Abstract
Semiconductor quantum dots are becoming valuable analytical tools for biomedical applications. Indeed, their unique photophysical properties offer the opportunity to design luminescent probes for imaging and sensing with unprecedented performance. In this context, we have identified operating principles to transduce the supramolecular association of complementary receptor–substrate pairs into an enhancement in the luminescence of sensitive quantum dots. Our mechanism is based on the electrostatic adsorption of cationic quenchers on the surface of anionic quantum dots. The adsorbed quenchers suppress efficiently the emission character of the associated nanoparticles on the basis of photoinduced electron transfer. In the presence of target receptors able to bind the quenchers and prevent electron transfer, however, the luminescence of the quantum dots is restored. Thus, complementary receptor–substrate pairs can be identified with luminescence measurements relying on our design logic. In fact, we have demonstrated with a representative example that our protocol can be adapted to signal protein–ligand interactions.
Keywords: electron transfer, luminescent chemosensors, nanoparticles, protein–ligand interactions
Semiconductor quantum dots are inorganic nanoparticles with remarkable photophysical properties (1–5). In particular, their one- and two-photon absorption cross-sections, luminescence lifetimes, and photobleaching resistances are significantly greater than those of conventional organic fluorophores. Furthermore, their broad absorption bands extend continuously from the UV to the visible region of the electromagnetic spectrum and, therefore, offer a vast selection of possible excitation wavelengths. Instead, their narrow emission bands can be positioned precisely within the visible and near-infrared regions with fine adjustments of their physical dimensions. In fact, pools of quantum dots with different diameters can be designed to emit in parallel at different wavelengths after excitation at a single wavelength, offering the opportunity to implement unprecedented multichannel assays.
Organic dyes do not offer the unique collection of attractive photophysical properties associated with semiconductor quantum dots. Indeed, it is becoming apparent that these inorganic nanoparticles can complement, if not replace, their organic counterparts in a diversity of biomedical applications (6–12). Nonetheless, decades of intensive investigations on the structure and properties of organic chromophores have indicated valuable strategies to design sensitive fluorescent probes able to signal the presence of target analytes with changes in emission intensity (13–16). Their operating principles generally rely on the covalent connection of a fluorescent component to a receptor unit. The receptor is engineered to quench the emission of the fluorophore on the basis of either electron or energy transfer. The supramolecular association of the receptor with a complementary analyte, however, suppresses the quenching mechanism and leads to a significant enhancement in fluorescence intensity. Under these conditions, the presence of the target analyte is transduced into a detectable fluorescence signal. At the stage of their development, however, it is not entirely clear whether and how these strategies can successfully be extended to quantum dots.
Promising studies demonstrate that semiconductor quantum dots can donate energy to complementary partners (17). In fact, clever assays for the recognition of various analytes are starting to be designed on the basis of energy transfer processes (18–34). Instead, mechanisms based on electron transfer still remain to be explored (35, 36). In this context, we have designed a competitive binding assay based on the photoinduced transfer of electrons from quantum dots to organic acceptors. Our method relies on the electrostatic association of a quencher on the surface of a quantum dot (Fig. 1). Under these conditions, the photoinduced transfer of electrons from the excited nanoparticles to the adsorbed acceptors is expected to translate into an efficient luminescence quenching. The luminescence of the quantum dots, however, can be restored by adding a receptor able to sequester and remove the quencher from the surface of the nanoparticles. Thus, the presence of the receptor can be transduced into a luminescent enhancement on the basis of this mechanism. Here, we demonstrate that these operating principles can indeed be implemented experimentally and that they can even be extended to signal protein–ligand interactions.
Fig. 1.
The supramolecular association of quencher and receptor prevents the electron transfer process and activates the luminescence of the quantum dot.
Results and Discussion
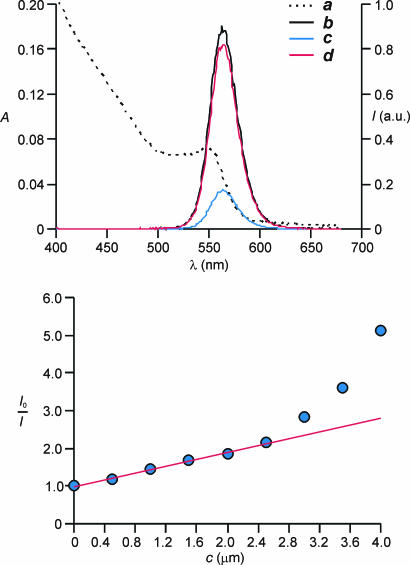
To demonstrate the viability of our design, we prepared CdSe–ZnS core–shell quantum dots coated with tri-n-octylphosphine oxide ligands, adapting literature procedures (37, 38). Then, we exchanged the hydrophobic ligands with hydrophilic mercaptoacetate groups (39).† The resulting nanoparticles are soluble in water, absorb at 548 nm (line a in Fig. 2Upper), and emit at 563 nm (line b in Fig. 2 Upper). The addition of increasing amounts of methyl viologen (1 in Fig. 3), however, results in a significant decrease in luminescence intensity (line c in Fig. 2 Upper).‡ Indeed, bipyridinium dications are known to accept electrons from excited CdSe and CdSe–ZnS core–shell quantum dots, quenching their luminescence (40, 41). The corresponding Stern–Volmer plot (Fig. 2 Lower) deviates from linearity at quencher concentrations greater than ≈2.5 μM. This behavior indicates that the quenching mechanism is predominantly static below this particular concentration with a Stern–Volmer constant of ≈0.45 μM−1, whereas dynamic terms contribute significantly to quenching only at higher concentrations. These observations suggest that the dicationic quencher is presumably adsorbed on the surface of the hydrophilic quantum dots as a result of electrostatic interactions with their anionic carboxylate groups.
Fig. 2.
Influence of 1 and 3 on the emission behavior of hydrophilic CdSe–ZnS core–shell quantum dots. (Upper) Absorption spectrum (line a) of hydrophilic CdSe–ZnS core–shell quantum dots (1.5 μM, sodium phosphate buffer; pH 7.8, 20°C). Emission spectra (λEx = 350 nm) of the same solution recorded before (line b) and after the consecutive addition of 1 (4.1 μM; line c) and 3 (21 μM; line d) are shown. (Lower) Stern–Volmer plot of hydrophilic CdSe–ZnS core–shell quantum dots (1.5 μM, sodium phosphate buffer; pH 7.8, 20°C; λEx = 350 nm) upon addition of increasing amounts of 1.
Fig. 3.
The bipyridinium-based quenchers 1 and 2 and the macrocyclic receptor 3.
The change imposed on the luminescence of the hydrophilic quantum dots by the quencher 1 can be reversed with the addition of an excess of cucurbituril (3 in Fig. 3). Indeed, this particular macrocyclic receptor is known to bind bipyridinium dications with high association constants in aqueous solutions (42–44). Consistently, the emission spectrum (line d in Fig. 2 Upper) recorded in the presence of an excess of 3 relative to 1 closely resembles the one recorded before the addition of 1 (line b in Fig. 2 Upper).§ Thus, the receptor 3 binds the quencher, prevents the electron transfer process, and restores the ability of the hydrophilic quantum dots to emit light. In principle, the very same mechanism can be adapted to signal protein–ligand interactions with luminescence changes. For example, a ligand able to recognize a complementary protein can covalently be attached to the quencher (Fig. 4). In the absence of the protein, the quencher can adsorb on the surface of a hydrophilic quantum dot and suppress its ability to emit light as a result of photoinduced electron transfer. However, the supramolecular association of the ligand with the protein can remove the quencher from the nanoparticle surface and switch on its luminescence.
Fig. 4.
The supramolecular association of protein and ligand prevents the electron transfer process and activates the luminescence of the quantum dot.
To test the potential of our operating principles to signal protein–ligand interactions, we designed a compound (2 in Fig. 3) integrating a bipyridinium quencher and a biotin ligand within the same molecular skeleton and prepared this molecule in two synthetic steps (see Fig. 7, which is published as supporting information on the PNAS web site). Once again, the luminescence of the hydrophilic quantum dots decreases upon exposure to 2 (lines a and b in Fig. 5Upper).‡ As observed for 1, the corresponding Stern–Volmer plot (Fig. 5 Lower) is linear at relatively low quencher concentrations. It deviates from linearity only above ≈2.5 μM, indicating that dynamic terms contribute significantly to quenching above this quencher concentration. The Stern–Volmer constant derived from the linear region of the plot, however, is only ≈0.22 μM−1. Presumably, the biotin tail of 2 disturbs the interaction between the appended quencher and the hydrophilic quantum dots, leading to a decrease in the Stern–Volmer constant of ≈0.23 μM−1 relative to 1.
Fig. 5.
Influence of 2 and streptavidin on the emission behavior of hydrophilic CdSe–ZnS core–shell quantum dots. (Upper) Emission spectra of hydrophilic CdSe–ZnS core–shell quantum dots (1.5 μM, sodium phosphate buffer; pH 7.8, 20°C; λEx = 350 nm) recorded before (line a) and after the consecutive addition of 2 (4.1 μM; line b) and streptavidin (21 μM; line c). (Lower) Stern–Volmer plot of hydrophilic CdSe–ZnS core–shell quantum dots (1.5 μM, sodium phosphate buffer; pH 7.8, 20°C; λEx = 350 nm) upon addition of increasing amounts of 2.
The addition of increasing amounts of streptavidin to a mixture of the hydrophilic quantum dots and 2 leads to a luminescence enhancement (line c in Fig. 5 Upper).§ Instead, the addition of BSA has no influence on the emission spectrum under otherwise identical conditions (see Fig. 8, which is published as supporting information on the PNAS web site). Indeed, only streptavidin can bind the biotin ligand of 2 and, therefore, alter the quenching efficiency of the bipyridinium appendage (45, 46). Nonetheless, the original emission intensity is not fully restored even in the presence of a large excess of streptavidin. The luminescence increases only by 30% after the association of 2 and streptavidin, whereas it grows by 80% after the interaction of 1 and 3. The different behavior is presumably a result of the different binding modes of the two receptors. Although streptavidin can only bind the biotin appendage of 2, the macrocycle 3 encapsulates the bipyridinium dication of 1 in its cavity, suppressing very effectively its quenching ability. Presumably, an adjustment in the length of the aliphatic spacer connecting the quencher to the ligand in 2 can be invoked to improve the luminescence enhancement upon streptavidin binding.
In summary, we have identified a mechanism to signal receptor–substrate interactions based on photoinduced electron transfer. Our method relies on the electrostatic adsorption of cationic quenchers on the surface of anionic quantum dots. The supramolecular association of the quenchers with target receptors prevents the electron transfer process and turns on the luminescence of the inorganic nanoparticles. In fact, this protocol can be adapted to probe protein–ligand interactions with luminescent measurements. Thus, our operating principles and choice of materials can eventually lead to the development of valuable binding assays for biorelevant targets relying on the unique photophysical properties of semiconductor quantum dots.
Materials and Methods
General Procedures.
Chemicals were purchased from commercial sources with the exception of 1-methyl-4,4′-pyridylpyridinium iodide (Fig. 7), which was synthesized following a literature protocol (47). Infrared absorption spectra were recorded with a PerkinElmer Spectrum One Fourier transform spectrometer. Visible absorption spectra were recorded with a Varian Cary 100 Bio spectrometer, using quartz cells with a path length of 0.5 cm. Emission spectra were recorded with a Varian Cary Eclipse spectrometer in aerated solutions. Fast atom bombardment mass spectra (FABMS) were recorded with a VG Mass Lab Trio-2 in a 3-nitrobenzyl alcohol matrix. NMR spectra were recorded with Bruker Avance 400 and 500 spectrometers.
Hydrophobic CdSe–ZnS Core–Shell Quantum Dots.
A mixture of CdO (51 mg, 0.4 mmol), tetra-n-decylphosphonic acid (223 mg, 0.8 mmol), and tri-n-octylphosphine oxide (3.78 g, 9.8 mmol) was heated at 320°C under Ar until a clear solution was obtained. Then, the temperature was lowered to 220°C, and a solution of Se (41 mg, 0.5 mmol) in tri-n-octylphosphine (2.4 ml) was added. After the addition, the mixture was maintained at 200°C for 40 min. Then, the temperature was lowered to 120°C, and a solution of ZnEt2 (1.6 ml, 0.16 mmol) and hexamethyldisilathiane (0.30 ml, 1.4 mmol) in tri-n-octylphosphine (5 ml) was added dropwise. After the addition, the mixture was maintained at 70°C for 5 h. After cooling down to ambient temperature, MeOH (200 ml) was added, and the resulting precipitate was filtered and dissolved in CHCl3 (50 ml). This procedure was repeated three more times, and then the solvent was distilled off under reduced pressure to afford the CdSe–ZnS core–shell quantum dots (367 mg) as a reddish powder.
Hydrophilic CdSe–ZnS Core–Shell Quantum Dots.
A solution of CdSe–ZnS core–shell quantum dots (25 mg) and mercaptoacetic acid (2 ml) in CHCl3 (30 ml) was heated under reflux for 3 h. After cooling down to ambient temperature, the mixture was subjected to centrifugation. The residue was suspended in CHCl3 (15 ml) and subjected to centrifugation. This treatment was repeated four additional times. The resulting solid was suspended in MeOH (15 ml) and subjected to centrifugation. This treatment was repeated two additional times. The residue was dried under reduced pressure and suspended in H2O (5 ml). Aqueous KOH (0.1 M) was added dropwise until a clear solution was obtained. After the addition of Me2CO (10 ml), the mixture was subjected to centrifugation to afford the modified CdSe–ZnS core–shell quantum dots (20 mg) as a reddish powder.
5-(2-Oxohexahydrothieno[3,4-d]imidazol-6-yl)-Pentanoic Acid 3-Iodopropyl Ester (4).
A solution of biotin (125 mg, 0.5 mmol), 3-iodo-1-propanol (150 μl, 1.6 mmol), and TsOH (10 mg, 0.05 mmol) in PhMe (50 ml) was heated under reflux and Ar for 3 d in a Dean–Stark apparatus. After cooling down to ambient temperature, the mixture was filtered, and the solvent was distilled off under reduced pressure. The residue was dissolved in CH2Cl2 (30 ml) and washed with H2O (5 ml). The organic phase was dried (MgSO4) and concentrated under reduced pressure to yield 4 (75 mg, 36%) as a white solid. Fast atom bombardment mass spectra (FABMS): m/z = 413 [M + H]+; 1H NMR (400 MHz, CDCl3): δ = 1.40–1.45 (2H, m), 1.61–1.75 (4H, m), 2.10–2.16 (2H, m), 2.30 (2H, t, 7 Hz), 2.79 (1H, d, 13 Hz), 2.88 (1H, dd, 5 and 13 Hz), 3.15–3.18 (1H, m), 3.22 (2H, t, 7 Hz), 4.13 (2H, t, 6 Hz), 4.42–4.45 (1H, m), 4.61–4.65 (1H, m), 7.22 (1H, d, 8 Hz), 7.72 (1H, d, 8 Hz); 13C NMR (100 MHz, CDCl3): δ = 25.0, 28.4, 28.8, 32.6, 34.2, 40.5, 55.8, 61.8, 63.6, 64.4, 164.5, 173.9.
1-Methyl-1′-(3-(5-(2-Oxohexahydrothieno[3,4-d]imidazol-6-yl)-pentanoxy)-propyl)-4,4′-Bipyridinium Bisiodide (2).
A solution of 4 (37 mg, 0.09 mmol) and 1-methyl-4,4′-pyridylpyridynium iodide (9 mg, 0.03 mmol) in MeCN (15 ml) was heated under reflux and Ar for 10 d. After cooling down to ambient temperature, the solvent was distilled off, and the residue was washed with MeCN (4 ml) to yield 2 (6 mg, 28%) as an orange solid. Fast atom bombardment mass spectra (FABMS): m/z = 457 [M − 2I]+; 1H NMR (500 MHz, CD3OD): δ = 1.45–1.49 (2H, m), 1.53–1.71 (4H, m), 2.29–2.31 (2H, m), 2.49–2.51 (2H, m), 2.73 (1H, dd, 4 and 12 Hz), 2.93 (1H, dd, 5 and 12Hz), 3.20–3.23 (1H, m), 4.28–4.32 (3H, m), 4.48–4.52 (2H, m), 4.54 (3H, s), 4.77 (2H, d, 8 Hz), 8.69 (2H, d, 6 Hz), 8.73 (2H, d, 6 Hz), 9.21 (2H, d, 6 Hz), 9.34 (2H, d, 7 Hz).
Supplementary Material
Acknowledgments
This work was supported by National Science Foundation CAREER Award CHE-0237578 and the University of Miami.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
The infrared spectra recorded before (line b in Fig. 6, which is published as supporting information on the PNAS web site) and after (line c in Fig. 6) treatment of the CdSe–ZnS core–shell quantum dots with mercaptoacetic acid and potassium hydroxide show the disappearance of the [C H] stretching vibrations at 2,800–3,000 cm−1 (line a in Fig. 6) for the hydrophobic ligands and the appearance of the [C
H] stretching vibrations at 2,800–3,000 cm−1 (line a in Fig. 6) for the hydrophobic ligands and the appearance of the [C O] stretching vibrations at 1,300–1,700 cm−1 (line d in Fig. 6) for the hydrophilic ones.
O] stretching vibrations at 1,300–1,700 cm−1 (line d in Fig. 6) for the hydrophilic ones.
The addition of either 1 or 2 to a solution of the quantum dots has no influence on the visible region of the absorption spectrum.
The addition of 3, streptavidin, or BSA to a solution of the hydrophilic quantum dots has no influence on the emission spectrum in the absence of bipyridinium quenchers.
References
- 1.Bawendi M. G., Steigerwald M. L., Brus L. E. Annu. Rev. Phys. Chem. 1990;41:477–496. [Google Scholar]
- 2.Alivisatos A. P. Science. 1996;271:933–937. [Google Scholar]
- 3.Efros A. L., Rosen M. Annu. Rev. Mater. Sci. 2000;30:475–521. [Google Scholar]
- 4.Yoffe A. D. Adv. Phys. 2001;50:1–208. [Google Scholar]
- 5.Burda C., Chen X. B., Narayana R., El-Sayed M. A. Chem. Rev. 2005;105:1025–1102. doi: 10.1021/cr030063a. [DOI] [PubMed] [Google Scholar]
- 6.Niemeyer C. M. Angew. Chem. Int. Ed. 2003;42:5796–5800. doi: 10.1002/anie.200301703. [DOI] [PubMed] [Google Scholar]
- 7.Willner I., Katz E. Angew. Chem. Int. Ed. 2004;43:6042–6108. doi: 10.1002/anie.200400651. [DOI] [PubMed] [Google Scholar]
- 8.Alivisatos A. P. Nat. Biotechnol. 2004;22:47–52. doi: 10.1038/nbt927. [DOI] [PubMed] [Google Scholar]
- 9.Rosi N. L., Mirkin C. A. Chem. Rev. 2005;105:1547–1562. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- 10.Gao X., Yang L., Petros J. A., Marshall F. F., Simons J. W., Nie S. Curr. Opin. Biotechnol. 2005;16:63–72. doi: 10.1016/j.copbio.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 11.Medintz I. G., Uyeda H. T., Goldam E. R., Mattoussi H. Nat. Mater. 2005;4:435–446. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- 12.Michalet X., Pinaud F. F., Bentolila L. A., Tsay J. M., Doose S., Li J. J., Sundaresan G., Wu A. M., Gambhir S. S., Weiss S. Science. 2005;307:538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ricco A. J., Crooks R. M., editors. Acc. Chem. Res. 1998;31:199–324. [Google Scholar]
- 14.Ellis A. B., Walt D. R., editors. Chem. Rev. 2000;100:2477–2738. doi: 10.1021/cr990025k. [DOI] [PubMed] [Google Scholar]
- 15.Fabbrizzi L., editor. Coord. Chem. Rev. 2000;205:1–232. [Google Scholar]
- 16.de Silva A. P., Tecilla P., editors. J. Mater. Chem. 2005;15:2617–2976. [Google Scholar]
- 17.Clapp A. R., Medintz I. L., Mattoussi H. Chem. Phys. Chem. 2006;7:47–57. doi: 10.1002/cphc.200500217. [DOI] [PubMed] [Google Scholar]
- 18.Willard D. M., Carillo L. L., Jung J., Van Orden A. Nano. Lett. 2001;1:469–474. [Google Scholar]
- 19.Wang S., Mamedova N., Kotov N. A., Chen W., Studer J. Nano. Lett. 2002;2:817–822. [Google Scholar]
- 20.Tran P. T., Goldman E. R., Anderson G. P., Mauro J. M., Mattoussi H. Phys. Status Solidi. 2002;229:427–432. [Google Scholar]
- 21.Medintz I. L., Clapp A. R., Mattoussi H., Goldman E. R., Fisher B., Mauro J. M. Nat. Mater. 2003;2:630–638. doi: 10.1038/nmat961. [DOI] [PubMed] [Google Scholar]
- 22.Medintz I. L., Clapp A. R., Melinger J. S., Deschamps J. R., Mattoussi H. Adv. Mater. 2005;17:2450–2455. [Google Scholar]
- 23.Goldman E. R., Medintz I. L., Whitley J. L., Hayhurst A., Clapp A. R., Uyeda H. T., Deschamps J. R., Lassman M. E., Mattoussi H. J. Am. Chem. Soc. 2005;127:6744–6751. doi: 10.1021/ja043677l. [DOI] [PubMed] [Google Scholar]
- 24.Patolsky F., Gill R., Weizmann Y., Mokari T., Banin U., Willner I. J. Am. Chem. Soc. 2003;125:13918–13919. doi: 10.1021/ja035848c. [DOI] [PubMed] [Google Scholar]
- 25.Gill R., Willner I., Shweky I., Banin U. J. Phys. Chem. B. 2005;109:23715–23719. doi: 10.1021/jp054874p. [DOI] [PubMed] [Google Scholar]
- 26.Nagasaki N., Ishii T., Sunaga Y., Watanabe Y., Otsuka H., Kataoka K. Langmuir. 2004;20:6396–6400. doi: 10.1021/la036034c. [DOI] [PubMed] [Google Scholar]
- 27.Hildebrandt N., Charbonnière L. J., Beck M., Ziessel R. F., Löhmannsröben H.-G. Angew. Chem. Int. Ed. 2005;44:1–5. doi: 10.1002/anie.200501552. [DOI] [PubMed] [Google Scholar]
- 28.Geissbuehler I., Hovius R., Martinez K. L., Adrian M., Thampi K. R., Vögel H. Angew. Chem. Int. Ed. 2005;44:1388–1392. doi: 10.1002/anie.200461491. [DOI] [PubMed] [Google Scholar]
- 29.Dyadyusha L., Yin H., Jaiswal S., Brown T., Baumberg J. J., Booy F. P., Melvin T. Chem. Commun. 2005:3201–3203. doi: 10.1039/b500664c. [DOI] [PubMed] [Google Scholar]
- 30.Hohng S., Ha T. Chem. Phys. Chem. 2005;6:956–960. doi: 10.1002/cphc.200400557. [DOI] [PubMed] [Google Scholar]
- 31.Oh E., Hong M.-Y., Lee D., Nam S.-H., Yoon H. C., Kim H.-S. J. Am. Chem. Soc. 2005;127:3270–3271. doi: 10.1021/ja0433323. [DOI] [PubMed] [Google Scholar]
- 32.Bakalova R., Zhelev Z., Ohba H., Baba Y. J. Am. Chem. Soc. 2005;127:11328–11335. doi: 10.1021/ja051089h. [DOI] [PubMed] [Google Scholar]
- 33.Zhang C.-Y., Yeh H.-C., Kuroki M. T., Wang T.-H. Nat. Mater. 2005;4:826–831. doi: 10.1038/nmat1508. [DOI] [PubMed] [Google Scholar]
- 34.Chen C.-Y., Cheng C.-T., Lai C.-W., Wu P.-W., Wu K.-C., Chou P.-T., Chou Y.-H., Chiu H.-T. Chem. Commun. 2006:263–265. doi: 10.1039/b512677k. [DOI] [PubMed] [Google Scholar]
- 35.Sandros M. G., Gao D., Benson D. E. J. Am. Chem. Soc. 2005;127:12198–12199. doi: 10.1021/ja054166h. [DOI] [PubMed] [Google Scholar]
- 36.Palaniappan K., Xue C., Arumugam G., Hackney S. A., Liu J. Chem. Mater. 2006;18:1275–1280. [Google Scholar]
- 37.Dabbousi B. O., Rodriguez-Viejo J., Mikulec F. V., Heine J. R., Mattoussi H., Ober R., Jensen K. F., Bawendi M. G. J. Phys. Chem. B. 1997;101:9463–9475. [Google Scholar]
- 38.Peng Z. A., Peng X. J. Am. Chem. Soc. 2001;123:183–184. doi: 10.1021/ja003633m. [DOI] [PubMed] [Google Scholar]
- 39.Chan W. C. W., Nie S. Science. 1998;281:2016–2018. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- 40.Burda C., Green T. C., Link S., El-Sayed M. A. J. Phys. Chem. B. 1999;103:1783–1788. [Google Scholar]
- 41.Yildiz I., Raymo F. M. J. Mater. Chem. 2006;16:1118–1120. [Google Scholar]
- 42.Ong W., Gómez-Kaifer M., Kaifer A. E. Org. Lett. 2002;4:1791–1794. doi: 10.1021/ol025869w. [DOI] [PubMed] [Google Scholar]
- 43.Ong W., Kaifer A. E. J. Org. Chem. 2004;69:1383–1385. doi: 10.1021/jo035030+. [DOI] [PubMed] [Google Scholar]
- 44.Kim H.-J., Keon W. S., Ko Y. H., Kim K. Proc. Natl. Acad. Sci. USA. 2002;99:5007–5011. doi: 10.1073/pnas.062656699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hendrickson W. A., Pahler A., Smith J. L., Satow Y., Merritt E. A., Phizackerley R. P. Proc. Natl. Acad. Sci. USA. 1989;86:2190–2194. doi: 10.1073/pnas.86.7.2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weber P. C., Ohlendorf D. H., Wendoloski J. J., Salemme F. R. Science. 1989;243:85–88. doi: 10.1126/science.2911722. [DOI] [PubMed] [Google Scholar]
- 47.Feng D.-J., Li X.-Q., Wang X.-Z., Jiang X.-K., Li Z.-T. Tetrahedron. 2004;60:6137–6144. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.