Abstract
Bacterial chemoreceptors are transmembrane homodimers that can form trimers, higher order arrays, and extended clusters as part of signaling complexes. Interactions of dimers in oligomers are thought to confer cooperativity and cross-receptor influences as well as a 35-fold gain between ligand binding and altered kinase activity. In addition, higher order interactions among dimers are necessary for the observed patterns of assistance in adaptational modification among different receptors. Elucidating mechanisms underlying these properties will require defining which receptor functions can be performed by dimers and which require specific higher order interactions. However, such an assignment has not been possible. Here, we used Nanodiscs, an emerging technology for manipulating membrane proteins, to prepare small particles of lipid bilayer containing one or only a few chemoreceptor dimers. We found that receptor dimers isolated in individual Nanodiscs were readily modified, bound ligand, and performed transmembrane signaling. However, they were hardly able to activate the chemotaxis histidine kinase. Instead, maximal activation and thus full-range control of kinase occurred preferentially in discs containing approximately three chemoreceptor dimers. The sharp dependence of kinase activation on this number of receptors per dimer implies that the core structural unit of kinase activation and control is a trimer of dimers. Thus, our observations demonstrate that chemoreceptor transmembrane signaling does not require oligomeric organization beyond homodimers and implicate a trimer of dimers as the unit of downstream signaling.
Keywords: membrane protein, nanoparticle, self-assembly, transmembrane receptor, transmembrane signaling
Motile bacteria move to favorable chemical environments through chemotaxis. The phenomenon and its mechanisms have been extensively characterized in Escherichia coli and its relatives (1–3). Transmembrane chemoreceptors form signaling complexes with the autophosphorylating histidine kinase CheA and coupling protein CheW. Phospho-CheA mediates phosphorylation of response regulator CheY. Phospho-CheY binds to the flagellar rotary motor, causing rotational reversal, which creates tumbles that alter swimming direction. Formation of signaling complexes activates CheA autophosphorylation and places that activity under the control of chemoreceptors. The sensory system directs cells toward favorable environments by modulating kinase activity, thus controlling the probability of tumbles and resulting directional changes.
Transmembrane Signaling
Transmembrane signaling by chemoreceptors (3) couples binding of stimulant molecules by its periplasmic domain with changes in its cytoplasmic domain that alter activation of the kinase and change the propensity for covalent modification of that domain, specifically methylation and demethylation at several glutamyl residues (four to six, depending on the specific receptor). Methylation is catalyzed by methyltransferase CheR. Demethylation is catalyzed by methylesterase CheB, an enzyme activated by CheA-mediated phosphorylation. Two receptor modification sites are synthesized as glutamines and deamidated by CheB to create methyl-accepting glutamates. Attractant binding to the receptor periplasmic domain reduces kinase activation by the cytoplasmic domain. Reduced kinase activity lowers cellular phospho-CheY and phospho-CheB. In addition, the transmembrane signal enhances the propensity of the receptor for methylation and reduces propensity for demethylation and deamidation. These changes result in increasing receptor methylation until modification balances ligand occupancy, restoring null-state kinase activity and modification propensities.
Chemoreceptor Interactions
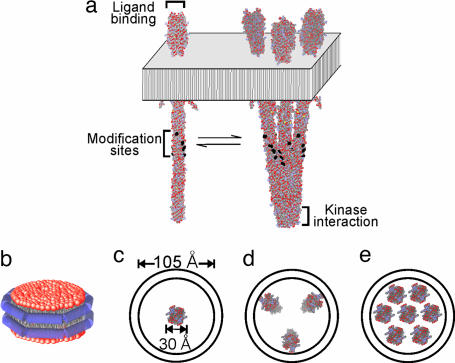
Chemoreceptors are elongated coiled-coil homodimers (4, 5) that bind attractant at sites on the membrane-distal tip of the periplasmic domain and associate with CheA and CheW at the membrane-distal tip of the cytoplasmic domain. Methyl-accepting glutamates are exposed on the surface of the coiled-coil cytoplasmic domain approximately halfway between the membrane and the kinase-binding tip (Fig. 1a). Homodimers can form trimers by interactions at the distal tips of their cytoplasmic domains (4, 6). They can also form higher order complexes and extended arrays by themselves or in complex with CheA and CheW (5, 7, 8). The interactions that create chemoreceptor clusters have not been defined.
Fig. 1.
Chemoreceptor oligomeric organization and insertion into Nanodiscs. (a) Diagram of a membrane-embedded chemoreceptor dimer (Left) and trimer of dimers (4) (Right) in equilibrium. (b) Diagram of a Nanodisc with two copies of a membrane scaffold protein surrounding the hydrocarbon side chains of a lipid bilayer. (c–e) Diagrams of Nanodiscs with scaffold protein MSP1D1E3(−) (13) containing one (c), three (d), or seven (e) chemoreceptor dimers.
Interactions of dimers in oligomers are thought to confer cooperativity and cross-receptor influences (9), as well as a 35-fold gain linking ligand binding and kinase activity (10). In addition, higher order interactions among dimers are necessary for the observed patterns of assistance in adaptational modification among different receptors (11). Elucidating mechanisms underlying these properties will require defining which receptor functions can be performed by dimers and which require specific higher order interactions. However, such an assignment has not been possible. Here, we used Nanodiscs (Fig. 1b), an emerging technology for manipulating membrane proteins (12, 13), to prepare small particles of lipid bilayer containing one or only a few chemoreceptor dimers (Fig. 1 c–e). We found that Nanodisc-isolated dimers were readily modified, bound ligand, and performed transmembrane signaling. However, they were essentially unable to activate the chemotaxis histidine kinase. Instead, maximal activation and thus full-range control of kinase occurred preferentially in discs containing approximately three chemoreceptor dimers. The sharp dependence of kinase activation on this number of receptors per dimer implied that the core structural unit of kinase activation and control is a trimer of dimers. Thus, our observations demonstrate that chemoreceptor transmembrane signaling does not require oligomeric organization beyond homodimers and implicate a trimer of dimers as the unit of downstream signaling.
Results
Nanodisc-Embedded Chemoreceptors.
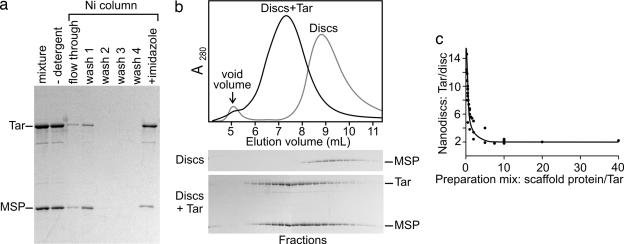
Nanodiscs form spontaneously when detergent is removed from a mixture of detergent-solubilized lipid and scaffold protein. If detergent-solubilized membrane protein is present, it is also incorporated (14, 15). We optimized this procedure for Tar, a well studied chemoreceptor of E. coli. Purified, detergent-solubilized Tar carrying a carboxyl-terminal six-histidine tag (Tar-6H) was added to a mixture of the scaffold protein MSP1D1E3(−) (13) and detergent-solubilized E. coli lipids, detergent was removed, and a Nanodisc was allowed to form. Those Nanodiscs containing Tar-6H were purified with a Ni column (Fig. 2a).
Fig. 2.
Preparation of Nanodisc-embedded chemoreceptor Tar-6H. (a) Coomassie-stained SDS-polyacrylamide gel of samples (equivalent proportions of the total material at each stage) from stages of Nanodisc preparation with chemoreceptor (Tar; 60 kDa) and MSP (30 kDa) indicated. (b) Size-exclusion chromatography of Nanodiscs containing only lipid (Discs) or lipid plus chemoreceptor (Discs + Tar). (Upper) Continuous recordings of absorbance. (Lower) Coomassie-stained SDS-polyacrylamide gels of samples from fractions collected. (c) Tar polypeptides per disk as a function of preparation ratio of scaffold protein to receptor determined by quantification of bands like those in a.
Size-exclusion chromatography of Ni column-purified Nanodiscs showed that these discs, purified from empty discs by the interaction of the histidine tag on Tar-6H, eluted in a roughly symmetrical peak of A280, ahead of the position for Nanodiscs alone (Fig. 2b Upper). The elution position indicated that the Ni column-purified discs were larger than empty discs, consistent with the insertion of receptor protein. The symmetry indicated a relatively homogenous population of Tar-6H-containing particles. This result was confirmed by the coelution of chemoreceptor and scaffold protein at roughly a constant ratio over most of the peak (Fig. 2b Lower). The modest enrichment of scaffold protein at greater elution volumes indicated a small proportion of empty discs.
We varied the preparation ratio of scaffold protein to Tar-6H and used a Ni column to purify the receptor-containing Nanodiscs formed at each ratio. The relative amounts of Tar-6H and membrane scaffold protein in each purified preparation were determined by analyzing stained gels like those in Fig. 2a by using densitometry and standard curves of purified proteins for which concentrations had been determined by quantitative amino acid analysis. We could use the Tar-6H per scaffold protein ratio to calculate the Tar-6H per disk ratio for each purified preparation because of previous studies (12, 13, 16, 17) of the properties of membrane scaffold proteins and the Nanodiscs formed by them. Determinations of lipid content, protein content, and diameters of Nanodiscs made with membrane scaffold proteins of different lengths, with or without inserted transmembrane protein, have been performed (12, 13, 16, 17). Those studies established that the family of membrane scaffold proteins of which MSP1D1E3(−) is a member form disk particles of a defined size, characteristic of the length of the particular membrane scaffold protein, in which a specific number of lipids is surrounded by exactly two copies of the scaffold protein. Using this ratio, we calculated the output ratios of Tar per disk and plotted them versus the scaffold protein/receptor ratio in the preparation mixture (Fig. 2c). With excess scaffold protein in the preparation mixture (8–40 scaffold proteins per Tar), the value for receptor polypeptides per disk reached a plateau, a constant, lower limit of two, indicating that the core structural unit incorporated into a disk was a receptor homodimer (1, 3, 18) and that each disk contained a single dimer. Nanodiscs produced from preparation mixtures with less scaffold protein per Tar contained, on the average, more than one receptor dimer in the same 105-Å diameter plug of bilayer (12). There was no preference evident for integer values of these ratios (Figs. 2c and 3c), arguing that detergent-solubilized dimers incorporated individually into Nanodiscs and that the number of receptors per disk represented a mean among a population with different integral numbers of dimers per disk.
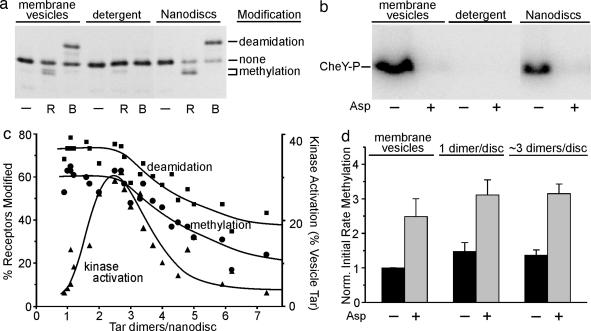
Fig. 3.
Activity assays of Nanodisc-embedded chemoreceptor Tar-6H. (a) Adaptational modification. Tar-6H contained in native membrane vesicles (membrane vesicles), solubilized in 25 mM cholate (detergent) or reconstituted in Nanodiscs at approximately one dimer per disk (Nanodiscs) was incubated without further addition (−) or with methyltransferase (R) or activated methylesterase (B) in conditions for maximal modification and analyzed by SDS/PAGE and immunoblotting with anti-Tar. Electrophoretic positions are indicated for gene-encoded (two-glutamine) Tar with no modification (none), one or two methylations (methylation) or two deamidations (deamidation). (b) Kinase activation and its control by ligand. Tar-6H as described for a, except Nanodiscs contained ≈three dimers per disk, were tested in the absence (−) or presence (+) of 1 mM aspartate (Asp) for activation of kinase CheA in a coupled assay producing phosphorylated CheY (CheY-P) as detected by phosphorimaging of an SDS-polyacrylamide gel, in conditions in which Tar-6H in native membrane vesicles produced its maximal kinase activation. All Nanodisc preparations exhibited some kinase activation (see c), and aspartate reduced that activation to the same background level illustrated in this figure. (c) Receptor activities as a function of dimers per Nanodisc. Receptor content and activities (a and b) were quantified for 21 preparations of Nanodisc Tar-6H. Activities are the percentage of the receptor population modified or percentage of the kinase activation observed for the same amount of Tar-6H embedded in native membrane vesicles with cytoplasmic domains exposed to the solvent (determined by percentage of deamidation in conditions of maximal modification). The curve was drawn to aid the eye. (d) Effect of ligand occupancy on initial rate of methylation. Labels are as for b. Values are averages with standard deviations of three independent determinations normalized to membrane vesicles in the absence of aspartate. Very similar results were obtained for two other Nanodisc preparations with approximately one and two with approximately three dimers per disk.
Functional Activities.
We tested each preparation of Nanodisc-embedded Tar-6H for CheR-catalyzed methylation and phospho-CheB-catalyzed deamidation (Fig. 3a), and for kinase activation (Fig. 3b). In our native membrane vesicles, orientation of Tar-6H was approximately random. Thus, only half the cytoplasmic domains were accessible to enzymes added to the suspension, resulting in 50% deamidation at the two susceptible glutamines and somewhat less methylation at the two susceptible glutamates. Detergent solubilization essentially eliminated these activities, but Nanodisc-incorporated Tar regained them (Fig. 3a). A larger proportion of receptor was modified in discs than in native vesicles, consistent with accessibility on both sides of the bilayer. A fraction of Nanodisc-incorporated Tar was not modified, perhaps because some receptors were no longer functional as the result of the experimental manipulations. Like modification, the ability of Tar to activate the chemotaxis kinase CheA was eliminated by detergent solubilization and reappeared in Nanodisc-inserted receptor (Fig. 3b).
Receptor Activity as a Function of Dimers per Disk.
Varying the number of Tar dimers per disk had strikingly different effects on adaptational modification and kinase activation (Fig. 3c). Deamidation and methylation were at the same high level for preparations with one to three dimers per disk and decreased gradually at higher ratios. In contrast, kinase activation was minimal at one dimer per disk, exhibited a distinct maximum at approximtely three dimers per disk, and dropped sharply at higher ratios. The patterns indicated that effective adaptational modification did not require more than one dimer, but kinase activation did.
We examined the ability of Nanodisc-embedded Tar to perform transmembrane signaling by determining the effect of a saturating concentration of the Tar-recognized attractant aspartate on activities mediated by the cytoplasmic domain. Tar embedded in native membrane vesicles couples aspartate binding in the periplasmic domain to a drastic reduction in activity of kinase associated with the cytoplasmic domain (Fig. 3b, leftmost two lanes). For every Nanodisc preparation, across the entire range of dimers per disk, saturating aspartate reduced kinase activity to the same background level observed for aspartate-occupied Tar in native membrane vesicles (Fig. 3b). This comprehensive effect indicated that receptor dimers were competent for transmembrane signaling independent of the number of dimers per disk and demonstrated that the kinase activation we observed was the result of physiologically relevant interaction with receptor. However, for preparations with approximately one dimer per disk, the difference between the low extent of receptor-mediated kinase activation and background activity was modest. Thus, we used an additional assay for transmembrane signaling, the coupling of ligand occupancy to propensity for adaptational modification. Saturating aspartate increased the initial rate of methylation ≈2-fold for Tar-6H in native vesicles (19) and had quantitatively the same effect in Nanodisc preparations with approximately one dimer and approximately three dimers per disk (Fig. 3d). This qualitatively similar effect demonstrated that single Tar dimers isolated in Nanodiscs were as efficient at transmembrane signaling as receptors in native membrane or in Nanodiscs with multiple dimers per disk. Aspartate also reduced the rate of deamidation for Tar in Nanodiscs with approximately one dimer and approximately three dimers per disk, as it did for the receptor in native vesicles (data not shown). The influence of ligand on modification rate for approximately one dimer per disk preparations indicated that chemoreceptor dimers performed functionally relevant ligand binding, a result consistent with binding of aspartate to isolated periplasmic domains (18).
Discussion
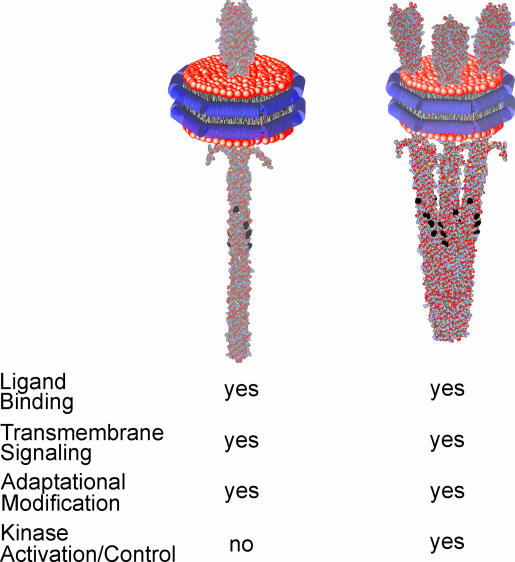
The ability of Nanodisc technology to produce small, defined-sized patches of lipid bilayer containing one or a few transmembrane proteins in biochemically relevant amounts allowed us to address the relationship between chemoreceptor activities and oligomeric organization. The issue was challenging because these transmembrane receptors are thought to form not only trimers of dimers but also higher order arrays and clusters of unknown and perhaps variable stoichiometry (20–22). We know little about the equilibrium between dimers and higher order oligomers, and there was no way to control or define the distribution of oligomeric forms in a conventional membrane vesicle, which would contain many molecules of receptor dimers available to interact in a single, continuous membrane. The solution was the small area of a Nanodisc-enclosed lipid bilayer, which provided a native environment but restricted the number of molecules that could interact. The observations we made are summarized in Fig. 4.
Fig. 4.
Functional activities of Nanodisc-embedded chemoreceptor. Diagrams of a single homodimer (Left) or a trimer of dimers (Right) inserted into a Nanodisc are shown above a summary of respective activities.
Different Dependencies on Oligomeric State for Adaptational Modification and Kinase Activation.
Using Nanodiscs to limit the number of potentially interacting receptor dimers produced the striking observation that adaptational modification and kinase activation exhibited distinctly different dependencies on the number of dimers per disk (Fig. 3c). Receptor dimers were efficiently modified in disk preparations containing one to three dimers per disk, and efficiency decreased gradually at higher ratios. In contrast, there was a distinct peak of kinase activation for preparations with approximately three dimers per disk. What can be concluded from these quite different patterns? For modification, effective activity in preparation of approximately one dimer per disk clearly demonstrates that isolated dimers are readily acted on by both enzymes of adaptational modification. There is no indication of a requirement for higher order interactions among dimers for this important receptor function. We do not know why modification effectiveness decreases at higher dimers per disk, but it is plausible that increasing numbers of receptors in a defined-sized disk would create increasing steric constraints on enzyme accessibility (Fig. 1e).
For kinase activation, the low activity for one dimer per disk preparations and the sharp maximum at approximately three dimers per Nanodisc (Fig. 3c) indicated that more than one dimer is necessary for effective activation. The tantalizing implication is that the effective combination is three dimers, presumably as a trimer of dimers inserted in the same, parallel orientation in a disk (4, 6). However, we do not yet have sufficient information about receptors inserted into Nanodiscs to make definitive interpretations. For instance, it is not clear why the decrease in activity at higher values of dimer per disk is significantly sharper for kinase activation than for modification, placing the peak at a value just less than three dimers per disk. Perhaps a fourth dimer in a disk has a drastic negative effect, for instance, through steric hindrance that blocks productive interaction among the three other dimers in the disk. Also, it is not clear why maximal activation by Nanodisc-embedded receptor was only 30% of the activation created by the same number of receptors embedded in native membrane (Fig. 3c), of which only 50% would have had cytoplasmic domains facing the solvent and thus exposed to kinase. However, 30% is close to what would be expected by the following reasoning. All Nanodisc-embedded dimers should have cytoplasmic domains exposed to solvent, but only 25% of Nanodiscs with three randomly inserted dimers would contain three parallel dimers capable of forming trimers, and modification assays indicated that 60–75% of inserted dimers were in a sufficiently native state to be modified. Thus, kinase activation by Nanodisc-embedded receptors would be predicted to be (100%/50%) × 25% × (60–75%) = 30–38% of the activity of the same number of receptors in native membrane vesicles. Alternatively, maximal kinase activation might require interactions beyond trimers not possible in the small area of a Nanodisc.
Preparations with approximately one receptor dimer per Nanodisc activated kinase at 5–10% the level of receptors in native membrane vesicles. This finding might reflect a reduction in the ability to bind CheA and CheW or to activate CheA once it is bound. Alternatively, individual receptor dimers might have no ability to bind or activate CheA, and the activation detected is the result of (i) a small proportion of discs with multiple inserted receptors or (ii) dimers in different discs associating in a complex with CheA and CheW. These possibilities need to be investigated.
Transmembrane Signaling Requires only a Receptor Dimer.
Transmembrane signaling by individual dimers isolated in Nanodiscs was as effective as Tar signaling in native membrane (Fig. 3d). This effective signaling indicated that the fundamental conformational change of ligand-induced transmembrane signaling required no greater structural complexity than the homodimer. This conclusion implies that changes in trimer or higher order interactions that have been observed or postulated to be related to signaling (21–24) are not necessary for coupling of ligand occupancy in the periplasmic domain to conformational change in the cytoplasmic domain. The notion of transmembrane signaling occurring within the structure of single dimers is consistent with the large body of data that identifies the conformational change of transmembrane signaling in the periplasmic and transmembrane domains as the piston sliding of a single signaling helix of a ligand-occupied receptor dimer (3, 25).
Small plugs of bilayers formed by E. coli lipids in Nanodiscs limited the potential for oligomerization of chemoreceptor dimers embedded in a two-dimensional bilayer, a limitation not possible in traditional vesicle systems. Application of this technology allowed us to determine the relationship between activity and oligomerization. We expect Nanodiscs will be equally useful in characterization of other membrane proteins, receptors, and enzymes (17).
Materials and Methods
Membrane Vesicle-Borne and Purified Tar-6H.
Cytoplasmic membrane vesicles were prepared, essentially as described (26), from RP3098 harboring pAL67 (19) grown in Luria broth plus 100 μg/ml ampicillin at 35°C to ≈2 × 108 cells per ml. Isopropyl-thio-β-d-galactoside was added to 1 mM, and the culture was chilled in an ice bath 3.5 h later. Membranes were suspended in 50 mM Tris·HCl (pH 7.5) and 10% wt/vol glycerol and were stored at −70°C after freezing in liquid N2. Protein and Tar-6H concentrations were determined by the bicinchinonic acid assay and quantitative immunoblotting (27), respectively. Membranes were thawed on ice with addition of room temperature buffer to yield ≤10 mg/ml protein; the suspension was brought to 2 μM leupeptin, 2 μM pepstatin, 1 mM PMSF, and 100 μM Nα-p-tosyl-l-lysine chloromethyl ketone hydrochloride (TLCK); β-octyl glucoside was added at 10 mg/mg protein, incubated for 10 min on ice, and clarified by centrifugation at 100,000 rpm, 15 min, 4°C in a TLA 100.4 rotor (435,680 × g). The supernatant was loaded on a Ni-NTA agarose column (Qiagen) equilibrated in 50 mM Tris·HCl (pH 9.0), 10% wt/vol glycerol, 100 mM NaCl, 25 mM sodium-cholate, and 15 mM imidazole (column buffer),and the column was washed with 5 column volumes of buffer and eluted with 5 volumes of buffer containing 300 mM imidazole. The latter fractions were concentrated ≈10-fold in a 30,000 molecular weight cutoff (MWCO) Centriprep, dialyzed in 12,000–14,000 MWCO Spectra/Por tubing against 3 × 100 volumes of 50 mM Tris·HCl (pH 7.5), 10% wt/vol glycerol, 25 mM sodium-cholate 8–12 h each, frozen in aliquots in liquid N2, and stored at −70°C. Receptor was quantified by densitometry of Coomassie-stained bands on SDS/PAGE by using Tar-6H standards.
Lipids.
Samples of E. coli lipids in chloroform (Avanti) were placed in glass tubes, solvent was removed with a gentle stream of N2 while rotating the tube to create a thin film, and samples were placed overnight in a vacuum desiccator. We added 50 mM Tris·HCl (pH 7.5) and 100 mM sodium-cholate to the original volume, and the tubes were flushed with N2 and sealed with Parafilm. To hydrate lipid, tubes were shaken for 2 h at 35°C and 200 rpm, and suspensions were sonicated 6 × 5 s with 25-s pauses by using a Tekmar TM-250 sonic disruptor equipped with a microtip at output 2. The suspension was passed through a 0.2-μm filter, flushed with N2, and stored at −70°C.
Nanodisc Preparation.
Lyophilized powder of scaffold protein MSP1D1E3(−) (13) was solubilized in H2O to yield a solution of ≈4.5 mg/ml in 20 mM Tris·HCl (pH 7.4), 100 mM NaCl, 0.5 mM EDTA, and 0.01% NaN3, passed through a 0.2-μm filter, and stored at 4°C. Membrane scaffold protein (MSP) concentration was determined spectrophotometrically by using ε280 = 0.868 ml mg−1·cm−1. Ten micromolar purified Tar-6H, MSP, and lipids were mixed, supplemented with 1 mM PMSF, 1 μM leupeptin, and 1 μM pepstatin, and incubated with gentle rocking for 1 h at room temperature with cholate concentration ≥25 mM. The MSP:lipid molar ratio was 1:120, and the ratio of Tar-6H to Nanodisc components varied as indicated. Two-thirds volume BioBeads SM-2 (Bio-Rad) hydrated in H2O was added to one volume of assembly mixture, incubated with gentle rocking for 1 h at room temperature and centrifuged for 1 min at 3,700 rpm in a GH 3.8 rotor (3,200 × g). The supernatant was loaded on a Ni-NTA agarose column equilibrated in 50 mM Tris·HCl (pH 9.0), 10% wt/vol glycerol, 100 mM NaCl, 15 mM imidazole with a bed volume equal to the assembly mixture volume, washed with 12–15 column volumes of equilibration buffer, and eluted with 5 column volumes of the buffer containing 300 mM imidazole. Eluted Nanodiscs were concentrated in a 30,000 molecular weight cutoff (MWCO) Amicon Ultra 4 concentrator, dialyzed in 12,000–14,000 MWCO Spectra/Por tubing against 3 × 100 volumes of 50 mM Tris·HCl (pH 7.5), 10% wt/vol glycerol, 100 mM NaCl, 0.5 mM EDTA for 8–12 h each, frozen in liquid N2, and stored at −70°C. Tar-6H and MSP were quantified by densitometry of Coomassie-stained bands on SDS/PAGE by comparison with standards of each purified protein, the concentrations of which had been determined by quantitative amino acid analysis.
Size-Exclusion Chromatography.
Chromatography was performed on a TSK G5000 PWXL column [7.8-mm inner diameter × 30 cm (Tosoh Haas)] in 50 mM Tris·HCl (pH 7.5), 10% wt/vol glycerol, 100 mM NaCl, and 0.5 mM EDTA at 19°C at 0.7 ml/min with 0.3-ml fractions.
Activity Assays.
Conditions were based on those described (28, 29). Tests of maximal modification used 2.5 μM receptor, 5 μM CheR in a lysate or 5 μM CheB and 50 mM phosphoramidate (30) for 30 min. These conditions are sufficient to overcome the modest effects of the carboxyl-terminal histidine tag on rates of receptor modification (19). Initial rates were determined by using 0.15 μM enzyme and 2.5 μM available receptor. In the context of interpreting the lack of activity for cholate-solubilized Tar-6H, we have not tested the effect of 25 mM cholate on the activities of CheR, CheB, or CheA, but 1% octyl glucoside, generally harsher on enzyme activity than cholate, allows CheR activity (31) but inhibits CheA (32). There is no information available about effects of detergents on CheB activity.
Acknowledgments
We thank Linda L. Randall for constant encouragement and advice throughout the course of this work, as well as for discussions and aid in chromatography, and Wing-Cheung Lai for help in manuscript preparation. This work was supported by National Institutes of Health Grants GM29963 (to G.L.H.) and GM33775 (to S.G.S.).
Abbreviation
- MSP
membrane scaffold protein
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Wadhams G. H., Armitage J. P. Nat. Rev. Mol. Cell Biol. 2004;5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- 2.Sourjik V. Trends Microbiol. 2004;12:569–576. doi: 10.1016/j.tim.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Falke J. J., Hazelbauer G. L. Trends Biochem. Sci. 2001;26:257–265. doi: 10.1016/s0968-0004(00)01770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim K. K., Yokota H., Kim S.-H. Nature. 1999;400:787–792. doi: 10.1038/23512. [DOI] [PubMed] [Google Scholar]
- 5.Park S.-Y., Borbat P. P., Gonzalez-Bonet G., Bhatnagar J., Pollard A. M., Freed J. H., Bilwes A. M., Crane B. R. Nat. Struct. Mol. Biol. 2006;13:400–407. doi: 10.1038/nsmb1085. [DOI] [PubMed] [Google Scholar]
- 6.Studdert C. A., Parkinson J. S. Proc. Natl. Acad. Sci. USA. 2004;101:2117–2122. doi: 10.1073/pnas.0308622100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weis R. M., Hirai T., Chalah A., Kessel M., Peters P. J., Subramaniam S. J. Bacteriol. 2003;185:3636–3643. doi: 10.1128/JB.185.12.3636-3643.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Francis N. R., Wolanin P. M., Stock J. B., DeRosier D. J., Thomas D. R. Proc. Natl. Acad. Sci. USA. 2004;101:17480–17485. doi: 10.1073/pnas.0407826101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sourjik V., Berg H. C. Nature. 2004;428:437–441. doi: 10.1038/nature02406. [DOI] [PubMed] [Google Scholar]
- 10.Sourjik V., Berg H. C. Proc. Natl. Acad. Sci. USA. 2002;99:123–127. doi: 10.1073/pnas.011589998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li M., Hazelbauer G. L. Mol. Microbiol. 2005;56:1617–1626. doi: 10.1111/j.1365-2958.2005.04641.x. [DOI] [PubMed] [Google Scholar]
- 12.Bayburt T. H., Grinkova Y. V., Sligar S. G. Nano Lett. 2002;2:853–856. [Google Scholar]
- 13.Denisov I. G., Grinkova Y. V., Lazarides A. A., Sligar S. G. J. Am. Chem. Soc. 2004;126:3477–3487. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- 14.Bayburt T. H., Sligar S. G. Protein Sci. 2003;12:2476–2481. doi: 10.1110/ps.03267503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duan H., Civjan N. R., Sligar S. G., Schuler M. A. Arch. Biochem. Biophys. 2004;424:141–153. doi: 10.1016/j.abb.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 16.Milligan D. L., Koshland D. E., Jr. J. Biol. Chem. 1988;263:6268–6275. [PubMed] [Google Scholar]
- 17.Bayburt T. H., Grinkova Y. V., Sligar S. G. Arch. Biochem. Biophys. 2006;450:215–222. doi: 10.1016/j.abb.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 18.Milligan D. L., Koshland D. E., Jr. J. Biol. Chem. 1993;268:19991–19997. [PubMed] [Google Scholar]
- 19.Lai W.-C., Hazelbauer G. L. J. Bacteriol. 2005;187:5115–5121. doi: 10.1128/JB.187.15.5115-5121.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shimizu T. S., Le Novere N., Levin M. D., Beavil A. J., Sutton B. J., Bray D. Nat. Cell Biol. 2000;2:792–796. doi: 10.1038/35041030. [DOI] [PubMed] [Google Scholar]
- 21.Baker M. D., Wolanin P. M., Stock J. B. BioEssays. 2006;28:9–22. doi: 10.1002/bies.20343. [DOI] [PubMed] [Google Scholar]
- 22.Kim S.-H., Wang W., Kim K. K. Proc. Natl. Acad. Sci. USA. 2002;99:11611–11615. doi: 10.1073/pnas.132376499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Homma M., Shiomi D., Homma M., Kawagishi I. Proc. Natl. Acad. Sci. USA. 2004;101:3462–3467. doi: 10.1073/pnas.0306660101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vaknin A., Berg H. C. Proc. Natl. Acad. Sci. USA. 2006;103:592–596. doi: 10.1073/pnas.0510047103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lai W.-C., Beel B. D., Hazelbauer G. L. Mol. Microbiol., 2006 doi: 10.1111/j.1365-2958.2006.05296.x. in press. [DOI] [PubMed] [Google Scholar]
- 26.Lin L. N., Li J., Brandts J. F., Weis R. M. Biochemistry. 1994;33:6564–6570. doi: 10.1021/bi00187a025. [DOI] [PubMed] [Google Scholar]
- 27.Li M., Hazelbauer G. L. J. Bacteriol. 2004;186:3687–3694. doi: 10.1128/JB.186.12.3687-3694.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barnakov A. N., Barnakova L. A., Hazelbauer G. L. J. Bacteriol. 1998;180:6713–6718. doi: 10.1128/jb.180.24.6713-6718.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barnakov A. N., Barnakova L. A., Hazelbauer G. L. Proc. Natl. Acad. Sci. USA. 1999;96:10667–10672. doi: 10.1073/pnas.96.19.10667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheridan R., McCullough J., Wakefield Z. Inorg. Syntheses. 1971;13:23–26. [Google Scholar]
- 31.Bogonez E., Koshland D. E., Jr. Proc. Natl. Acad. Sci. USA. 1985;82:4891–4895. doi: 10.1073/pnas.82.15.4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Borkovich K. A., Simon M. I. Methods Enzymol. 1991;200:205–214. doi: 10.1016/0076-6879(91)00140-r. [DOI] [PubMed] [Google Scholar]