Abstract
3-Hydroxy-3-methylglutaryl CoA synthase (HMGS) catalyzes the first committed step in the mevalonate metabolic pathway for isoprenoid biosynthesis and serves as an alternative target for cholesterol-lowering and antibiotic drugs. We have determined a previously undescribed crystal structure of a eukaryotic HMGS bound covalently to a potent and specific inhibitor F-244 [(E,E)-11-[3-(hydroxymethyl)-4-oxo-2-oxytanyl]-3,5,7-trimethyl-2,4-undecadienenoic acid]. Given the accessibility of synthetic analogs of the F-244 natural product, this inhibited eukaryotic HMGS structure serves as a necessary starting point for structure-based methods that may improve the potency and species-specific selectivity of the next generation of F-244 analogs designed to target particular eukaryotic and prokaryotic HMGS.
Keywords: cholesterol-lowering, F-244, inhibition, x-ray structure, mevalonate
The mevalonic acid metabolic pathway found in all eukaryotic organisms, some Gram-positive bacteria, and the cytosol of plants provides isoprenoid precursors used for the synthesis of essential natural compounds including sterols such as cholesterol, heme, ubiquinones, dolichols, farnesylated and geranylgeranylated proteins, isoprenoid-derived hormones, and vitamin D (1, 2). The most studied enzyme in the mevalonic acid pathway is HMG–CoA reductase (HMGR), which is the target of clinically important cholesterol-lowering drugs known as statins (3, 4). Less studied but equally important for isoprenoid biosynthetic pathways, 3-hydroxy-3-methylglutaryl–CoA synthase (HMGS) catalyzes the condensation of acetoacetyl–CoA (AcAc-CoA) with acetyl–CoA (Ac-CoA) to form 3-hydroxy-3-methylglutaryl–CoA (HMG-CoA) plus one free CoA. Recent crystal structures of bacterial HMGS highlight the conserved fold of this family of biosynthetic enzymes that includes α-keto-thiolases, fatty acid synthases, and polyketide synthases (5–7). All members of this extended enzyme family use a catalytically essential cysteine-derived thiolate nucleophile.
HMGS from the higher plant Brassica juncea exhibits only 29% amino acid sequence identity with the published bacterial HMGS structures (5, 7) and is more closely related to human HMGS (49% amino acid sequence identity to the human cytosolic HMGS). Recently, detailed kinetic studies of wild-type and mutant forms of one of the four B. juncea HMGS (BjHMGS), BjHMGS1, demonstrated that (E,E)-11-[3-(hydroxymethyl)-4-oxo-2-oxytanyl]-3,5,7-trimethyl-2,4-undecadienenoic acid (F-244), alternatively known as 1233A, L-659,699, or hymeglusin, acts as a potent inhibitor with an IC50 of 35 nM (8–12). F-244 is a polyketide from Fusarium sp. and Scopulariopsis sp. and contains a β-lactone ring likely mediating HMGS inhibition (Fig. 1A). Moreover, F-244 was previously shown to inhibit HMGS from rat liver (IC50 of 0.12 μM), as well as from human (IC50 of 54 nM) (12, 13). Rat and human HMGS share 94% sequence identity and show strict conservation of all of the residues in the active site and located at the active site entrance (data not shown).
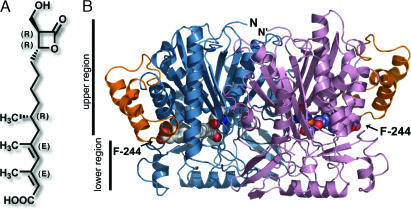
Fig. 1.
F-244 structure and overall architecture of HMGS. (A) Structure and stereochemistry of F-244. (B) The HMGS homodimeric architecture (blue and rose) and the extra 30-residue insertion forming a helical subdomain (gold) are shown. F-244 is depicted as van der Waals spheres with oxygen in red, carbon in gray in the left monomer, and carbon in blue in the right monomer.
Here, we present a previously undescribed structure of a eukaryotic HMGS and show, by using protein x-ray crystallography, that BjHMGS1 through its catalytic Cys-117 covalently attaches to a ring-opened form of F-244 resulting in a well ordered and stereochemically defined conformation of the highly functionalized natural product in the HMGS active site. This thioester adduct with Cys-117 leads to a “dead-end” complex incapable of initiating and completing the HMG-CoA forming reaction. The structures of the apoform of BjHMGS1, BjHMGS1 covalently linked to an acetyl group forming an acetylated Cys-117 and acetyl-CoA, and BjHMGS1 bound covalently and noncovalently to its product HMG-CoA are also presented. In total, these structures provide a starting point to rationally engineer F-244 analogs to improve the efficacy of new cholesterol-lowering agents or antibiotics, and they also clarify the final thioesterase-mediated enzymatic reaction leading to HMG-CoA release.
Results and Discussion
Overall Structure of a Eukaryotic HMGS.
Crystallization of BjHMGS1 required either substrate, Ac-CoA or AcAc-CoA, or product, HMG-CoA, to be present during screens (Table 1). In the apo-HMGS structure, whereas HMG-CoA was present during crystallization trials, the resultant electron density showed no HMG-CoA or cleaved products (reverse reaction) bound in the HMGS active site. The overall structure revealed a homodimeric quaternary structure (Fig. 1B). This dimeric arrangement and the fold of the monomer units are similar to the recently published bacterial HMGS crystal structures [rmsd = 1.6 Å compared with Protein Data Bank (PDB) ID code 1XPK using all Cα]. The αβαβα core is conserved among all condensing enzymes and thiolases, reflecting their common evolutionary lineage. As described for the reported bacterial HMGS structures, the architecture of the thiolase fold enzymes can be divided into two regions (upper and lower; Fig. 1B) with the catalytic machinery located at the interface of these two subdomains (5, 7). Moreover, the dimer interface between monomers encompasses a relatively large flat surface burying ≈19% of the available surface area of each monomer. This oligomeric interface is composed of both polar and nonpolar interactions.
Table 1.
Data collection and refinement statistics
| Statistic | HMGS·apo | HMGS·F-244 | HMGS·Ac-CoA | HMGS·HMG-CoA* |
|---|---|---|---|---|
| Data collection | ||||
| Space group | P6(1)22 | P6(1)22 | P6(1)22 | P6(1)22 |
| Cell dimensions | ||||
| a, b, c, Å | 61.27, 61.27, 435.70 | 61.04, 61.04, 411.77 | 61.34, 61.34, 409.87 | 61.19, 61.19, 410.93 |
| α, β, γ, ° | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 |
| Resolution, Å | 50–2.1 (2.22–2.1) | 50–2.5 (2.65–2.50) | 50–2.5 (2.66–2.5) | 50–2.49 (2.64–2.49) |
| Rsym or Rmerge | 6.7 (28.4) | 10.0 (40.0) | 11.3 (48.9) | 13.0 (62.1) |
| I/σI | 20.16 (4.82) | 19.93 (4.90) | 16.26 (3.10) | 17.46 (3.04) |
| Completeness, % | 98.0 (89.5) | 96.8 (83.4) | 97.4 (85.6) | 96.7 (86.4) |
| Redundancy | 8.05 | 11.6 | 7.9 | 13.4 |
| Refinement | ||||
| Resolution, Å | 2.1 | 2.5 | 2.5 | 2.5 |
| No. reflections | 27,963 | 15,746 | 15,876 | 16,104 |
| Rwork/Rfree | 0.19/0.24 | 0.17/0.24 | 0.19/0.28 | 0.19/0.24 |
| No. atoms | ||||
| Protein | 3,510 | 3,510 | 3,510 | 3,510 |
| Ligand/ion | 0 | 22 | 53 | 57, 57* |
| Water | 154 | 272 | 133 | 99 |
| B factors | ||||
| Protein | 34.15 | 23.64 | 30.97 | 29.36 |
| Ligand/ion | 30.32 | 48.23 | 31.84 | |
| Water | 37.88 | 28.01 | 33.00 | 26.13 |
| rmsd | ||||
| Bond lengths, Å | 0.015 | 0.011 | 0.014 | 0.021 |
| Bond angles, ° | 1.55 | 1.35 | 1.55 | 2.29 |
One crystal was used for each structure summarized above. Numbers in parentheses represent the highest-resolution shell.
*Two ligands of the same size refined with the occupancy of each set to 50%.
Eukaryotic HMGS differ from bacterial HMGS by an ≈30-aa insertion between helices α8 and α9 of Staphylococcus aureus HMGS (5). This insert forms a folded subdomain composed of α-helices (Fig. 1B). It is notable that sequence alignments and 3D modeling support the conservation of this folded module in other eukaryotic HMGS including the human cytosolic (responsible for five carbon isoprenoid precursor biosynthesis; GenBank accession no. Q01581) and human mitochondrial (involved in ketogenesis; GenBank accession no. P54868) HMGS. This extra eukaryote-specific domain sits at the entrance of the active site tunnel, further stabilizing CoA binding through electrostatic interaction of Arg-296 (corresponding to Arg-313 in human cytosolic HMGS) with the ribose 3′-phosphate of CoA (Fig. 2). Given the presence of this domain in eukaryotic HMGS and its absence from prokaryotic HMGS, it may play a eukaryotic-specific role in HMGS, possibly including modulation of access to the active site and/or participation in formation of protein–protein interaction surfaces mediating flux through the essential mevalonic acid metabolic pathway.
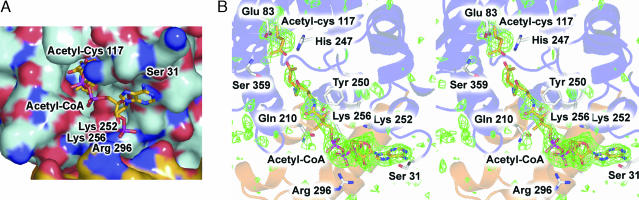
Fig. 2.
Overview of HMGS in complex with acetyl–Cys-117 and Ac-CoA. (A) Electrostatic surface of HMGS in complex with acetyl–Cys-117 and Ac-CoA. Red and blue indicate negatively and positively charged surface features, respectively. Acetyl–Cys-117 and Ac-CoA are shown as color-coded bonds with red for oxygen, blue for nitrogen, and gold for carbon. (B) Stereoview of the HMGS active site for the acetyl–Cys-117 and Ac-CoA complexes. Secondary structure is shown as ribbons with the conserved thiolase fold colored light blue and the eukaryotic-specific helical subdomain in gold. Color-coding of acetyl–Cys-117 and Ac-CoA is identical to that in A. The SIGMAA-weighted Fo − Fc electron density map is shown in green, contoured at 2.7σ, and calculated by using model phases with acetyl–Cys-117 and Ac-CoA omitted from refinement and phase calculations.
Ring-Opened Form of F-244 Binds to the Catalytic Cys.
The well defined electron density exhibited by the ring-opened form of F-244 shows that the carbonyl carbon of the β-lactone ring of F-244 and Cys-117 form a covalent thioester bond (Fig. 3). This thioester-mediated adduct resembles the covalent adduct accompanying acetylation of Cys 117 and elongation to form the HMG-CoA moiety. Furthermore, unlike what was observed for the HMG-CoA bound form of BjHMGS1 where the electron density suggests that a mixture of covalently bound and free HMG-CoA results, the electron density associated with the F-244 inhibited form of BjHMGS1 does not reflect a reversible adduct (12). In vitro studies with purified rat liver cytosolic HMGS (8, 14) and with recombinant hamster HMGS (8) demonstrate the covalent and irreversible acylation of the active-site Cys residue. The ring-opened form of F-244 presents an ideal set of hydrogen-bonding (H-bonding) groups to the catalytic machinery at the BjHMGS1 active site, including H-bonds of the oxygen atoms of F-244 to Glu-83, His-247, and Asn-326. Moreover, the stereochemical complexity of ring-opened F-244 exploits additional backbone contacts to Ser-359 and Cys-117, the latter of which form an oxyanion hole necessary for catalysis (Fig. 4).
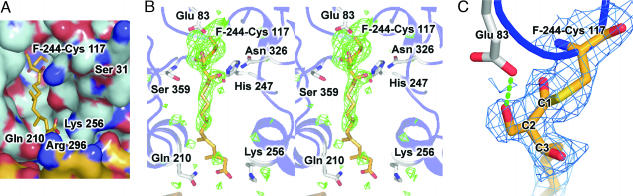
Fig. 3.
Overview of HMGS in complex with F-244. (A) Electrostatic surface of HMGS covalently bound to ring-opened F-244. Color-coding is identical to Fig. 2A. The inhibitor F-244 does not occupy the same part of the pantothenate-binding tunnel as the CoA tail shown in Fig. 2A. (B) Stereoview of the HMGS active site for the ring-opened F-244 covalent complex. Color-coding and map calculations are identical to Fig. 2B. The first eight carbons of the acyl tail of F-244 are well ordered; however, the position of the remaining six carbons display much weaker electron density as the tail protrudes out of the active site entrance. (C) Close-up view of the HMGS active site for the ring-opened F-244 covalent complex. The SIGMAA-weighted 2Fo − Fc electron density map is shown in blue, contoured at 1σ. A H-bond between Glu-83 and the 2-hydroxymethyl moiety of ring-opened F-244 is shown as rendered green cylinders.
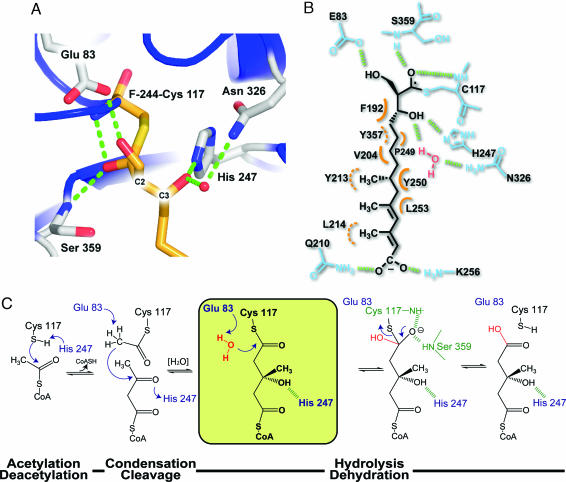
Fig. 4.
Close-up view of F-244 covalently modifying the BjHMGS1 active site and a posited reaction mechanism based on the F-244 complex. (A) Secondary structure is shown as ribbons colored as in Fig. 1. Side chains and F-244 are depicted as half-colored bonds with red for oxygen, blue for nitrogen, and gold and gray for carbons on F-244 and HMGS side chains, respectively. The oxygen atoms of the ring-opened form of F-244 form H-bonds (green dashes) with Glu-83, His-247, and Asn-326 through a water molecule (red sphere) and backbone amides of Ser-359 and Cys-117. (B) Schematic representation of F-244 tethered to Cys-117 highlighting all intermolecular interactions. H-bonds are depicted as green dashes. Orange half circles depict van der Waals contacts with dashed curves specifying residues sitting behind the plane formed by F-244. (C) Putative reaction mechanism of HMGS. The proposed role of Glu-83 in the last hydrolytic step necessary for release of HMG-CoA is shown in a yellow box. The residues implicated in formation of the oxyanion hole are depicted in green, and H-bonds are shown as dashed bonds.
The stereochemistry (2R, 3R) of the β-lactone ring (Fig. 1A) is optimal for all active-site interactions upon lactone ring opening (Fig. 4A). Indeed, the structure confirms previous observations that other synthetically derived enantiomers and diasteromers of F-244 homologs are considerably less potent (8). Inhibition assays carried out using four chiral F-244 homologs (DU-6622) showed that alteration of the stereochemical arrangement of carbon-3 substituents of the β-lactone ring compared with carbon-2 lead to more drastic losses of overall inhibitory potency. This observation highlights the therapeutic need for coupling chirally defined inhibitors with high-resolution structural studies to guide the design and synthesis of improved pharmacophores. Moreover, the loss of potency accompanying a stereochemical change at carbon-3 as opposed to carbon-2 can be explained structurally. A stereochemical inversion at carbon-2 would still allow for H-bonding between the 2-hydroxylmethyl moiety and the Glu-83 side chain albeit with less-than-optimal geometry. In contrast, the stereochemical inversion at carbon-3 would dramatically alter the orientation of the inhibitor tail, leading to severe steric clashes between the F-244 acyl tail and the backbone atoms of HMGS.
Reaction Mechanism of HMGS.
The multistep reaction catalyzed by HMGS consists of three catalytic steps using a ping-pong kinetic mechanism (15, 16) (Fig. 4C). (i) CoA carries a reactive acetyl group into the catalytic tunnel and juxtaposes the acetyl moiety for efficient trans-acetylation of Cys-117 [Cys-129 in hamster (8) and human cytosolic HMGS (13)]. (ii) After release of CoA, the next substrate, AcAc-CoA, again localized by its CoA tether, positions itself for the Claisen condensation reaction leading to a covalently bound HMG-CoA di-thioester intermediate. (iii) A putative water-mediated cleavage of the Cys-117–HMG-CoA thioester bond releases HMG-CoA in a manner reminiscent of thioesterases that employ a transient hydroxide anion nucleophile (Fig. 4C).
By cocrystallization of BjHMGS1 with Ac-CoA, we trapped an acetylated Cys-117 adduct, although the putative thioesterase cleavage machinery likely reduced the occupancy of this covalent enzyme intermediate significantly (Fig. 2). The CoA part of Ac-CoA exhibits near stoichiometric occupancy of the CoA-binding tunnel. In BjHMGS1, Cys-117, His-247, and Glu-83 form a catalytic triad common in HMGS that is positioned to carry out the proposed three-step reaction to form HMG-CoA (Fig. 4C) (5, 7). In step one, His-247 serves as a base to activate Cys-117, or by analogy to type III polyketide synthases such as chalcone synthase, serves as a H-bond donor and counterion to a preformed thiolate anion (17). The resultant thiolate anion initiates acetyl group transfer through nucleophilic attack on the carbonyl carbon of Ac-CoA (Fig. 4C). CoA resides next to the catalytic machinery by virtue of van der Waals contacts of the pantothenate unit of CoA with the HMGS CoA-binding tunnel. Furthermore, CoA is anchored by conserved electrostatic interactions between the ribose phosphates of CoA and Lys-252, Lys-256, and Arg-296 (situated on the eukaryotic specific helical subdomain), as well as a H-bond between the adenine moiety of CoA to the Ser-31 hydroxyl side chain (Fig. 2B). In step two, the acetyl-Cys adduct through Glu-83 mediated activation of the methyl moiety of the acetyl group serves as a carbanion nucleophile to attack the thioester activated carbonyl carbon of AcAc-CoA releasing CoA and forming a HMG-CoA adduct. In the presence of excess HMG-CoA, an equilibrium mixture of covalently bound and free HMG-CoA forms in the active site of crystalline HMGS. As observed in the acetyl-CoA cocrystallizations, CoA is positioned and anchored by the same conserved electrostatic interactions as the CoA derived from acetyl-CoA.
A remaining question surrounding the thioesterase machinery necessary for activating the nucleophilic hydroxide anion to initiate release of HMG-CoA remains. The F-244–inhibited structure of BjHMGS1 offers some possible clues as to the nature of this thioesterase activity. Interestingly, this inhibited structure of BjHMGS1 was obtained after soaking of F-244 into previously cocrystallized HMG-CoA·HMGS. In all of the crystals analyzed, only F-244 was visible in the active site, indicating a nearly stoichiometric replacement of excess F-244 for HMG-CoA or cleavage products derived from it, including CoA. This result is not unexpected given the specificity and potency of F-244–mediated inhibition of HMGS as well as the excess of F-244 present during stabilization and soaking of preformed crystals.
Clarification of Thioesterase Activity of HMGS.
It is important to address why the normal thioesterase machinery inherent in HMGS would not result in cleavage of the F-244 thioester bond. Part of the explanation may reside in the hydroxyl-bearing side chain at carbon-2 (2R) of F-244. This substituent forms a well ordered H-bond with the side chain of Glu-83 (Figs. 3C and 4A). If Glu-83 serves as a general base for the catalytic generation of a hydroxide nucleophile from water, which would be necessary for thioesterase-mediated cleavage of the HMG-CoA·HMGS covalent adduct, then the 2R-hydroxyl moiety on F-244 would effectively displace such a water molecule at this spatial location. Notably, a water molecule at this site would be ideally positioned for attack on the carbonyl carbon of the Cys-117–tethered HMG-CoA adduct (Fig. 4C). Indeed, this observation would then posit that Glu-83 not only serves as a general base to activate the methyl group of the initial acetyl group attached to Cys-117, but that Glu-83 also would serve as the final general base to trigger the thioesterase machinery of HMGS. This latter activity would first require loss of a proton from the free acid form of Glu-83 that forms immediately after activation of the acetyl moiety. Although no ordered water molecule H-bonded to Glu-83 is observed in the apo-, substrate-bound, or product-bound structures, the active site appears quite accessible to bulk solvent, suggesting that water can easily access the active site during the multistep reactions leading to HMG-CoA.
Curiously, the stereochemical arrangement of Glu-83 relative to the F-244 ring-opened adduct bound to BjHMGS mimics, in part, the proposed thioesterase machinery found in the plant polyketide synthase, stilbene synthase (18). In fact, position 83 of BjHMGS1 spatially overlaps with Thr-132 found in Scots Pine stilbene synthase (STS), the latter of which tethers a putative nucleophilic water molecule in STS that ultimately severs a thioester-linked polyketide product in aldol forming type III polyketide synthases (18). The role of Glu-83 in the thioesterase machinery is supported by the recently published structure of a prokaryotic HMGS substrate/inhibitor complex. In the absence of an initial source of Ac-CoA, AcAc-CoA can serve as an effective HMGS inhibitor. However, unlike F-244, the inhibitory effect of AcAc-CoA is reversible (7). In the AcAc–enzyme structure in which AcAc is covalently bound to the catalytic Cys, Glu-79 (corresponding to Glu-83 in BjHMGS1) does not participate in any additional H-bonding interactions with the active site or with the AcAc moiety unlike observed here for the F-244 inhibited structure. This arrangement for the reversible AcAc-CoA inhibitor could therefore allow for Glu-79–mediated cleavage of the thioester bound of the AcAc-Cys complex leading to the reversible inhibitory effect. This hypothesis is also supported by the observation that the F-244 homologs, Ebelactone A and B, both in which the hydroxyl group projecting from carbon-2 of the β-lactone ring is replaced by an aliphatic chain (methyl and ethyl groups, respectively), do not exhibit any HMGS-inhibitory activity (9).
Foundation for the Design of Potent and Species-Specific HMGS Inhibitors.
Although the first eight carbons of the acyl tail of F-244 are well ordered and reside within the largely hydrophobic pantothenate-binding tunnel, the position of the remaining six carbons of the inhibitor tail display much weaker electron density (Fig. 3B). There is a paucity of interactions along this portion of F-244 with the more hydrophilic exit of the CoA-binding tunnel. Finally, the terminal carboxylate moiety of F-244 may interact with a H-bonding/basic patch (Gln-210, Lys-256, and Arg-296) at the tunnel exit (Fig. 4 A and B). Significantly, it was shown that F-244 derivatives with free carboxyl groups show more potent inhibitory activity against HMGS than the corresponding methyl esters (19).
Furthermore, the inhibitor tail does not occupy the same part of the pantothenate-binding tunnel leading to the active site of HMGS as the CoA cofactor (Figs. 2 and 3). Also, the CoA-bound structures (Ac-CoA or HMG-CoA) exhibit a narrower conformation of the active-site tunnel entrance to either the apoform or the F-244–bound form of BjHMGS. Specifically, a consistent displacement (2.8 Å) of the HMGS backbone (Pro-202 to Tyr-213) upon binding CoA leads to creation of new intramolecular H-bonding networks among the side chains of Gln-210, Tyr-250, and Lys-256 and between Tyr-151 and -328, both of which reside near the catalytic residues, including Cys-117 and His-247.
As described above, it is now clear why the specific 2R,3R stereochemistry of F-244 presents an ideal set of intermolecular H-bonds to the HMGS catalytic machinery upon lactone ring opening (Fig. 4A and B). Therefore, modification to this reactive core of F-244 will compromise inhibitory potency. On the contrary, the lack of observable electron density for the last six carbons of the F-244 hydrocarbon tail, indicative of a paucity of F-244–HMGS interactions, suggests that rationally designed synthetic analogs of F-244 could exploit the architectural features of HMGS in the wide active-site exit for improved potency in a general sense and for species-specific inhibition that exploits intermolecular features not generally shared among HMGS. In these latter cases, one can now employ rational structure-based design during the elaboration of synthetic analogs of F-244 by referring to published prokaryotic HMGS structures (5, 7) and to the structures of the eukaryotic HMGS reported here. This rational approach could lead to the creation of new classes of antimicrobial, herbicidal, or cholesterol-lowering compounds.
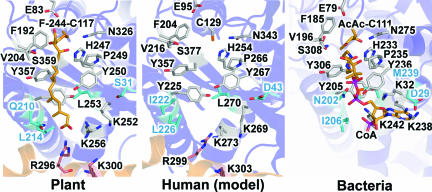
Specifically, comparison of the HMGS active sites among plant (B. juncea; this work), cytosolic human (model based on the highly conserved BjHMGS1 sequence), and bacterial (Enterococcus faecalis) forms of HMGS show strict conservation of the residues abutting the catalytic Cys (Cys-117 in B. juncea, Cys-129 in human, and Cys-111 in E. faecalis). In contrast, the residues clustered around the entrance of the active-site tunnel are more variable. For example, Gln-210 in BjHMGS1 is replaced by Ile-222 in human cytosolic HMGS and by Asn-202 in E. faecalis HMGS. In addition, Leu-253 and -270 in BjHMGS1 and human cytosolic HMGS, respectively, are replaced by Met-239 in E. faecalis HMGS (Fig. 5).
Fig. 5.
Comparison of F-244 covalently modifying the BjHMGS1 active site to human and bacterial HMGS. Active sites of HMGS from B. juncea (Left), from modeled human cytosolic (Center; five carbon isoprenoid precursors; model based on GenBank accession no. Q01581), and from Enterococcus faecalis (Right; PDB ID code 1YSL). The extra-helical domain, found in the eukaryotic HMGS, but not in prokaryotic HMGS, sequence is colored gold with Arg-296 (Arg-299 in human cytosolic HMGS) involved in H-bonding with the 3′-phosphate of CoA shown as sticks along with other residues lining the pantothenate-binding tunnel. The active-site residues, which differ among the various species, are depicted as blue bonds with blue labels.
The most prominent architectural feature distinguishing prokaryotic and eukaryotic HMGS that also resides near the F-244 hydrocarbon tail bound to HMGS is the eukaryotic-specific helical domain located at the periphery of the active-site entrance (Fig. 1B). Eukaryotic-specific inhibitors with greater potency could exploit interactions with this extra domain through extension and conformational restriction of the carboxylate-bearing tail of F-244. An anionic moiety at this location may favorably interact with the basic path encompassing Arg-296 (Arg-299 in human) and Lys-300 (Lys-303 in human) found in the eukaryotic HMGS. On the other hand, prokaryotic-specific F-244 analogs would likely exploit differences discussed above that line the CoA-binding pocket nearer to the catalytic machinery. Generally, more potent analogs lacking species specificity could exploit the greater volume available to the F-244 tail as it nears the exit of the active-site tunnel.
Although F-244 is a polyketide-derived natural product with multiple stereocenters (9), synthetic analogs are readily accessible (12, 19–21). Whereas a great deal of synthetic attention has been paid to the stereochemically rich and reactive lactone core of F-244, the hydrocarbon tail has not been exploited to develop structure–activity relationships for the evolution of more potent HMGS inhibitors. Determination of the high-resolution x-ray crystal structure of F-244–inhibited HMGS is a starting point for rational design of the next generation of synthetic F-244–like compounds. The utility of this rational, structurally guided approach extends to multiple organisms. In short, specifically designed rational analogs could ultimately serve as novel antibiotics targeting HMGS found in pathogenic bacteria such as Streptococcus pneumonia and S. aureus, as novel cholesterol-lowering drugs in humans, and as fungicides and plant growth regulators for agriculture (see Fig. 6, which is published as supporting information on the PNAS web site).
Materials and Methods
Protein Expression and Purification.
BjHMGS1 (GenBank accession no. AAF69804) was cloned, expressed, and purified as described (11). The N-terminal hexahistidine tag was cleaved by thrombin digestion during overnight dialysis against 100 mM NaCl/25 mM Hepes (pH 7.5) at 4°C. The cleaved product was further purified by gel-filtration chromatography on a Superdex-S200 FPLC column (Amersham Pharmacia Biosciences) equilibrated in 100 mM NaCl/25 mM Hepes, pH 7.5/2 mM DTT. Peak fractions containing homodimeric BjHMGS1 were collected, concentrated to 15 mg/ml, and stored at −80°C.
Crystallization of BjHMGS1.
Crystals were grown by vapor diffusion at 4°C in 2-μl hanging drops, consisting of a 1-μl reservoir [17% (wt/vol) PEG 20,000/0.25 M trimethylamine N-oxide/100 mM Pipes (pH 6.5)/2 mM DTT] and 1 μl of protein solution (BjHMGS1 preincubated with 10 mM Ac-CoA, AcAc-CoA, or HMG-CoA). The BjHMGS1 complex with F-244 was obtained by soaking BjHMGS1 crystals (cocrystallized with HMG-CoA) in 2-μl drops containing 10 mM F-244. The apoform was obtained by cocrystallization with HMG-CoA; however, no density attributable to HMG-CoA was visible in the active site. Before freezing in liquid nitrogen, crystals were soaked for 5 min in a cryoprotectant identical to the crystallization buffer except for the substitution of 21% (wt/vol) PEG 20,000 and the addition of 20% (wt/vol) 2-methyl-2,4-pentanediol.
Structure Determination.
Diffraction data were collected on the FIP-BM30A, ID29, and ID14 beamlines at the European Synchrotron Radiation Facility (Grenoble, France). Data were indexed, integrated, and scaled with XDS (22). BjHMGS1 crystallized in the P6 (1)22 space group with average unit cell dimensions of a = b = 60 Å; c varies from 409.87 to 435.70 Å, with one molecule in the asymmetric unit (Table 1). Data collections, because of the long c axis, required the use of a focused beam and 0.2° to 0.5° oscillations. Phase determination was carried out by molecular replacement using MOLREP (23), part of the CCP4 suite (24). We used as a search model the recently published structure of Staphyloccocus aureus HMGS (PDB ID code 1TXT) (5). After refinement, the structure of the apoform of BjHMGS1 was used as the starting model for solving additional BjHMGS1–small-molecule complexes. The initial molecular-replacement models were manually adjusted in O (25) and refined first with CNS (26) then in final rounds with REFMAC5 (27) (Table 1). The refined structures were evaluated with PROCHECK (28). The HMGS complex bound to ring-opened F-244 had 90.0%, 9.2%, 0.2%, and 0.5% of residues in the most favored, allowed, generously allowed, and disallowed regions of the Ramachandran plot, respectively (HMGS-apo, 90.8%, 8.7%, 0.2%, and 0.2%, respectively; HMGS–HMG-CoA: 87.2%, 11.8%, 1%, and 0%, respectively; HMGS–Ac-CoA, 87.6%, 11.4%, 0.5%, and 0.5%, respectively). The two residues consistently found in the disallowed region but boarding the generously allowed region of the Ramachandran plot, Ala-116 and Tyr-328, reside in well defined electron density and were previously observed in the same conformations in the bacterial structures (5, 7). The final structure coordinates and structure factors were deposited in the PDB [PDB ID codes 2F82, 2FA3, 2FA0, and 2F9A for the apo, acetyl-Cys-117·Ac-CoA, HMG-CoA, and F-244 complexes, respectively]. Structure figures were prepared with PyMol (www.pymol.org).
Supplementary Material
Acknowledgments
We thank Dr. Sheo B. Singh (Merck Research Laboratories) for kindly providing F-244. This work was supported by National Institutes of Health Grant AI51438 (to J.P.N.) and by a Deutsche Forschungsgemeinschaft postdoctoral fellowship (to F.P.). J.P.N. is a Howard Hughes Medical Institute Investigator.
Abbreviations
- Ac-CoA
acetyl–CoA
- AcAc-CoA
acetoacetyl–CoA
- F-244
(E,E)-11-[3-(hydroxymethyl)-4-oxo-2-oxytanyl]-3,5,7-trimethyl-2,4-undecadienenoic acid
- HMG
3-hydroxy-3-methylglutaryl
- HMGS
HMG–CoA synthase(s)
- HMGR
HMG–CoA reductase
- BjHMGS
B. juncea HMGS
- PDB
Protein Data Bank
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The atomic coordinates and structure factors were deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 2F82 (apocomplex), 2FA3 (acetyl-Cys-117·Ac-CoA complex), 2FA0 (HMG-CoA complex), and 2F9A (F-244 complex)].
References
- 1.Edwards P. A., Ericsson J. Annu. Rev. Biochem. 1999;68:157–185. doi: 10.1146/annurev.biochem.68.1.157. [DOI] [PubMed] [Google Scholar]
- 2.Holstein S. A., Hohl R. J. Lipids. 2004;39:293–309. doi: 10.1007/s11745-004-1233-3. [DOI] [PubMed] [Google Scholar]
- 3.Vaughan C. J., Gotto A. M., Jr. Circulation. 2004;110:886–892. doi: 10.1161/01.CIR.0000139312.10076.BA. [DOI] [PubMed] [Google Scholar]
- 4.Istvan E. Atheroscler. Suppl. 2003;4:3–8. doi: 10.1016/s1567-5688(03)00003-5. [DOI] [PubMed] [Google Scholar]
- 5.Campobasso N., Patel M., Wilding I. E., Kallender H., Rosenberg M., Gwynn M. N. J. Biol. Chem. 2004;279:44883–44888. doi: 10.1074/jbc.M407882200. [DOI] [PubMed] [Google Scholar]
- 6.Theisen M. J., Misra I., Saadat D., Campobasso N., Miziorko H. M., Harrison D. H. Proc. Natl. Acad. Sci. USA. 2004;101:16442–16447. doi: 10.1073/pnas.0405809101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steussy C. N., Vartia A. A., Burgner J. W., 2nd, Sutherlin A., Rodwell V. W., Stauffacher C. V. Biochemistry. 2005;44:14256–14267. doi: 10.1021/bi051487x. [DOI] [PubMed] [Google Scholar]
- 8.Tomoda H., Ohbayashi N., Morikawa Y., Kumagai H., Omura S. Biochim. Biophys. Acta. 2004;1636:22–28. doi: 10.1016/j.bbalip.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 9.Tomoda H., Kumagai H., Tanaka H., Omura S. Biochim. Biophys. Acta. 1987;922:351–356. [PubMed] [Google Scholar]
- 10.Kumagai H., Tomoda H., Omura S. J. Antibiot. 1992;45:563–567. doi: 10.7164/antibiotics.45.563. [DOI] [PubMed] [Google Scholar]
- 11.Nagegowda D. A., Bach T. J., Chye M. L. Biochem. J. 2004;383:517–527. doi: 10.1042/BJ20040721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenspan M. D., Yudkovitz J. B., Lo C. Y., Chen J. S., Alberts A. W., Hunt V. M., Chang M. N., Yang S. S., Thompson K. L., Chiang Y. C., et al. Proc. Natl. Acad. Sci. USA. 1987;84:7488–7492. doi: 10.1073/pnas.84.21.7488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rokosz L. L., Boulton D. A., Butkiewicz E. A., Sanyal G., Cueto M. A., Lachance P. A., Hermes J. D. Arch. Biochem. Biophys. 1994;312:1–13. doi: 10.1006/abbi.1994.1273. [DOI] [PubMed] [Google Scholar]
- 14.Greenspan M. D., Bull H. G., Yudkovitz J. B., Hanf D. P., Alberts A. W. Biochem. J. 1993;289:889–895. doi: 10.1042/bj2890889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miziorko H. M., Clinkenbeard K. D., Reed W. D., Lane M. D. J. Biol. Chem. 1975;250:5768–5773. [PubMed] [Google Scholar]
- 16.Miziorko H. M., Lane M. D. J. Biol. Chem. 1977;252:1414–1420. [PubMed] [Google Scholar]
- 17.Jez J. M., Noel J. P. J. Biol. Chem. 2000;275:39640–39646. doi: 10.1074/jbc.M008569200. [DOI] [PubMed] [Google Scholar]
- 18.Austin M. B., Bowman M. E., Ferrer J. L., Schröder J., Noel J. P. Chem. Biol. 2004;11:1179–1194. doi: 10.1016/j.chembiol.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 19.Sunazuka T., Tsuzuki K., Kumagai H., Tomoda H., Tanaka H., Nagashima H., Hashizume H., Omura S. J. Antibiot. 1992;45:1139–1147. doi: 10.7164/antibiotics.45.1139. [DOI] [PubMed] [Google Scholar]
- 20.Tomoda H., Kumagai H., Ogawa Y., Sunazuka T., Hashizume H., Nagashima H., Omura S. J. Org. Chem. 1997;62:2161–2165. doi: 10.1021/jo9621433. [DOI] [PubMed] [Google Scholar]
- 21.Hashizume H., Ito H., Morikawa T., Kanaya N., Nagashima H., Usui H., Tomoda H., Sunazuka T., Kumagai H., Omura S. Chem. Pharm. Bull. (Tokyo) 1994;42:2097–2107. doi: 10.1248/cpb.42.2097. [DOI] [PubMed] [Google Scholar]
- 22.Kabsch W. XDS in International Tables for Crystallography. Dordrecht, The Netherlands: Kluwer Academic; 2001. [Google Scholar]
- 23.Vagin A. A., Isupov M. N. Acta Crystallogr. D. 2001;57:1451–1456. doi: 10.1107/s0907444901012409. [DOI] [PubMed] [Google Scholar]
- 24.CCP4 Acta Crystallogr. D. 1999;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 25.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 26.Brunger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., et al. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 27.Vagin A. A., Steiner R. A., Lebedev A. A., Potterton L., McNicholas S., Long F., Murshudov G. N. Acta Crystallogr. D. 2004;60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 28.Laskowski R. A. Methods Biochem. Anal. 2003;44:273–303. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.