the morphology of the cell. Furthermore, the compression in wall PG synthesis patterns suggests that Mbl plays an indirect role in directing PG synthesis through its control of cell length. Nevertheless, our results do not support the proposal that Mbl is essential for the incorporation of sidewall PG.
If Mbl does not direct cylindrical PG synthesis along the sidewalls of B. subtilis, then what does? Several other morphological proteins, including MreB and MreC, are involved in cell shape determination and may play roles. It was recently reported that in Caulobacter crescentus both MreB and MreC are required for the correct localization of PBP2, which was also found to form a helical pattern in this organism (35, 36). In addition, it is possible that localization of PG synthesis is correlated with the localization of protein transport machinery. Campo et al. (37) have reported that the core components of the Sec machinery, which is a major protein transport system in B. subtilis, are localized at the poles, the septa, and along the cylindrical walls in a helicoid structure that is independent of both MreB and Mbl helices. Several of the penicillin-binding proteins are exported by the Sec machinery. Perhaps the location of the machinery for PG synthesis is determined initially by where key components are transported, and then maintained by other processes, including interactions with other proteins on the cell surface and with the nascent PG chains and completed PG layers. Colocalization experiments with small-molecule probes of PG synthesis and candidate morphological proteins may shed more light on these issues.
Materials and Methods
Reagents.
Ramoplanin (2a) and vancomycin (1a) were gifts from Oscient Pharmaceuticals (Waltham, MA) and Merck, respectively. TMA-DPH, Van-BDP (1d), BodipyFL-SE, and 6-(fluorescein-5-carboxamido)hexanoic acid succinimidyl ester (fluorescein-C6-NHS) were purchased from Invitrogen–Molecular Probes. FITC and reagents for Edman degradation were purchased from Sigma–Aldrich.
Synthesis of Compounds 1b, 1c, 2b, 2c, and 2d.
Derivatives 2b, 2c, and 2d were prepared following reported procedures (26). Derivative 1b was prepared by mixing 1:1 vancomycin/fluorescein-C6-NHS with 7 μl of triethylamine in 0.4 ml of dimethylformamide (DMF). After 20 h of stirring at room temperature, 1b was purified by reverse-phase HPLC. 1c was prepared by mixing desleucyl vancomycin (25) and FITC in a 1:1 ratio with 15 μl of triethylamine in 1 ml of DMF. After 15 h of stirring at room temperature, the desired product was purified by reverse-phase HPLC.
Microscopy.
B. subtilis PY79 WT or mbl− cells grown in LB or CH growth medium (38) to an OD600 of 0.5 were centrifuged and incubated with probe solution in 1 ml of PBS buffer for 5–10 min, then washed three times with PBS. TMA-DPH was added to the centrifuged cells at a final concentration of 0.01 mM to stain the membrane. Cells were spotted on glass slides and immobilized with poly-Lys-treated coverslips. Fluorescence microscopy was performed with an Olympus BX61 microscope, equipped with phase-contrast and epifluorescence optics. Images were captured with a monochrome CoolSnapHQ digital camera (Photometrics, Tucson, AZ) and analyzed with metamorph 6.1 (Molecular Devices–Universal Imaging, Downingtown, PA).
For additional details on gene disruption, MIC measurements, probe synthesis, compound characterization, and microscopy, see Supporting Methods.
Supplementary Material
Acknowledgments
We thank Oscient Pharmaceuticals for ramoplanin, Merck for vancomycin, and A. D. Grossman (Massachusetts Institute of Technology, Cambridge) for the vector pKL147. This work was supported by National Institutes of Health Grant AI50855 (to S.W.) and National Institutes of Health Training Grant T32 GM007598 (to M.B.L.).
Abbreviations
- PG
peptidoglycan
- MIC
minimum inhibitory concentration
- Van-FL
fluorescent derivative of vancomycin
- Van-BDP
BodipyFL vancomycin
- TMA-DPH
1-(4-trimethylammoniumphenyl)-6-phenyl-1,3,5-hexatriene p-toluenesulfonate.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Archibald A. R., Hancock I. C., Harwood C. R. In: Bacillus subtilis and Other Gram-Positive Bacteria, Biochemistry, Physiology, and Molecular Genetics, Hoch J. A., Losick R., editors. Washington, DC: Am. Soc. Microbiol; 1993. pp. 381–410. [Google Scholar]
- 2.Holtje J.-V. Microbiol. Mol. Biol. Rev. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walsh C. Antibiotics: Actions, Origins, Resistance. Washington, DC: Am. Soc. Microbiol; 2003. [Google Scholar]
- 4.Young K. D. Mol. Microbiol. 2003;49:571–580. doi: 10.1046/j.1365-2958.2003.03607.x. [DOI] [PubMed] [Google Scholar]
- 5.Popham D. L., Young K. D. Curr. Opin. Microbiol. 2003;6:594–599. doi: 10.1016/j.mib.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 6.Bramhill D. Annu. Rev. Cell Dev. Biol. 1997;13:395–424. doi: 10.1146/annurev.cellbio.13.1.395. [DOI] [PubMed] [Google Scholar]
- 7.Scheffers D.-J., Jones L. J. F., Errington J. Mol. Microbiol. 2004;51:749–764. doi: 10.1046/j.1365-2958.2003.03854.x. [DOI] [PubMed] [Google Scholar]
- 8.Murray T., Popham D. L., Pearson C. B., Hand A. R., Setlow P. J. Bacteriol. 1998;180:6493–6502. doi: 10.1128/jb.180.24.6493-6502.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schlaeppi J. M., Schaefer O., Karamata D. J. Bacteriol. 1985;164:130–135. doi: 10.1128/jb.164.1.130-135.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mobley H. L. T., Koch A. L., Doyle R. J., Streips U. N. J. Bacteriol. 1984;158:169–179. doi: 10.1128/jb.158.1.169-179.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Den Blaauwen T., Aarsman M. E., Vischer N. O., Nanninga N. Mol. Microbiol. 2003;47:539–547. doi: 10.1046/j.1365-2958.2003.03316.x. [DOI] [PubMed] [Google Scholar]
- 12.de Pedro M. A., Quintela J. C., Holtje J. V., Schwarz H. J. Bacteriol. 1997;179:2823–2834. doi: 10.1128/jb.179.9.2823-2834.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burman L. G., Raichler J., Park J. T. J. Bacteriol. 1983;155:983–988. doi: 10.1128/jb.155.3.983-988.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daniel R. A., Errington J. Cell. 2003;113:767–776. doi: 10.1016/s0092-8674(03)00421-5. [DOI] [PubMed] [Google Scholar]
- 15.Charpentier X., Chalut C., Remy M. H., Masson J. M. J. Bacteriol. 2002;184:3749–3752. doi: 10.1128/JB.184.13.3749-3752.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schiffer G., Holtje J.-V. J. Biol. Chem. 1999;274:32031–32039. doi: 10.1074/jbc.274.45.32031. [DOI] [PubMed] [Google Scholar]
- 17.Vollmer W., von Rechenberg M., Holtje J.-V. J. Biol. Chem. 1999;274:6726–6734. doi: 10.1074/jbc.274.10.6726. [DOI] [PubMed] [Google Scholar]
- 18.Holtje J.-V. Microbiology. 1996;142:1911–1918. doi: 10.1099/13500872-142-8-1911. [DOI] [PubMed] [Google Scholar]
- 19.Romeis T., Holtje J.-V. J. Biol. Chem. 1994;269:21603–21607. [PubMed] [Google Scholar]
- 20.Zijderveld C. A., Aarsman M. E., den Blaauwen T., Nanninga N. J. Bacteriol. 1991;173:5740–5746. doi: 10.1128/jb.173.18.5740-5746.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walker S., Chen L., Hu Y., Rew Y., Shin D., Boger D. L. Chem. Rev. 2005;105:449–476. doi: 10.1021/cr030106n. [DOI] [PubMed] [Google Scholar]
- 22.Fang X., Tiyanont K., Zhang Y., Wanner J., Boger D., Walker S. Mol. BioSyst. 2006;2:69–76. doi: 10.1039/b515328j. [DOI] [PubMed] [Google Scholar]
- 23.Pinho M. G., Errington J. Mol. Microbiol. 2003;50:871–881. doi: 10.1046/j.1365-2958.2003.03719.x. [DOI] [PubMed] [Google Scholar]
- 24.Kahne D., Leimkuhler C., Lu W., Walsh C. Chem. Rev. 2005;105:425–448. doi: 10.1021/cr030103a. [DOI] [PubMed] [Google Scholar]
- 25.Allen N. E., LeTourneau D. L., Hobbs J. N., Jr., Thompson R. C. Antimicrob. Agents Chemother. 2002;46:2344–2348. doi: 10.1128/AAC.46.8.2344-2348.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Helm J. S., Chen L., Walker S. J. Am. Chem. Soc. 2002;124:13970–13971. doi: 10.1021/ja021097n. [DOI] [PubMed] [Google Scholar]
- 27.Hu Y., Helm J. S., Chen L., Ye X. Y., Walker S. J. Am. Chem. Soc. 2003;125:8736–8737. doi: 10.1021/ja035217i. [DOI] [PubMed] [Google Scholar]
- 28.Merchante R., Pooley H. M., Karamata D. J. Bacteriol. 1995;177:6176–6183. doi: 10.1128/jb.177.21.6176-6183.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Pedro M. A., Young K. D., Holtje J. V., Schwarz H. J. Bacteriol. 2003;185:1147–1152. doi: 10.1128/JB.185.4.1147-1152.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones L. J., Carballido-Lopez R., Errington J. Cell. 2001;104:913–922. doi: 10.1016/s0092-8674(01)00287-2. [DOI] [PubMed] [Google Scholar]
- 31.Cabeen M. T., Jacobs-Wagner C. Nat. Rev. Microbiol. 2005;3:601–610. doi: 10.1038/nrmicro1205. [DOI] [PubMed] [Google Scholar]
- 32.Fuchs-Cleveland E., Gilvarg C. Proc. Natl. Acad. Sci. USA. 1976;73:4200–4204. doi: 10.1073/pnas.73.11.4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ward J. B., Perkins H. R. Biochem. J. 1973;135:721–728. doi: 10.1042/bj1350721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ge M., Chen Z., Onishi H. R., Kohler J., Silver L. L., Kerns R., Fukuzawa S., Thompson C., Kahne D. Science. 1999;284:507–511. doi: 10.1126/science.284.5413.507. [DOI] [PubMed] [Google Scholar]
- 35.Divakaruni A. V., Loo R. R., Xie Y., Loo J. A., Gober J. W. Proc. Natl. Acad. Sci. USA. 2005;102:18602–18607. doi: 10.1073/pnas.0507937102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dye N. A., Pincus Z., Theriot J. A., Shapiro L., Gitai Z. Proc. Natl. Acad. Sci. USA. 2005;102:18608–18613. doi: 10.1073/pnas.0507708102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Campo N., Tjalsma H., Buist G., Stepniak D., Meijer M., Veenhuis M., Westermann M., Muller J. P., Bron S., Kok J., et al. Mol. Microbiol. 2004;53:1583–1599. doi: 10.1111/j.1365-2958.2004.04278.x. [DOI] [PubMed] [Google Scholar]
- 38.Nicholson W. L., Setlow P. In: Molecular Biological Methods for Bacillus, Harwood C. R., Cutting S. M., editors. New York: Wiley; 1990. pp. 391–450. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.