Abstract
It was, until now, not entirely clear how the nervous system attains its cellular phenotypic diversity and wired complexity during development. Here we describe how environmental interactions alone can modify the development of neurogenic precursor cells. Upon evaluating distinct growth-permissive substrates in an embryonic stem cell–neurogenesis assay, we found that laminin, fibronectin, and gelatin instruct neural fate and alter the functional specification of neurons when applied at distinct stages of development. Changes in phenotypic, electrophysiological, and molecular characteristics could resemble cellular events and interactions in the early embryonic brain and may explain why these extracellular matrix components transiently demarcate certain developing brain structures.
Keywords: cell culture, extracellular matrix, fibronectin, laminin, sonic hedgehog
The extracellular matrix (ECM) assembles three-dimensional templates around functional units of developing brain and spinal cord before neurons complete their final positioning and alignment. These structures contain large numbers of extracellular molecules that disappear soon after synaptic stabilization (1, 2). It is believed that transient ECM molecules act as “boundaries” that instruct afferent fiber ingrowth through adhesive and repulsive cues and thus play a significant role during CNS pattern formation (3). The function of the ECM might extend beyond morphogenetic effects, because loss or disarrangement of boundaries is observed in CNS diseases and malformations (1, 4–6) and because the ECM is involved in regulating synaptic plasticity in the adult (7). However, early embryonic lethality in some animal knockout models (8, 9) and only minor phenotypic abnormalities in others (10) puzzles the study of individual matrix molecules and their receptors during critical periods of neural development in vivo. ES cells used as a model system for neurogenesis offer an alternative approach in vitro. Neural differentiation of pluripotent ES cells can be directed in culture mimicking the entire temporal sequence of CNS development (11–13). Series of defined media and growth factors yield highly enriched multipotent precursors, and, in subsequent culture steps, a variety of molecules and growth factors influence neural subtype specification. For instance, morphogenetic factors [e.g., sonic hedgehog (SHH), FGF8, FGF4, and retinoic acid] direct the differentiation of different rostral and caudal CNS neuron groups (14–16). The focus of the present study was on potential roles for ECM and cell-substrate interactions during fate specification and differentiation of ES cell-derived neural precursors (ESNPs).
Results
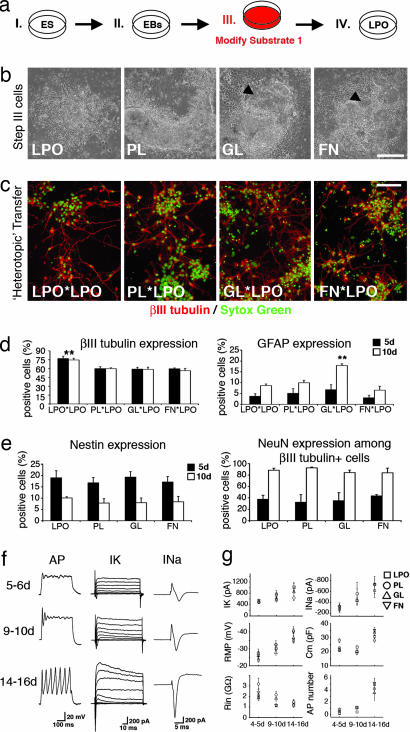
In an attempt to optimize the efficacy of generating neurons and glia from ES cells in vitro, we compared distinct growth-permissive dish coatings in a controlled four-step neural induction protocol. In this paradigm, ES cells grow attached to culture dishes in step III (generation of neural precursors) and IV (maturation of neural phenotypes), thus permitting the selective exposure to substrates in early and late developmental stages (Fig. 1a, see also Materials and Methods). At the end of culture step III, we noted the highest densities of neural precursors on laminin substrates used with laminin poly-l-ornithine (LPO). More nonneural phenotypes were present on plastic (PL), gelatin (GL) (a mixture of water-soluble collagen components), and fibronectin (FN) (Fig. 1b). “Homotopic” transfer of precursor cells into the same environmental condition in culture step IV further accelerated the differences of neural lineage enrichment. Interestingly, however, substrate-specific neuronal morphologies developed with time of differentiation in step IV (Fig. 5, which is published as supporting information on the PNAS web site). In contrast, “heterotopic” transfer of step III PL, GL, and FN populations to LPO conditions in culture step IV resulted in a robust increase of βIII tubulin+ cells (2.3, 4.7, and 5.9×, respectively), which were morphologically indistinguishable from one condition to another (Fig. 1c). These findings provided evidence for the common view that laminin enhances adhesion and survival of most types of neurons and glia. However, quantification of newborn neurons and glia on LPO in culture step IV revealed that, additionally, lineage decisions of neural precursors were influenced by substrate exposure in culture step III. LPO significantly increased the yield of βIII tubulin+ cells, and GL induced an exclusive, 2-fold rise of glial cell commitments (Fig. 1d). These findings, in combination with the apparent flexibility of neural cell morphologies, indicated that cell–substrate interactions in early stages of neurogenesis could influence neural fate, challenging a detailed analysis of substrate-specific cell development in protracted stages of differentiation.
Fig. 1.
Culture step III substrate modifications. (a) Experimental layout. (b) Phase-contrast images of cells at the end of step III. Note the nearly pure ESNP populations on LPO (arrowheads point to ESNP islands in GL and FN conditions). (c and d) Developing neurons appear morphologically uniform after transfer of step-III cells onto LPO in step VI (c), but quantification of neuronal and glial phenotypes reveals that, in step III, LPO increases neuronal and GL induces glial fate decisions (d). (e) Quantification of nestin and NeuN expression reveals a similar developmental time course in all investigated cell populations. (f) Single-cell patch-clamp analysis of differentiating neurons demonstrates typical progression of functional properties. (g) Stereotypic development of neuronal membrane biophysics on LPO, regardless of substrate pretreatment in culture step III. ∗∗, P < 0.001. (Scale bars, 150 μm.)
To evaluate the consequences of altered fate decisions, substrates were modified in culture step III, and neural cells were analyzed on LPO in step IV (Fig. 1a). Gradual maturation of neural phenotypes was observed in each condition, with neuronal commitment preceding glial differentiation. At 5 days, immunofluorescence analysis revealed distinct populations of young βIII tubulin+ neurons and immature nestin+ neuroepithelial phenotypes coexisting with a small number of glial fibrillary acidic protein-expressing radial glia-like cells (<4% per condition; data not shown). At 10 days, immature neuroepithelial rosettes had disappeared, most neurons expressed NeuN, and nestin was found in low levels restricted to young glial fibrillary acidic protein+ type I astrocytes (Fig. 6, which is published as supporting information on the PNAS web site). Only a few oligodendrocytes appeared on any substrate [<1% O4+ or 2′,3′-cyclic nucleotide-3′-phosphodiesterase (CNPase)+ cells; data not shown]. Quantification of decreasing nestin expression and increasing NeuN expression showed a similar developmental time course for all substrate-specific cell populations (Fig. 1e). We then used electrophysiology at 5 and 6, 9 and 10, and 14–16 days to additionally investigate the progression of neuronal membrane properties. Similar to previously reported findings in ES cell-derived neurons after transplantation (17, 18), over time, a gradual increase of voltage-dependent outward potassium (IK) and inward sodium (INa) current amplitudes was noted, as well as increasing numbers of action potentials (APs) in response to current injection (Fig. 1f). In vitro patch-clamp recordings were performed blinded to substrate conditions (5–13 cells per time point and condition; n = 129 cells) and revealed a remarkable stereotypy of developing biophysical profiles in all four groups of neurons. IK, INa, resting membrane potentials (RMPs), capacitances, input resistances, and maximum AP numbers were not significantly different at any stage of maturation (Fig. 1g). Thus, phenotypic and functional analysis alike suggested that, although substrates could alter neuronal vs. glial lineage decisions at early stages of development, the subsequent course of precursor cell maturation remained unaffected.
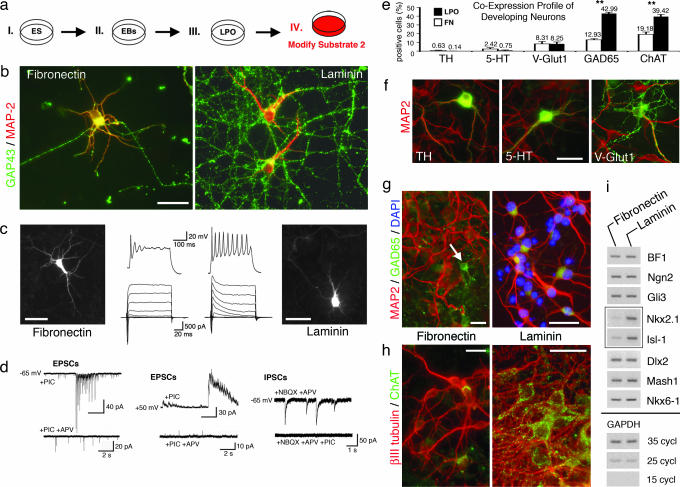
We were next curious to isolate the influence of cell–substrate interactions in later stages of neurogenesis. Neural precursors were generated on LPO in culture step III and then exposed to either LPO or FN in culture step IV (Fig. 2a). After 14–16 days of maturation, substrate-specific neuronal cell morphologies became apparent. In both conditions, neurons possessed polarized cell bodies with ramified microtubule-associated protein (MAP)-2+ dendritic trees but, consistent with previous reports (19, 20), enhanced outgrowth and branching of growth-associated protein (GAP)-43+ axons were observed on LPO substrates (Fig. 2b). Whole-cell patch-clamp recordings further revealed differences of intrinsic membrane characteristics at this stage. Neurons developing on FN showed a significant, ≈68%, increase in sodium channel densities but lacked the repetitive firing patterns typically observed in LPO conditions (Fig. 2c). Only 1/16 FN neurons fired more than five APs in response to 300-ms current injection, in comparison with 9/26 LPO neurons with repetitive discharge behaviors. These distinct functional and morphological profiles could not simply be attributed to different levels of cellular maturity. We examined this possibility and found that neuronal differentiation proceeded without significant difference on LPO and FN substrates. Within 4 or 5 and 9 or 10 days after plating in either condition, βIII tubulin+ cells demonstrated similar increasing levels of NeuN-expression (from ≈40% to >85%, respectively) and decreasing uptake of BrdU (from ≈15% to <7%, respectively) (data not shown). Also, at 14–16 days of maturation, developmentally regulated biophysical membrane characteristics (IK, RMPs, capacitances, and input resistances) were similar for FN and LPO neurons (Table 1). Furthermore, analysis of inhibitory postsynaptic currents (IPSCs) and excitatory postsynaptic currents (EPSCs) at this stage demonstrated that neurons on LPO (n = 10) and FN (n = 12) matured and successfully established functional GABAergic and glutamatergic synaptic transmission (Fig. 2d). However, these examinations exposed an additional remarkable finding. Spontaneous NMDA-mediated, tetrodotoxin-sensitive burst-like events were commonly observed in FN neurons plated at 7.5–10 × 104 cells per cm2. Similar activity in LPO conditions required 1.5–2 times more plated cells (≥15 × 104 cells per cm2). Immunofluorescence analysis tied this finding to a substrate-induced alteration of neuronal subtype specification. Fourteen to 16 days of exposure to LPO increased the number of cholinergic and glutamate decarboxylase 65 (GAD65)-expressing inhibitory cells significantly in comparison with FN conditions at comparable cell densities (105 cells per cm2) (Fig. 2 e, g, and h), and because similar quantities of vesicularglutamate transporter 1 excitatory neurons were generated in either condition, the ratio of glutamatergic/GABAergic cells was altered from 1:1.6 on FN to 1:5.2 on LPO substrates. Differentiation into other neurotransmitter phenotypes was not substrate-dependent. Proportions of tyrosine hydroxylase dopaminergic and 5-hydroxytryptamine catecholamine/serotoninergic neurons were not significantly different (Fig. 2f).
Fig. 2.
Culture step IV substrate modifications. (a) Experimental layout. (b) Axo-dendritic appearance of neurons on FN vs. LPO substrates. (c) Whole-cell patch-clamp recordings reveal repetitive firing behavior of LPO neurons and significantly increased Na+ currents and singular APs on FN substrates. Images reflect biocytin immunofluorescence of individual recorded cells. (d) Analysis of spontaneous synaptic activity demonstrating ubiquitous excitatory [combined α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA)- and NMDA-mediated EPSCs (Left); isolated NMDA components (Center)] and inhibitory events (Right). Note that 2-amino-5-phosphonovaleric acid (APV) blocks NMDA-mediated burst-like activity (lower EPSC traces). (e) Quantification of substrate-specific neuronal subtype antigen profiles. (f–h) Fluorescence photomicrographs depict transmitter antigens (green) of phenotypes without (f) or with (g and h) differential neuronal fate choice in LPO vs. FN conditions. Note varicosities that may represent GABAergic boutons arising from a GAD65+ cell as marked by the arrow (g Left). (i) Semiquantitative RT-PCR demonstrates a selective increase of the transcription factors Nkx2.1 and Isl-1 on LPO substrates. ∗∗, P < 0.001. (Scale bars: in b and f–h, 50 μm; in c, 30 μm.)
Table 1.
Comparison of neuronal membrane properties after substrate-dependent maturation for 14–16 days
| INa, nA | IK, nA | Cm, pF | RMP, mV | Rin, GΩ | Ra, MΩ | No. of APs | |
|---|---|---|---|---|---|---|---|
| LPO | 0.79 ± 0.1 (21)* | 1.35 ± 0.2 (21) | 34.0 ± 1.5 (26) | −37.2 ± 1.4 (26) | 1.36 ± 0.1 (26) | 10.8 ± 0.8 (26) | 4.3 ± 0.6 (26) |
| FN | 1.33 ± 0.2 (14) | 1.58 ± 0.2 (14) | 34.5 ± 1.3 (16) | −39.1 ± 1.1 (16) | 1.35 ± 0.1 (16) | 12.4 ± 3.1 (16) | 2.7 ± 0.3 (16) |
| P value | 0.0064† | 0.4857 | 0.8101 | 0.3516 | 0.9567 | 0.073 | 0.0496† |
INa, maximum sodium currents; IK, maximum potassium currents; Cm, capacitance; RMP, resting membrane potential; Rin, input resistance; Ra, series resistance; AP, maximum number of action potentials in response to 200 ms of current injection.
*The number of recorded cells is listed in parentheses.
†Significantly different.
Semiquantitative RT-PCR was used to additionally compare the expression of a broad list of transcription factors known to represent distinct developmental regions in the maturing CNS (21–24). At 14–16 days of differentiation, we found a surprisingly exclusive increase in the expression of Nkx2.1 and Isl-1 on LPO substrates, whereas other intrinsic regulators of neural development along the dorsoventral and rostrocaudal axes seemed unaffected (Fig. 2i). Interestingly, levels of Nkx2.1 and Isl-1 remained low when cells grew in the presence of LPO in culture step III and then were transferred onto FN in step IV. In contrast, Nkx2.1 and Isl-1 expression was strong when FN was present in step III and LPO was present in culture step IV (Fig. 7, which is published as supporting information on the PNAS web site). Thus, stimulation of Nkx2.1 and Isl-1 expression critically depends on the timing of LPO application in later stages of neural precursor cell development.
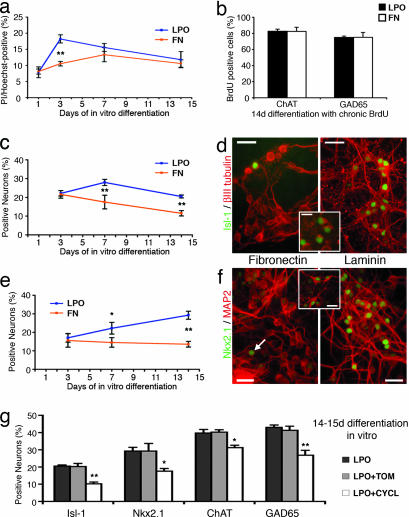
To evaluate potential mechanisms for differential neuronal fate choice in culture step IV LPO conditions, we first determined the levels of dying cells at various time points during substrate-specific differentiation (Fig. 3a). A significantly increased presence of propidium iodide+ cells in LPO was noted at 3 days of differentiation, indicating a selection process at this time. Continuous BrdU application, however, did not reveal a substrate-specific proliferation of either GABAergic or cholinergic cell populations (Fig. 3b). Fig. 3 c–f shows a characteristic developmental progression of Isl-1 and Nkx2.1 neuronal phenotypes with significant substrate-specific differences occurring not earlier than 7 days of in vitro differentiation. This finding suggests that induction in addition to selection is responsible for differential neuronal fate choice under LPO growth conditions. Colabeling studies at 14 days of differentiation in culture demonstrated that most Nkx2.1- and Isl-1-expressing cells are choline acetyltransferase (ChAT)+ cholinergic neurons (84 ± 7% and 83 ± 2%, respectively; see Fig. 3 d and f Insets). Finally, to evaluate a potential mechanism of LPO-mediated modulation (e.g., increase) of SHH signaling as seen in vivo (21), pharmacological blockade of SHH pathways was found to significantly reduce all phenotypes that we observed to increase in LPO conditions (Fig. 3g).
Fig. 3.
Evaluating mechanisms for differential neuronal fate choice in culture step IV. (a) Ratio of propidium iodide (Pi)/Hoechst indicates levels of dead cells at certain time points of in vitro differentiation. (b) Evaluation of BrdU uptake indicates no substrate-specific selective proliferation of ChAT+ or GAD65+ precursors throughout culture step IV. (c and d) Developmental profile of Isl-1 expression in LPO vs. FN conditions (c) and examples of Isl-1+/βIII tubulin+ cells at 14-days differentiation (d). (d Inset) Isl-1+/ChAT+ neurons. (e and f) Similar profile and examples of Nkx2.1-expressing cells (arrow in f points to one of these cells). (f Inset) Nkx2.1+/ChAT+ neurons. (g) Blocking SHH pathways with cyclopamine (CYCL) in LPO conditions significantly decreases the quantity of Isl-1+, Nkx2.1+, ChAT+, and GAD65+ neurons compared with control levels [tomatidine (TOM), a structural homologue of cyclopamine] without SHH-inhibiting activity. ∗, P < 0.05. ∗∗; P < 0.01. (Scale bars: d and f, 50 μm; Insets, 25 μm.)
All of these data together support a notion that differentiating neural progeny might be influenced by LPO conditions in a temporally restricted manner to induce characteristic cellular phenotypes as seen in the developing medial ganglionic eminence, as discussed below.
Discussion
It is generally accepted that phenotypic diversity is established during development as the result of a combination of genetic and epigenetic factors (25). However, little is known about environmental interactions during CNS development. Extracellular signaling is not essential to acquire neural identities (26), but interactions with ECM through integrin receptors can determine the developmental fate of pluripotent cells (27). On these grounds, we investigated underlying principles of transiently expressed ECM in the embryonic brain by using an in vitro model of ES cell-derived neurogenesis. We show here that, by modifying culture dish coating protocols from those originally described by Okabe et al. (11), one can indeed affect neural fate choice and that the effects of environmental signaling strongly depend on the developmental stage of immature neural cells. When multipotent precursors are generated in our culture step III, substrates such as GL and LPO influence glial vs. neuronal fate decisions. Our evaluations demonstrate that these early interactions do not affect later stages of differentiation. Regardless of pretreatment in step III, developing neural progeny display similar characteristics during maturation on LPO. In contrast, we demonstrate that neural subtype specification can be altered when substrates interact with developmentally more committed precursor cells; two distinct neuronal cell populations appear as result of exposure to FN or LPO in culture step IV.
These molecules represent prominent members in the family of developmentally regulated ECM components, and some of our findings can be correlated with cellular events and interactions in the early embryonic brain. During formation of the telencephalon, transient expression of FN, and distribution around radial glia in the preplate was suggested to serve as positional cues for young neurons that form the cortical plate (28). Interestingly, and similar to developing cortical plate cells in vivo (29), patch-clamp recordings of culture step-IV FN neurons demonstrate a remarkable increase of Na channel densities upon maturation. These neurons also show restricted axonal outgrowth and branching resembling patterns present in the subplate, where FN helps determine whether afferent fibers have to wait or can pass through into the cortical plate (30). In contrast, LPO substrates stimulate the functional maturation of GAD65+ neurons in our neurogenesis assay. It is tempting to speculate that this specific phenotypic induction observed in vitro corresponds to a prominent early population of GABAergic cells (31) appearing simultaneously with a transient laminin expression in the subplate (32). However, the unexpected selective increase of Nkx2.1 and Isl-1expression levels and the concomitant increase of ChAT+ cholinergic neurons might indicate a previously underestimated, temporally restricted phenotypic induction of, e.g., medial ganglionic eminence identity that is mediated through LPO conditions. It is extremely tempting to speculate that the GABAergic neuronal phenotypes observed under our LPO growth conditions reflect a medial ganglionic eminence origin. However, at present, we do not have enough data to support such a hypothesis. Among the potential mechanisms that might underlie a role for an ECM molecule during the induction of forebrain identity, the results from cell death and cell birth studies, along with SHH pathway blockade, together teach us that both selection and induction must take place. Although the precise sequence of cellular events and interactions during formation of the forebrain remains to be resolved (33), it appears that transient patterns of ECM could encrypt substrate- and time-specific cues for fate and function of developing neural precursor cells in vitro and in vivo. It is known that ECM interacts with, binds to, and helps present growth factors to neurons and their precursors (3), and ECM could thus potentially select desired phenotypes by promoting their proliferation or by eliminating unwanted progeny. Although direct evidence is lacking that laminin–receptor (e.g., integrin) interactions with SHH is involved in forebrain cell fate decisions, it is known that SHH does bind to laminin and depends on βI integrin expression (34). Furthermore, there are findings that suggest a possible link between ECM and SHH during ventral patterning throughout the neuraxis (35).
In conclusion, the presented evidence for morphogenic roles of ECM during ES cell-derived neural and neuronal subtype and fate specification now further supports a notion that surface does matter. In the advent of stem cell technology, large-scale assays (36, 37) could soon reveal other morphogenic adhesion molecules and further propel the exciting new perspective of directing ES cell differentiation toward highly specified neural lineages under the control of extracellular cues.
Materials and Methods
Cell Culture.
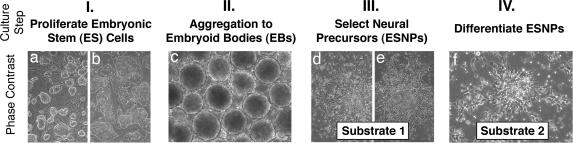
Experiments were performed using R1 mouse ES cells, and findings were verified by using ubiquitously GFP- and YFP-expressing R1 ES cells (A. Nagy, University of Toronto, Toronto, Canada), and τ EGFP knockin J1 ES cells (K. L. Tucker, University of Heidelberg, Heidelberg, Germany) (data not shown). Controlled neural differentiation was achieved by adopting a common four-step culture protocol (11) (Fig. 4). Undifferentiated ES cells (62.5 × 103 cells per cm2) were expanded on mitomycin C-inhibited (10 μg/ml for 2.5 h; ICN) embryonic fibroblasts and subsequently on GL (0.1% precoating, 30 min) in culture step I before differentiation was induced in step II by aggregating 20 cm2 cells (90% confluent) to free-floating embryoid bodies (EBs) (in 8 × 6-cm nonadhesive dishes; Nunc 4034). In step III, EB-derived cells grow surface-attached until neural precursors appear. Regardless of which culture dish coating was used, 5 μg FN per milliliter of culture media (ITSFn) (see Supporting Appendix, which is published as supporting information on the PNAS web site) was present in all experiments in culture step III. Cells are trypsinized, triturated through flame-polished Pasteur pipettes, and exposed to a second substrate in culture step IV. Because FGF-2 (10 ng/ml) was recently shown to induce ventral phenotypes in cultured neural precursors during prolonged exposure (≥3 days) (38, 39), FGF-2 application in culture step IV was limited to the first 24 h. FGF-2 withdrawal initiated the final stages of differentiation (designated as day 1). Media were changed every other day in all steps of the protocol (for more details and media formulations, see Supporting Appendix). Media, supplements, and growth factors were obtained from Invitrogen, Sigma, and R & D Systems.
Fig. 4.
Culture protocol. (a and b) In step I, pluripotent ES cells expand on embryonic fibroblasts (a) and GL (b). (c) Step II represents the first stage of differentiation, when ES cells aggregate for 3 days in suspension to form EBs. (d and e) Step III is a 7-day period in which ESNPs are selected in adhesive conditions. (d) One day after EB plating. (e) Last day of step III. (f) In step IV, selected cells differentiate spontaneously on a second substrate. In all experiments, neural progeny was evaluated at 5, 10, or 15 (±1) days after passage on substrate 2.
Surface and substrate conditions were standardized. Falcon plasticware (Becton Dickinson) was used in step III (#353803) and IV (#353002). LPO coating included incubation in 140 μl/cm2 LPO solution (15 μg/ml; P-3655; Sigma) at 37°C for 96 h followed by washes in Ca2+/Mg2+-free PBS and in DMEM/F12 before cells were plated in the presence of 1 μg/ml laminin-1 (23017-015; Invitrogen). Thus, our LPO coating varies from the originally described technique (11) in which dishes are precoated with mixtures of polyornithine and laminin before cells are plated in media with laminin. FN coating was carried out with 50 μl/cm2 FN (50 μg/ml; 33010-018; Invitrogen) at 37° for 1 h. Dishes were washed three times in DMEM/F12 before cell seeding. GL coating was performed for 30 min at room temperature by using 140 μl/cm2 GL (0.1%; G-1890; Sigma). GL was removed before the plating of cells. Proliferating cells were labeled with either 48-h pulses of 10 μM or 2 μM BrdU (Sigma) applied throughout the differentiation period in step IV. Cell death analysis was performed in living cultures, with 1 μg/ml propidium iodide (Sigma) for labeling dead cells and 1 μg/ml Hoechst 33342 (Sigma) for labeling living and dead cells. SHH blocking experiments required 10 μM cyclopamine (LC Laboratories, Woburn, MA) or 10 μM tomatidine (Sigma) (a structural homologue of cyclopamine not inhibiting SHH signaling) applied throughout culture step IV.
Electrophysiology.
Culture dishes were transferred to the stage of an upright, fixed-stage microscope (Axioskop FS; Zeiss) and perfused with artificial cerebrospinal fluid (1–2 ml/min, saturated with 95% O2/5% CO2) containing 124 mM NaCl, 26 mM NaHCO3, 1.25 mM NaH2PO4, 2.5 mM KCl, 2 mM CaCl2, 2 mM MgCl2, and 10 mM glucose (pH 7.4). Cells with typical neuronal morphology (i.e., large, triangular soma, single apical dendrite, located in a substantial neuritic network of neighboring cells) were targeted by using a ×40 water-immersion objective and infrared differential interference contrast microscopy optics. Immunocytochemistry was used in nine independent experiments to verify neuronal identity of recorded cells. All cells (n = 18; identified with fluorophore-conjugated avidin) expressed neuronal antigens (MAP-2 or βIII tubulin) and were negative for glial antigens (glial fibrillary acidic protein or S100β). Whole-cell recordings were obtained at room temperature (22°C) by using an Axopatch 1D amplifier (Axon Instruments, Foster City, CA). Patch electrodes had a resistance of 7–8 MΩ when filled with internal solution. The pipette solution contained 125 mM K-gluconate, 8 mM NaCl, 10 mM Hepes, 2 mM MgATP, 0.3 mM Na3GTP, 0.2 mM EGTA and 0.1% biocytin (pH 7.3, adjusted with KOH; 290–300 mOsm) for recording EPSCs and sodium and potassium currents, or 130 mM CsCl, 10 mM Hepes, 2 mM MgATP, 0.3 mM Na3GTP, 0.2 mM EGTA, and 0.1% biocytin (pH 7.3, adjusted with CsOH; 290–300 mOsm) for IPSCs. Neurons were voltage-clamped at −65 mV for both EPSC and IPSC recording. Series resistance was similar in all evaluated cells (8–18 MΩ), and recordings were rejected if series resistance changed >10%. NMDA receptor-mediated spontaneous EPSCs were evaluated at a holding potential of +40 or +50 mV by using Cs-gluconate internal solution in the presence of picrotoxin (50 μM) to block GABA-mediated synaptic responses. IPSCs were recorded in the presence of 2,3-dihydroxy-6-nitro-7-sulfamoylbenzo[f]quinoxaline (10 μM) and 2-amino-5-phosphonovaleric acid (50 μM) to block glutamatergic synaptic transmission. Chemicals were purchased from Sigma unless stated otherwise. For characterizing spiking properties, a series of current pulses (from −50 to +150 pA, in 50-pA increments) was applied in current-clamp mode, and maximum numbers and amplitudes of APs were valued. Amplitudes of maximum INa (occurred at +5 or +30 mV) and the steady (sustained) component of IK (at +55 mV) were determined by applying a series of voltage steps (from −80 to +50 mV, in 15-mV increments) under voltage-clamp mode.
Data were acquired and saved by using pclamp 8 (Axon Instruments) and analyzed by using clampfit 8 and origin 6 software. Values are expressed as the mean ± SEM. Student’s t test was used for evaluation of statistical significance.
Immunocytochemistry.
Cells were fixed for 30 min with 4% paraformaldehyde. The basic immunolabeling buffer contained PBS (Invitrogen), 10% FCS, and for intracellular antigens additionally 0.1% Triton X-100 (Sigma). After blocking unspecific antibody activity for 20 min in 5% goat serum, primary antibodies (see Table 2, which is published as supporting information on the PNAS web site, for details) were applied 1–4 h at room temperature or overnight at 4°C. BrdU detection required pretreatment for 2 h in SSC-formamide, 3 × 10 min washes in SSC, 30 min in 2M HCl, and 10 min in 0.1 M borate buffer. Antigens (including biocytin) were visualized by using corresponding secondary antibodies (Jackson ImmunoResearch or Molecular Probes). Cell nuclei were labeled for 10 min with either 0.8 μg/ml DAPI (Sigma) or green nucleic acid stain (Sytox green; 1:50,000; Molecular Probes). Fluorescence microscopy was performed on a Leica (Bannockburn, IL) DMLB upright microscope and images were captured with a Spot RT Color CCD camera (Diagnostic Instruments, Sterling Heights, MI). To quantify and compare antigen expression, cell counting was performed in three or four culture dishes (6-cm diameter) per condition from at least two independent experiments. At least 1,000 total cells (mean = 3,497 ± 1,915) were evaluated per dish for antigen expression from 15 separate, randomly chosen ×40 visual fields. Data are expressed as mean ± SEM, and Student’s t test was used for statistical evaluation.
RNA Extraction and RT-PCR Analysis.
Total cellular RNA was isolated by using the RNeasy Mini Kit (Qiagen, Valencia, CA), with DNase I treatment on the column. Eluted RNA was subjected to a second DNase I treatment using the Turbo DNA-free kit (Ambion, Woodward, TX). Total RNA concentration was measured by using a UV spectrophotometer (SmartSpec 3000; Bio-Rad). Isolated RNA had an A260/A280 ratio of 2; 3 μg total RNA per condition was used to synthesize cDNA according to the instructions of the SuperScript III reverse transcriptase kit (Invitrogen). cDNA samples were normalized based on GAPDH as a reference gene. PCRs were performed by using Platinum Taq polymerase (Invitrogen). The total number of cycles varied between 15 and 39, depending on the abundance of particular mRNA. The number of cycles was chosen on the linear portion of the reaction curve avoiding “saturation effects” of PCR. Single bands of the amplification products were confirmed in a 2% agarose gel (Invitrogen) containing ethidium bromide for visualization. Minus RT reactions were set up, and PCR negative controls were run for each pair of primers. Primer sequences, length of amplified products, and PCR conditions are summarized in Table 3, which is published as supporting information on the PNAS web site.
Supplementary Material
Acknowledgments
We thank Dirk Dietrich for discussion of findings and for critically reviewing the manuscript. This work was supported by National Institutes of Health/National Institute of Neurological Disorders and Stroke Grants NS37556, HL70143 (to D.A.S.), and NS46384 (to B.S.), Federal Ministry of Education and Research (BMBF) Grant 01GN0502 (to O.B.), and the Evelyn F. and William L. McKnight Brain Research Foundation Fund (B.S.).
Abbreviations
- AP
action potential
- ChAT
choline acetyltransferase
- EB
embryoid body
- ECM
extracellular matrix
- EPSC
excitatory postsynaptic currents
- ESNP
ES cell-derived neural precursor
- FN
fibronectin
- GL
gelatin
- IK
outward potassium current
- INa
inward sodium current
- IPSC
inhibitory postsynaptic currents
- LPO
laminin poly-l-ornithine
- MAP
microtubule-associated protein
- PL
plastic
- RMP
resting membrane potential
- SHH
sonic hedgehog
- GAD65
glutamate decarboxylase 65.
Footnotes
Conflict of interest statement: D.A.S. and B.S. are involved with a biotechnology start-up company, RegenMed, Inc. This company is involved with stem cell technology related to human therapeutics.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Steindler D. A. Annu. Rev. Neurosci. 1993;16:445–470. doi: 10.1146/annurev.ne.16.030193.002305. [DOI] [PubMed] [Google Scholar]
- 2.Letourneau P. C., Condic M. L., Snow D. M. J. Neurosci. 1994;14:915–928. doi: 10.1523/JNEUROSCI.14-03-00915.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Faissner A., Steindler D. Glia. 1995;13:233–254. doi: 10.1002/glia.440130402. [DOI] [PubMed] [Google Scholar]
- 4.Cooper N. G., Steindler D. A. Brain Res. 1989;489:167–176. doi: 10.1016/0006-8993(89)90020-6. [DOI] [PubMed] [Google Scholar]
- 5.Steindler D. A., Faissner A., Harrington K. L. Cereb. Cortex. 1994;4:129–137. doi: 10.1093/cercor/4.2.129. [DOI] [PubMed] [Google Scholar]
- 6.Scheffler B., Faissner A., Beck H., Behle K., Wolf H. K., Wiestler O. D., Blumcke I. Glia. 1997;19:35–46. [PubMed] [Google Scholar]
- 7.Dityatev A., Schachner M. Nat. Rev. Neurosci. 2003;4:456–468. doi: 10.1038/nrn1115. [DOI] [PubMed] [Google Scholar]
- 8.George E. L., Georges-Labouesse E. N., Patel-King R. S., Rayburn H., Hynes R. O. Development (Cambridge, U.K.) 1993;119:1079–1091. doi: 10.1242/dev.119.4.1079. [DOI] [PubMed] [Google Scholar]
- 9.Colognato H., Yurchenco P. D. Dev. Dyn. 2000;218:213–234. doi: 10.1002/(SICI)1097-0177(200006)218:2<213::AID-DVDY1>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 10.Steindler D. A., Settles D., Erickson H. P., Laywell E. D., Yoshiki A., Faissner A., Kusakabe M. J. Neurosci. 1995;15:1971–1983. doi: 10.1523/JNEUROSCI.15-03-01971.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okabe S., Forsberg-Nilsson K., Spiro A. C., Segal M., McKay R. D. Mech. Dev. 1996;59:89–102. doi: 10.1016/0925-4773(96)00572-2. [DOI] [PubMed] [Google Scholar]
- 12.Brustle O., Jones K. N., Learish R. D., Karram K., Choudhary K., Wiestler O. D., Duncan I. D., McKay R. D. Science. 1999;285:754–756. doi: 10.1126/science.285.5428.754. [DOI] [PubMed] [Google Scholar]
- 13.Bibel M., Richter J., Schrenk K., Tucker K. L., Staiger V., Korte M., Goetz M., Barde Y. A. Nat. Neurosci. 2004;7:1003–1009. doi: 10.1038/nn1301. [DOI] [PubMed] [Google Scholar]
- 14.Lee S. H., Lumelsky N., Studer L., Auerbach J. M., McKay R. D. Nat. Biotechnol. 2000;18:675–679. doi: 10.1038/76536. [DOI] [PubMed] [Google Scholar]
- 15.Kim J. H., Auerbach J. M., Rodriguez-Gomez J. A., Velasco I., Gavin D., Lumelsky N., Lee S. H., Nguyen J., Sanchez-Pernaute R., Bankiewicz K., McKay R. Nature. 2002;418:50–56. doi: 10.1038/nature00900. [DOI] [PubMed] [Google Scholar]
- 16.Wichterle H., Lieberam I., Porter J., Jessell T. Cell. 2002;110:385–397. doi: 10.1016/s0092-8674(02)00835-8. [DOI] [PubMed] [Google Scholar]
- 17.Benninger F., Beck H., Wernig M., Tucker K. L., Brustle O., Scheffler B. J. Neurosci. 2003;23:7075–7083. doi: 10.1523/JNEUROSCI.23-18-07075.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wernig M., Benninger F., Schmandt T., Rade M., Tucker K. L., Bussow H., Beck H., Brustle O. J. Neurosci. 2004;24:5258–5268. doi: 10.1523/JNEUROSCI.0428-04.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rogers S. L., Letourneau P. C., Palm S. L., McCarthy J., Furcht L. T. Dev. Biol. 1983;98:212–220. doi: 10.1016/0012-1606(83)90350-0. [DOI] [PubMed] [Google Scholar]
- 20.Tom V. J., Doller C. M., Malouf A. T., Silver J. J. Neurosci. 2004;24:9282–9290. doi: 10.1523/JNEUROSCI.2120-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kohtz J. D., Baker D. P., Corte G., Fishell G. Development (Cambridge, U.K.) 1998;125:5079–5089. doi: 10.1242/dev.125.24.5079. [DOI] [PubMed] [Google Scholar]
- 22.Anderson S., Mione M., Yun K., Rubenstein J. L. Cereb. Cortex. 1999;9:646–654. doi: 10.1093/cercor/9.6.646. [DOI] [PubMed] [Google Scholar]
- 23.Fode C., Ma Q., Casarosa S., Ang S. L., Anderson D. J., Guillemot F. Genes Dev. 2000;14:67–80. [PMC free article] [PubMed] [Google Scholar]
- 24.Backman M., Machon O., Mygland L., van den Bout C. J., Zhong W., Taketo M. M., Krauss S. Dev. Biol. 2005;279:155–168. doi: 10.1016/j.ydbio.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 25.Watt F. M., Hogan B. L. Science. 2000;287:1427–1430. doi: 10.1126/science.287.5457.1427. [DOI] [PubMed] [Google Scholar]
- 26.Tropepe V., Hitoshi S., Sirard C., Mak T. W., Rossant J., van der Kooy D. Neuron. 2001;30:65–78. doi: 10.1016/s0896-6273(01)00263-x. [DOI] [PubMed] [Google Scholar]
- 27.Czyz J., Wobus A. Differentiation (Berlin) 2001;68:167–174. doi: 10.1046/j.1432-0436.2001.680404.x. [DOI] [PubMed] [Google Scholar]
- 28.Sheppard A. M., Hamilton S. K., Pearlman A. L. J. Neurosci. 1991;11:3928–3942. doi: 10.1523/JNEUROSCI.11-12-03928.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Picken Bahrey H. L., Moody W. J. J. Neurophysiol. 2003;89:1761–1773. doi: 10.1152/jn.00972.2002. [DOI] [PubMed] [Google Scholar]
- 30.Chun J. J., Shatz C. J. J. Cell Biol. 1988;106:857–872. doi: 10.1083/jcb.106.3.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Del Rio J. A., Soriano E., Ferrer I. J. Comp. Neurol. 1992;326:501–526. doi: 10.1002/cne.903260403. [DOI] [PubMed] [Google Scholar]
- 32.Hunter D. D., Llinas R., Ard M., Merlie J. P., Sanes J. R. J. Comp. Neurol. 1992;323:238–251. doi: 10.1002/cne.903230208. [DOI] [PubMed] [Google Scholar]
- 33.Campbell K. Neuron. 2005;46:373–376. doi: 10.1016/j.neuron.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 34.Blaess S., Graus-Porta D., Belvindrah R., Radakovits R., Pons S., Littlewood-Evans A., Senften M., Guo H., Li Y., Miner J. H., et al. J. Neurosci. 2004;24:3402–3412. doi: 10.1523/JNEUROSCI.5241-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ericson J., Muhr J., Placzek M., Lints T., Jessell T. M., Edlund T. Cell. 1995;81:747–756. doi: 10.1016/0092-8674(95)90536-7. [DOI] [PubMed] [Google Scholar]
- 36.Anderson D. G., Levenberg S., Langer R. Nat. Biotechnol. 2004;22:863–866. doi: 10.1038/nbt981. [DOI] [PubMed] [Google Scholar]
- 37.Flaim C. J., Chien S., Bhatia S. N. Nat. Methods. 2005;2:119–125. doi: 10.1038/nmeth736. [DOI] [PubMed] [Google Scholar]
- 38.Gabay L., Lowell S., Rubin L. L., Anderson D. J. Neuron. 2003;40:485–499. doi: 10.1016/s0896-6273(03)00637-8. [DOI] [PubMed] [Google Scholar]
- 39.Hack M. A., Sugimori M., Lundberg C., Nakafuku M., Gotz M. Mol. Cell. Neurosci. 2004;25:664–678. doi: 10.1016/j.mcn.2003.12.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.