Abstract
Barth syndrome is an X-linked disease presenting with cardiomyopathy and skeletal muscle weakness. It is caused by mutations in tafazzin, a putative acyl transferase that has been associated with altered metabolism of the mitochondrial phospholipid cardiolipin. To investigate the molecular basis of Barth syndrome, we created Drosophila melanogaster mutants, resulting from imprecise excision of a P element inserted upstream of the coding region of the tafazzin gene. Homozygous flies for that mutation were unable to express the full-length isoform of tafazzin, as documented by RNA and Western blot analysis, but two shorter tafazzin transcripts were still present, although the expression levels of their encoded proteins were too low to be detectable by Western blotting. The tafazzin mutation caused an 80% reduction of cardiolipin and a diversification of its molecular composition, similar to the changes seen in Barth patients. Other phospholipids, like phosphatidylcholine and phosphatidylethanolamine, were not affected. Flies with the tafazzin mutation showed a reduced locomotor activity, measured in flying and climbing assays, and their indirect flight muscles displayed frequent mitochondrial abnormalities, mostly in the cristae membranes. Thus, tafazzin mutations in Drosophila generated a Barth-related phenotype, with the triad of abnormal cardiolipin, pathologic mitochondria, and motor weakness, suggesting causal links between these findings. We conclude that a lack of full-length tafazzin is responsible for the cardiolipin deficiency, which is integral to the disease mechanism, leading to mitochondrial myopathy.
Keywords: cardiolipin, mitochondria, myopathy
Barth syndrome (BTHS) is an X-linked multisystemic disorder presenting with dilated cardiomyopathy, skeletal myopathy, cyclic neutropenia, and growth retardation (1). The disease is caused by mutations in the tafazzin gene (2) that encodes a putative phospholipid acyltransferase (3). Accordingly, BTHS patients have phospholipid abnormalities, in particular, a reduced content and an altered composition of cardiolipin (4, 5). Among the many cardiolipin species that can be formed by various fatty acid combinations (6), BTHS specifically affects the concentration of tetralinoleoyl-cardiolipin (7, 8). This species is formed by remodeling of the four acyl groups of cardiolipin, suggesting that tafazzin is involved in the acyl group exchange (9). BTHS has generally been considered a mitochondrial disease because pathologic mitochondria have been found in tissue biopsies from BTHS patients (10, 11) and because cardiolipin is a specific mitochondrial lipid (6). However, the presence of multiple tafazzin transcripts (2, 12–14) raises the prospect that other cellular functions are affected as well.
Although it is plausible that BTHS myopathy is caused by mitochondrial dysfunction, which in turn is caused by cardiolipin deficiency, this pathogenetic link has never been actually demonstrated, mainly because of the lack of a suitable experimental model. To investigate the molecular mechanism of BTHS, we created a disease model in Drosophila melanogaster. We choose Drosophila because it has specialized flight muscles with superabundant mitochondria (15), and, like humans, it expresses several tafazzin transcripts (16). Thus, Drosophila combines a susceptible organ system with a complex tafazzin expression pattern, which makes it attractive for comparative studies. It promises to be a significant advancement over tafazzin-deficient yeast that until now has been the main model in BTHS research.
Results
We created tafazzin mutations in D. melanogaster by imprecise excision of the P element from strain KG02529. The P{SUPor-P}tafazzinKG02529 insertion was located upstream of the start codon in exon 1 of the tafazzin gene (Fig. 1A). In this location, it did not have any apparent effect on phenotype (data not shown). We mobilized the P element with transposase under conditions favoring deletions of adjacent DNA and screened a collection of 149 excision strains by PCR. The screen yielded three clones with significant deletions in the tafazzin gene (Fig. 1A). Flies that were homozygous for this deletion were referred to as TAZ−/−. They were compared with a control group that had undergone the same crosses but had a precise excision of the P element and, thus, had an intact tafazzin gene. This group was referred to as TAZ+/+.
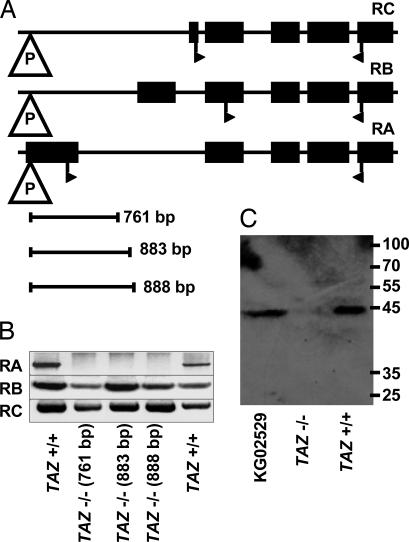
Fig. 1.
Tafazzin mutants of Drosophila. (A) Organization of the tafazzin gene, showing the exons of the three transcripts RA, RB, and RC with the insertion site of the P element P{SUPor-P}tafazzinKG02529. Start and stop codons are indicated by flags. Imprecise excision of the P element yielded three clones with genomic deletions of 761, 883, and 888 base pairs, respectively. (B) RT-PCR analysis of Drosophila clones generated by precise (TAZ+/+) and imprecise (TAZ−/−) excision of the P element. (C) Western blot analysis of mitochondrial preparations of TAZ+/+, TAZ−/−, and KG02529 by using a polyclonal antibody raised against Drosophila tafazzin. The size of the single band in TAZ+/+ and KG02529 corresponds to the protein product of RA.
First, we analyzed the expression of the three known transcripts of tafazzin, RA, RB, and RC. The TAZ−/− clones failed to express RA, the full-length tafazzin transcript, but they still produced the two shorter isoforms RB and RC (Fig. 1B). Next, we analyzed protein expression by using a polyclonal rabbit antibody raised against Drosophila tafazzin. This antibody was able to recognize the translation products of RA, RB, and RC, expressed in 293 embryonic human kidney cells, but it failed to detect any of the tafazzin proteins in total Drosophila homogenate, even when the amount of protein was increased to 0.2 mg per lane (data not shown). However, in purified Drosophila mitochondria, the antibody detected a single band that corresponded to the size of full-length tafazzin. We found this band in wild-type and TAZ+/+ mitochondria but not in mitochondria from TAZ−/− mutants (Fig. 1C). The data demonstrate the absence of full-length tafazzin in TAZ−/− flies.
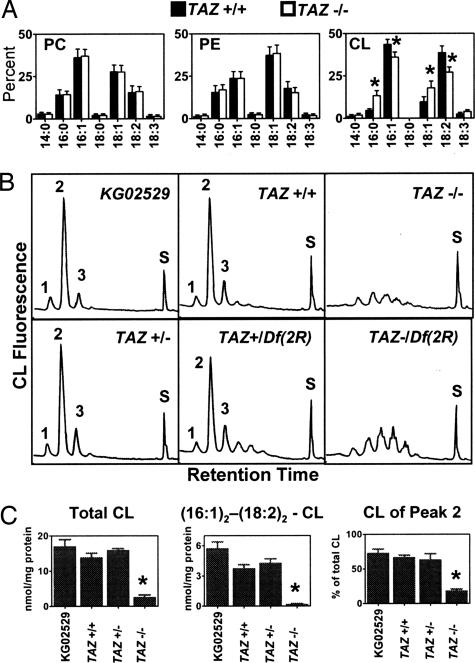
Because previous studies suggested that tafazzin is a phospholipid acyltransferase, we determined the fatty acid composition of phospholipids in Drosophila mutants. Deletion of full-length tafazzin caused a change in the fatty acid pattern of cardiolipin but not of phosphatidylcholine and phosphatidylethanolamine (Fig. 2A). HPLC analysis showed that only a single cardiolipin peak was affected by the mutation. This peak, which contains a mixture of cardiolipins with palmitoleoyl and linoleoyl residues, is the dominant component in normal Drosophila (17). Instead of this peak, TAZ−/− flies produced multiple minor species of cardiolipin (Fig. 2B). The total amount of cardiolipin was reduced by ≈80%, and the main molecular species, dipalmitoleoyl-dilinoleoyl-cardiolipin, was virtually absent (Fig. 2C). Heterozygous TAZ+/− flies had a normal cardiolipin profile (Fig. 2 B and C). Because part of the tafazzin gene was left intact in TAZ−/−, we wanted to know whether a more severe mutation would result in a more severe cardiolipin phenotype. We, therefore, generated flies that were heterozygous for the partial tafazzin deletion and a complete deletion of the chromosomal region containing the tafazzin gene [TAZ−/Df(2R)]. We found similar changes of cardiolipin in TAZ−/Df(2R) and TAZ−/−, indicating that the partial tafazzin deletion represented a functional “null” for the cardiolipin phenotype (Fig. 2B).
Fig. 2.
Lipid analysis of Drosophila mutants. (A) Fatty acid composition of phosphatidylcholine (PC), phosphatidylethanolamine (PE), and cardiolipin (CL) in mutants generated by precise (TAZ+/+) and imprecise (TAZ−/−) excision of the P element of strain KG02529. Data are given in mass percent. Asterisks indicate a significant difference between TAZ+/+ and TAZ−/− (n = 3, P < 0.05). (B) Molecular composition of cardiolipin in Drosophila mutants determined by fluorescence HPLC. The fluorescence yield is plotted against the retention time from 7–47 min. Peak 2 contains cardiolipins with palmitoleoyl (16:1) and linoleoyl (18:2) residues (see ref. 17). Peak S contains the internal standard. (C) Quantification of cardiolipin. Total cardiolipin and the proportion of peak 2 were measured by fluorescence-HPLC. Dipalmitoleoyl-dilinoleoyl-cardiolipin was measured by mass spectroscopy. Asterisks indicate a significant difference between TAZ+/+ and TAZ−/− (n = 3, P < 0.001).
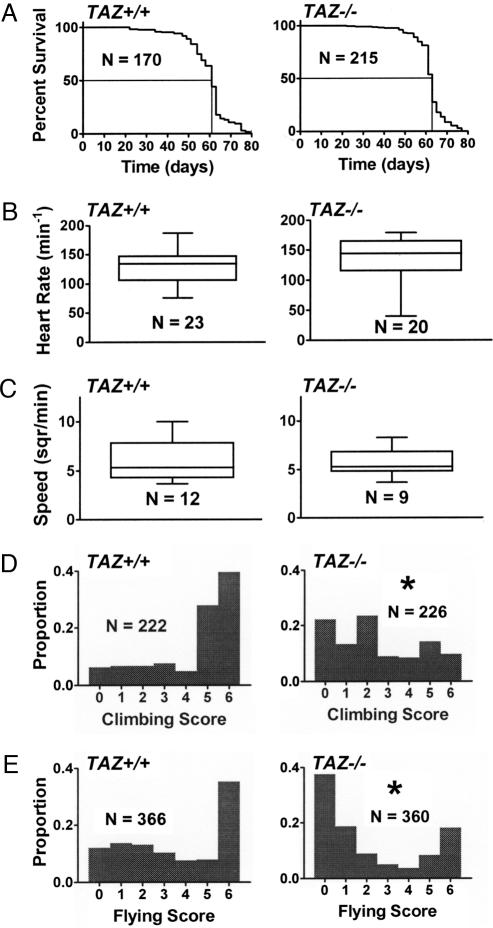
Next, we asked whether the tafazzin mutation has any functional phenotype in Drosophila. We found that TAZ−/− mutants had the same lifespan as TAZ+/+ controls (Fig. 3A). Likewise, the heart rate of TAZ−/− pupae (Fig. 3B) and the locomotor activity of TAZ−/− larvae (Fig. 3C) were normal. However, adult TAZ−/− flies showed moderate signs of motor weakness. Although motor weakness was not severe enough to be picked up by casual observation, quantitative assays clearly demonstrated the reduced ability of adult TAZ−/− flies to climb against gravity (Fig. 3D) and to fly (Fig. 3E).
Fig. 3.
Physiologic assays in TAZ+/+ and TAZ−/− mutants. (A) Kaplan–Meier analysis of longevity. Median survival is 61 and 63 days for TAZ+/+ and TAZ−/−, respectively. (B) Heart rate of early pupae. (C) Locomotor activity of larvae. The speed of larvae is expressed as 0.04 square inches crossed per minute. (D) Climbing activity of adults (1 week old). Histograms show distribution of the populations over climbing scores ranging from zero (lowest activity) to six (highest activity). (E) Flying activity of adults (1 week old). Histograms show distribution of the populations over flying scores ranging from zero (lowest activity) to six (highest activity). Asterisks indicate a significant difference between TAZ+/+ and TAZ−/− (P < 0.0001). N, number of flies tested.
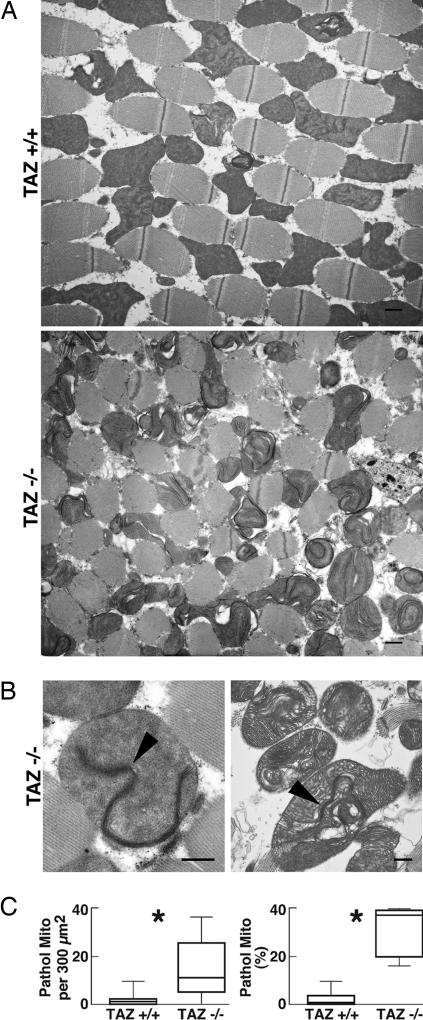
To further investigate the cause of motor weakness, we performed electron microscopy of the indirect wing muscles. Although no alteration of the myofibril structure was observed, TAZ−/− muscles contained many structurally abnormal mitochondria (Fig. 4A). The abnormality affected primarily the internal cristae membranes, which presented as hyperdense stacks that formed swirls, curls, or rings, in many cases extending through almost the entire mitochondrial length. Often, these changes were associated with swelling and disruption of the cristae (Fig. 4B). Abnormal mitochondria were not homogeneously distributed throughout the muscle fibers. Instead, they formed clusters that coexisted with zones of normal mitochondria. Nevertheless, the overall abundance of abnormal mitochondria was >10-fold higher in TAZ−/− than in TAZ+/+ (Fig. 4C). Tafazzin deficiency also caused an increase in the number of giant mitochondria (>3.5 μm2) from 1.2 ± 0.5 per 300 μm2 in TAZ+/+ to 3.2 ± 0.8 per 300 μm2 in TAZ−/− (P < 0.05), and abnormal cristae were more frequent in the giant mitochondria than in the total mitochondrial population (prevalence of 89% versus 30%; P < 0.0005). To identify cardiolipin changes specific to muscle, we dissected fly thoraces, in which muscles make up most of the tissue mass. However, the cardiolipin composition was similar in thoraces and whole flies, suggesting that muscle cardiolipin is not different from cardiolipin in other tissues (data not shown).
Fig. 4.
Mitochondrial ultrastructure in dorsal indirect flight muscles. (A) Electron micrographs of transversely sectioned muscles from TAZ+/+ and TAZ−/− mutants. (Scale bars: 1 μm.) (B) Electron micrographs of abnormal TAZ−/− mitochondria. Arrowheads point to bundles of stacked cristae with high electron density. (Scale bars: 0.5 μm.) (C) Morphometric analysis of crista abnormalities. The frequency of pathologic mitochondria is expressed as their absolute number per 300 μm2 and as their relative proportion in the total mitochondrial population. Nineteen random sections were analyzed in each group, containing a total of 959 (TAZ+/+) and 808 (TAZ−/−) mitochondria, respectively. Asterisks indicate a significant difference between TAZ+/+ and TAZ−/− (P < 0.005).
Discussion
Experimental models of BTHS have been developed in yeast (14, 18), cultured fibroblasts (4), and lymphoblasts (19). These models have provided important insights into the underlying molecular defect but have not been useful to study the organ-specific aspects of the disease. The present work with Drosophila establishes a BTHS model in an organism with differentiated tissues, among them the highly developed flight muscle apparatus.
We have generated Drosophila, which are unable to produce the full-length isoform of tafazzin, and found that these mutants have cardiolipin deficiency, abnormal muscle mitochondria, and poor motor performance. Replication in Drosophila of the specific pathologies of human BTHS strongly suggests a causal link between these features. We propose that aberrant cardiolipin induces mitochondrial malformations, which in turn causes the myopathy. The data show that full-length tafazzin is essential for the formation of normal cardiolipin and that the shorter tafazzin species are unable to compensate for the lack of full-length tafazzin. We have yet to demonstrate whether or not short tafazzins are actually expressed as proteins from the corresponding transcripts. Given the low expression level of tafazzin, this endeavor will require careful analysis of concentrated subcellular fractions from Drosophila at different developmental stages.
Like BTHS patients, tafazzin-deficient Drosophila mutants were unable to form the dominant molecular species of cardiolipin. However, the dominant molecular species are not identical in humans and in Drosophila (17), suggesting that tafazzin plays a general role in cardiolipin remodeling, rather than a specific role in the formation of any particular cardiolipin species. In both humans and Drosophila, tafazzin deficiency led to (i) a diversification of the cardiolipin pattern, i.e., formation of multiple species at the expense of the dominant species, and (ii) a decline of the total cardiolipin content.
This specific form of cardiolipin deficiency was associated with abnormalities of the cristae membranes, which bear some resemblance to abnormalities caused by age, hyperoxia (20), or certain mutations (21, 22). In all of these examples, an atypical pattern of cristae architecture emerged, albeit with variations in shape and form. For instance, in tafazzin-deficient flight muscle, we found characteristic stacks of hyperdense, closely apposed membranes, which were deposited inside the mitochondria. A very similar morphology was observed in heart biopsies from BTHS patients (11). According to our electron tomographic studies, these hyperdense structures may represent collapsed cristae with obliterated intracrista space (D. Acehan, Y.X., D. L. Stokes, M.S., unpublished data).
Physiologic consequences of tafazzin deficiency are only beginning to emerge from clinical and experimental studies (23). In cell cultures, poor coupling between respiration and phosphorylation has been observed (19, 24), but a general breakdown of energy metabolism is unlikely to be the pathogenetic mechanism of BTHS, because the symptoms are often subtle and confined to selected organ systems. Deletion of full-length tafazzin from Drosophila did not alter lifespan or heart rate, and the mutants appeared to be in good overall health. However, the tafazzin mutants may become vulnerable once they are removed from the optimal culture environment and exposed to wildlife conditions. This proposition may be tested by lifespan studies under conditions of stress.
Tafazzin deletion caused motor weakness, strongly supporting the specific role of this protein in muscle function. Muscle mitochondria may be susceptible to tafazzin deletion because they require a high concentration of cardiolipin and because they may not tolerate deviations from the normal cardiolipin composition. For instance, mammalian skeletal muscle contains cardiolipin with a very specific molecular composition, mainly consisting of tetralinoleoyl species (7). The requirement for specific cardiolipins may be related to the structural organization, high density, or tight association with myofibrils, which is characteristic of muscle mitochondria. The selective effect on muscle function makes Drosophila an appropriate animal model of BTHS, the further exploration of which should yield insights into the mechanism by which tafazzin mutations cause myopathy.
Materials and Methods
Strains, Crossbreedings, and Mutant Selection.
The fly stock, y[1]; P{y[+mDint2] w[BR.E.BR] = SUPor-P}tafazzin[KG02529]/SM1; ry[506], with a P element insertion (KG02529) in the tafazzin gene (CG8766), was obtained from the Bloomington Drosophila Stock Center. By crossing this stock with the balancer stock w; Sco/CyO, we obtained a KG02529 stock in white background, w; P{y[+mDint2] w[BR.E.BR] = SUPor-P}tafazzin[KG02529]/CyO, which was used in the following protocol, designed to achieve imprecise excision of the P element. Briefly, w; P{y[+mDint2] w[BR.E.BR] = SUPor-P}tafazzin[KG02529]/CyO were crossed with w; Sp/CyO; Δ2–3,Sb/TM6,Ubx, and their male progeny with the genotype w/Y; P{y[+mDint2] w[BR.E.BR] = SUPor-P}tafazzin[KG02529]/CyO; Δ2–3,Sb/+ were then individually crossed with five virgin females of w;Sco/CyO. From each latter cross, a single white-eye male progeny with the genotype w/Y; ΔKG02529/CyO was collected to establish a balanced P element excision stock by crossing it with the balancer stock w; Sco/CyO. We screened 149 independent P element excision stocks for deletions in the tafazzin gene by PCR analysis using various primer combinations around the P insertion site. Deletions detected by PCR were further characterized by DNA sequencing. Three independent strains with large deletions within the tafazzin gene were identified in this screen and used as the study group (TAZ−/−). Strains with intact tafazzin gene because of precise excision of the P element were used as control group (TAZ+/+). To collect homozygous TAZ−/− animals in the larval stage, excision stocks were rebalanced with CyO, GFP, and nonfluorescent larvae were selected. We also generated flies that were heterozygous for the tafazzin deletion and Df(2R)vg-C, a deficiency of a chromosomal region that includes the tafazzin locus (Bloomington Drosophila Stock Center).
Transcript Analysis.
Adult flies were homogenized and total Drosophila RNA was isolated and purified by using the RNeasy Mini kit and RNase-free DNase I from Qiagen (Valencia, CA). First-strand cDNA was produced from 5 μg RNA in the presence of 250 units SuperScript reverse transcriptase (Invitrogen; Carlsbad, CA). PCR was performed with 2 μl aliquots of the first-strand cDNA mixture in the presence of 2.75 mM MgCl2, 200 nM primers, 300 nM deoxyribonucleoside triphosphates, 10 mM Tris (pH 8.3), and thermostable DNA polymerase from the Expand Long Template PCR System supplied by Roche (Indianapolis, IN). Samples were heated to 94°C for 2 min and then cycled 25 times through the following temperature protocol: 94°C for 0.5 min, 54°C for 0.5 min, and 68°C for 1.5 min. The forward primers included 5′-CAA ACA TCA GTT GGA TGT AAA TGG CAA AAA A-3′ for transcript RA, 5′-CAT TAT ACA CGG AAG CGC TCG G-3′ for transcript RB, and 5′-GGC GTT TGG CTA GAC ACA AGC-3′ for transcript RC. The reverse primer was 5′-CAG GAT CGT CGA AGC AGG AGT AG-3′ in all reactions.
Tafazzin Antibody.
Full-length Drosophila tafazzin cDNA was inserted in-frame into the EcoRI-XhoI site of the vector pGEX-EF (Invitrogen). The vector was expressed in BL-21 Escherichia coli cells to produce tafazzin fused to glutathione S-transferase (GST). The fusion protein was purified from inclusion bodies by SDS/PAGE. Rabbits were immunized with GST-tafazzin by using the standard protocol of Invitrogen. Polyclonal anti-tafazzin antibodies were purified from rabbit serum 10 weeks after immunization by incubation with nitrocellulose strips carrying a fusion protein of tafazzin and the maltose-binding protein (MBP). Antibodies were eluted from strips by gentle shaking in 0.2 M glycine (pH 2.5) and stored in a neutralized solution containing 0.15 M glycine, 0.25 M Tris (pH 7.5), and 0.02% NaN3 at 4°C. The MBP-tafazzin fusion protein was expressed in BL21 E. coli cells with the pMAL-c2 vector that had full-length tafazzin cDNA inserted at the EcoRI-SalI site. MBP-tafazzin was affinity-purified on amylase resin (New England BioLabs; Boston, MA), from which it eluted in the presence of 10 mM maltose.
Isolation of Subcellular Membranes and Western Blot Analysis.
Adult flies were homogenized with a Teflon-glass homogenizer in ice-cold buffer containing 280 mM sucrose, 10 mM Hepes (pH 7.4), 1 mM EDTA, and protease inhibitor mixture from Roche. The homogenate was filtered through cheesecloth and centrifuged at 750 × g for 10 min to remove nuclei and unbroken cells. The postnuclear supernatant was centrifuged at 17,000 × g for 10 min to pellet crude mitochondria. Protein concentration was determined by the method of Lowry et al. (25). Mitochondrial fractions were run on SDS/PAGE at a concentration of 0.2 mg per lane and subsequently transferred to a nitrocellulose membrane. Nitrocellulose membranes were incubated with affinity-purified anti-tafazzin antibodies (1:1000) overnight at 4°C, followed by incubation with goat anti-rabbit antibody conjugated with horseradish peroxidase (1:3500; Bio-Rad) for 2 h at room temperature. Antibodies were visualized on x-ray films after incubation with the SuperSignal West Pico chemiluminescent substrate (Pierce) for 5 min.
Lipid Analysis.
Lipids were extracted (26) from adult Drosophila homogenates and separated by two-dimensional TLC on silica gel 60 developed by chloroform/methanol/water (65/25/4) in the first direction and chloroform/acetone/methanol/acetic acid/water (50/20/10/10/5) in the second direction. Lipids were briefly stained with iodine vapor. Silica gel spots were scraped off and subjected to transmethylation in methanol/0.5 M hydrochloric acid at 90°C for 10 h. Fatty acid methyl esters were extracted into hexane and analyzed by capillary gas chromatography (27). Analysis of cardiolipin was performed by HPLC with fluorescence detection (28). To this end, cardiolipin was methylated, derivatized with naphthyl-1-acetic anhydride, and purified by solid-phase extraction (28). Tristearoyl-oleoyl-cardiolipin was added to all samples as an internal standard. The concentration of dipalmitoleoyl-dilinoleoyl-cardiolipin was measured by electrospray-ionization mass spectroscopy in connection with HPLC as described (17).
Electron Microscopy.
Thoraces of 1-week-old flies were bisected longitudinally, immediately placed in 2% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4), and fixed for 2 h at room temperature. Subsequent procedures included postfixation with 2% osmium tetraoxide, in-block staining with uranyl acetate, dehydration through a graded series of alcohol solutions, treatment with propylene oxide, and embedding in epoxy resin. Semithin sections (0.5–1 μm) were mounted on glass slides, stained with toluidine blue, and observed under a light microscope to identify dorsal indirect flight muscles and to determine their anatomic orientation in the block. Thin sections (100 nm) were collected on carbon-coated formvar grids, contrasted with uranyl acetate and lead citrate, and observed under a JEOL LEM1200EXII transmission electron microscope at 80 kV. Images were recorded at 5,000-fold and 12,000-fold magnification.
Physiological Assays.
All assays were conducted at 25°C. For the longevity assay, age-synchronized fly populations were transferred to new vials every other day. The number of dead flies was recorded at each transfer. Heart rate was measured in the early pupal stage, immediately after becoming immobile and before optical transparency was lost (29). Pupae were placed on glass slides in a drop of saline, and their hearts were viewed through an inverted microscope. Heart rate was counted in quadruples of 15-s intervals for each animal. Locomotor activity of third instar larvae was measured on 5.5-inch agar dishes placed on a grid. We measured the number of 0.2×0.2-inch squares crossed per minute for each larva in triplicates. Climbing and flying assays were performed with 1-week-old adults. Multiple batches of 30–60 flies were used. The ability to climb against gravity was measured by countercurrent distribution (30). Flies, placed at the bottom of a tube, were given 20 s to climb a distance of 4.5 inches into an inverted tube placed on top. Flies that successfully climbed into the top tube were given another chance to climb and so forth. Flies scored one point for every two tubes they climbed out of. The ability to fly was assayed on a 30×30-inch airfield. The distance an animal crossed in its first attempt to fly was measured after it was placed in the center of the field. Flies scored one point for each unit of 2.5 inches up to a maximal score of 6.
Statistical Analysis.
Groups were compared by unpaired or, where appropriate, nonparametric t tests. Survival curves were analyzed by the method of Kaplan and Meier. Statistical significance was accepted at P values <0.05. N indicates the number of animals per group. Both sexes were present in about equal proportion in all groups. TAZ+/+ and TAZ−/− groups contained flies from three independent clones in about equal proportions. Statistical data were computed with the software PRISM 4 (GraphPad, San Diego, CA).
Acknowledgments
This work was supported in part by grants from the Barth Syndrome Foundation, Inc. (to Y.X. and M.R.), American Heart Association Grant 0350126N (to M.S.), and National Institutes of Health Grant 1 R01 HL078788-01 (to M.S.).
Abbreviation
- BTHS
Barth syndrome.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Barth P. G., Wanders R. J. A., Vreken P., Janssen E. A. M., Lam J., Baas F. J. Inherited Metab. Dis. 1999;22:555–567. doi: 10.1023/a:1005568609936. [DOI] [PubMed] [Google Scholar]
- 2.Bione S., D’Adamo P., Maestrini E., Gedeon A. K., Bolhuis P. A., Toniolo D. Nat. Genet. 1996;12:385–389. doi: 10.1038/ng0496-385. [DOI] [PubMed] [Google Scholar]
- 3.Neuwald A. F. Curr. Biol. 1997;7:R465–R466. doi: 10.1016/s0960-9822(06)00237-5. [DOI] [PubMed] [Google Scholar]
- 4.Vreken P., Valianpour F., Nijtmans L. G., Grivell L. A., Plecko B., Wanders R. J. A., Barth P. G. Biochem. Biophys. Res. Commun. 2000;279:378–382. doi: 10.1006/bbrc.2000.3952. [DOI] [PubMed] [Google Scholar]
- 5.Schlame M., Kelley R. I., Feigenbaum A., Towbin J. A., Heerdt P. M., Schieble T., Wanders R. J. A., DiMauro S., Blanck T. J. J. J. Am. Coll. Cardiol. 2003;42:1994–1999. doi: 10.1016/j.jacc.2003.06.015. [DOI] [PubMed] [Google Scholar]
- 6.Schlame M., Rua D., Greenberg M. L. Prog. Lipid Res. 2000;39:257–288. doi: 10.1016/s0163-7827(00)00005-9. [DOI] [PubMed] [Google Scholar]
- 7.Schlame M., Towbin J. A., Heerdt P. M., Jehle R., DiMauro S., Blanck T. J. J. Ann. Neurol. 2002;51:634–637. doi: 10.1002/ana.10176. [DOI] [PubMed] [Google Scholar]
- 8.Valianpour F., Wanders R. J. A., Overmars H., Vreken P., van Gennip A. H., Baas F., Plecko B., Santer R., Becker K., Barth P. G. J. Pediatr. 2002;141:729–733. doi: 10.1067/mpd.2002.129174. [DOI] [PubMed] [Google Scholar]
- 9.Xu Y., Kelley R. I., Blanck T. J. J., Schlame M. J. Biol. Chem. 2003;278:51380–51385. doi: 10.1074/jbc.M307382200. [DOI] [PubMed] [Google Scholar]
- 10.Barth P. G., Scholte H. R., Berden J. A., Van der Klei-Van Moorsel J. M., Luyt-Houwen I. E. M., Van’t Veer-Korthof E. T., Van der Harten J. J., Sobotka-Plojhar M. A. J. Neurol. Sci. 1983;62:327–355. doi: 10.1016/0022-510x(83)90209-5. [DOI] [PubMed] [Google Scholar]
- 11.Bissler J. J., Tsoras M., Goring H. H. H., Hug P., Chuck G., Tombragel E., McGraw C., Schlotman J., Ralston M. A., Hug G. Lab. Invest. 2002;82:335–344. doi: 10.1038/labinvest.3780427. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez I. L. Am. J. Med. Genet. 2005;134A:409–414. doi: 10.1002/ajmg.a.30661. [DOI] [PubMed] [Google Scholar]
- 13.Lu B., Kehler M. R., Lee D. P., Lewin T. M., Coleman R. A., Choy P. C., Hatch G. M. Biochem. Cell Biol. 2004;82:569–576. doi: 10.1139/o04-055. [DOI] [PubMed] [Google Scholar]
- 14.Vaz F. M., Houtkooper R. H., Valianpour F., Barth P. G., Wanders R. J. A. J. Biol. Chem. 2003;278:43089–43094. doi: 10.1074/jbc.M305956200. [DOI] [PubMed] [Google Scholar]
- 15.Sohal R. S. Gerontology. 1976;22:317–333. doi: 10.1159/000212146. [DOI] [PubMed] [Google Scholar]
- 16.Grumbling G., Strelets V. FlyBase Consortium. Nucleic Acids Res. 2006;34:D484–D488. doi: 10.1093/nar/gkj068. Available at http://flybase.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schlame M., Ren M., Xu Y., Greenberg M. L., Haller I. Chem. Phys. Lipids. 2005;138:38–49. doi: 10.1016/j.chemphyslip.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 18.Gu Z., Valianpour F., Chen S., Vaz F. M., Hakkaart G. A., Wanders R. J. A., Greenberg M. L. Mol. Microbiol. 2004;51:149–158. doi: 10.1046/j.1365-2958.2003.03802.x. [DOI] [PubMed] [Google Scholar]
- 19.Xu Y., Sutachan J. J., Plesken H., Kelley R. I., Schlame M. Lab. Invest. 2005;85:823–830. doi: 10.1038/labinvest.3700274. [DOI] [PubMed] [Google Scholar]
- 20.Walker D. W., Benzer S. Proc. Natl. Acad. Sci. USA. 2004;101:10290–10295. doi: 10.1073/pnas.0403767101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paumard P., Vaillier J., Coulary B., Schaeffer J., Soubannier V., Mueller D. M., Brethes D., di Rago J.-P., Velours J. EMBO J. 2002;21:221–230. doi: 10.1093/emboj/21.3.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.John G. B., Shang Y., Li L., Renken C., Mannella C. A., Selker J. M. L., Rangell L., Bennett M. J., Zha J. Mol. Biol. Cell. 2005;16:1543–1554. doi: 10.1091/mbc.E04-08-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barth P. G., Valianpour F., Bowen V. M., Lam J., Duran M., Vaz F. M., Wanders R. J. A. Am. J. Med. Genet. 2004;126A:349–354. doi: 10.1002/ajmg.a.20660. [DOI] [PubMed] [Google Scholar]
- 24.Ma L., Vaz F. M., Gu Z., Wanders R. J. A., Greenberg M. L. J. Biol. Chem. 2004;279:44394–44399. doi: 10.1074/jbc.M405479200. [DOI] [PubMed] [Google Scholar]
- 25.Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 26.Bligh E. G., Dyer W. J. Can. J. Biochem. Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 27.Patton G. M., Cann S., Brunnengraber H., Lowenstein J. M. Methods Enzymol. 1981;72:8–20. doi: 10.1016/s0076-6879(81)72004-4. [DOI] [PubMed] [Google Scholar]
- 28.Schlame M., Shanske S., Doty S., Konig T., Sculco T., DiMauro S., Blanck T. J. J. J. Lipid Res. 1999;40:1585–1592. [PubMed] [Google Scholar]
- 29.Wessells R. J., Bodmer R. BioTechniques. 2004;37:58–66. doi: 10.2144/04371ST01. [DOI] [PubMed] [Google Scholar]
- 30.Benzer S. Proc. Natl. Acad. Sci. USA. 1967;58:1112–1119. doi: 10.1073/pnas.58.3.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]