Abstract
Classical cadherin-mediated interactions between axons and dendrites are critical to target selection and synapse assembly. However, the molecular mechanisms by which these interactions are controlled are incompletely understood. In the Drosophila visual system, N-cadherin is required in both photoreceptor (R cell) axons and their targets to mediate stabilizing interactions required for R cell target selection. Here we identify the scaffolding protein Liprin-α as a critical component in this process. We isolated mutations in Liprin-α in a genetic screen for mutations affecting the pattern of synaptic connections made by R1–R6 photoreceptors. Using eye-specific mosaics, we demonstrate a previously undescribed, axonal function for Liprin-α in target selection: Liprin-α is required to be cell-autonomous in all subtypes of R1–R6 cells for their axons to reach their targets. Because Liprin-α, the receptor tyrosine phosphatase LAR, and N-cadherin share qualitatively similar mutant phenotypes in R1–R6 cells and are coexpressed in R cells and their synaptic targets, we infer that these three genes act at the same step in the targeting process. However, unlike N-cadherin, neither Liprin-α nor LAR is required postsynaptically for R cells to project to their correct targets. Thus, these two proteins, unlike N-cadherin, are functionally asymmetric between axons and dendrites. We propose that the adhesive mechanisms that link pre- and postsynaptic cells before synapse formation may be differentially regulated in these two compartments.
Keywords: axon, synapse formation, cadherin, cell adhesion
Developing axons make specific choices amongst alternate synaptic targets before initiating the assembly of pre- and postsynaptic components. Although a number of molecules involved in target selection and synapse assembly have been identified, fundamental questions remain regarding how these processes are coordinated. What are the molecular relationships between target selection and synapse formation? Here we demonstrate that Liprin-α, a regulator of synapse structure, also functions during target selection.
Liprin-α was initially identified through its biochemical interactions with the receptor protein tyrosine phosphatase LAR (1, 2). Further in vitro studies demonstrated that Liprin-α acts postsynaptically to regulate α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor insertion into dendrite membranes and presynaptically to form synaptic active zones (3–6). In Drosophila, both LAR and Liprin-α regulate synaptic bouton growth at the larval neuromuscular junction, suggesting that Liprin-α is required for LAR function (7). This work also demonstrated that these proteins can act independently, because LAR, but not Liprin-α, mutants display defects in axon guidance (7, 8). Together, these studies argue that Liprin-α acts as an evolutionarily conserved synaptic scaffold associated with LAR.
Are the functions of Liprin-α in synapse assembly linked to the earlier process of choosing a synaptic partner? In Drosophila, the classical cadherin N-cadherin plays a central role in axon targeting in many neurons (9, 10). In the visual system, N-cadherin regulates target specificity by stabilizing connections between photoreceptor axons and their postsynaptic targets (11–15). Axons from six photoreceptors, designated R1–R6, elaborate a complex, precise set of synaptic connections within a single optic ganglion, the lamina (16). Axons from two other R cell types, R7 and R8, terminate within distinct layers of a second optic ganglion, the medulla. Both patterns of connections require N-cadherin. Although R1–R6 axons lacking N-cadherin correctly reach the lamina, once there, they fail to make a short extension toward their synaptic target (11, 12). Similarly, R7 axons mutant for N-cadherin reach the medulla but terminate inappropriately in the layer normally associated with R8 (11, 13, 14).
Interestingly, R cells lacking LAR function display defects in target selection similar to those seen in N-cadherin mutations (17, 18). In particular, R1–R6 cells mutant for LAR fail to extend to their targets in the lamina, and R7 cells mistarget to the R8 layer in the medulla. In R7 cells, further developmental analysis reveals that this phenotypic similarity emerges in two steps: an initial process of axon extension to a temporary set of targets that depends on N-cadherin and a later process of stabilization that depends on both N-cadherin and LAR (13). Intriguingly, in culture, protein tyrosine phosphatases including LAR can modulate N-cadherin adhesive function and form complexes with cadherins (19–26).
Here we identify mutations in Liprin-α that disrupt the same process of R1–R6 target stabilization affected by mutations in N-cadherin and LAR. Using mosaic studies in single cells, we demonstrate that Liprin-α and LAR, unlike N-cadherin, are required presynaptically, not postsynaptically. We propose that in R1–R6 axons, Liprin-α and LAR work together with N-cadherin to mediate adhesive events between pre- and postsynaptic cells and that LAR and Liprin-α are not required postsynaptically for this adhesive interaction to occur.
Results
The Identification of Mutations in Liprin-α.
To identify genes involved in R cell target selection, we had undertaken a behavioral screen for mutations that caused defects in the optomotor response in eye-specific somatic mosaic animals (18). In this screen, photoreceptor cells were rendered homozygous for chromosomes of interest, whereas the rest of the fly was heterozygous and, presumably, phenotypically wild type. This behavioral response depends on R1–R6 function, and we surmised that a subset of mutations isolated in this way should affect photoreceptor connectivity. Histological analysis of this mutant collection by using an R cell-specific marker identified four additional mutations affecting R cell target selection on the left arm of chromosome 2. One of these four, designated Liprin-α1, was of particular interest.
To identify additional alleles of this locus, we undertook a lethal noncomplementation screen by using chemical mutagenesis. From ≈1,800 F1 lines, we identified two mutations that failed to complement the recessive lethal phenotype associated with our original mutation. These previously undescribed alleles were designated Liprin-αE and Liprin-αF. Three lines of evidence demonstrated that all three of our mutations affect Liprin-α. First, single-nucleotide polymorphism-based mapping identified a small region of chromosome 2L, including the Liprin-α locus, containing the lesion responsible for the cartridge phenotype observed in Liprin-α1. Second, DNA sequence analysis of all three alleles revealed premature stop codons in the Liprin-α coding sequence (Fig. 6, which is published as supporting information on the PNAS web site). Third, a previously described Liprin-α mutation, Liprin-αR60, caused indistinguishable targeting phenotypes in R cell axons.
Liprin-α Is Required for R Cell Target Selection.
All of our Liprin-α mutations displayed a specific pattern of disruptions in the structure of the cartridge, the synaptic unit in the lamina. In WT animals, axons from R1–R6 photoreceptors form regularly arrayed clusters of six axons. Using a marker specifically expressed in R1–R6 cells, cross-sectional views of the lamina revealed an array of circles surrounding the unlabeled processes of lamina neurons (Fig. 1A). R7 and R8 axons, labeled with an R cell specific marker, formed small profiles located in one corner of each cluster. In all of our Liprin-α alleles, as well as in the previously identified Liprin-αR60, this unit structure was broken: Some cartridges have either >6 or <6 R cell axons, and some adjacent cartridges fuse (Fig. 1 B–D). Similar defects were seen in R cells homozygous for N-cadherinΔ14 and LAR2127 (Fig. 1 E and F; refs. 10 and 14).
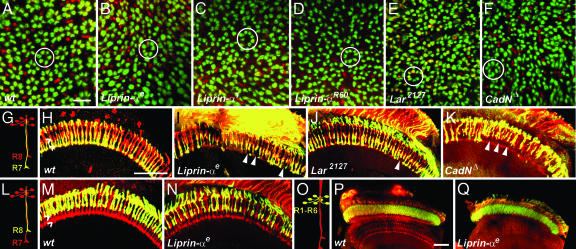
Fig. 1.
Liprin-α is required for both cartridge assembly and layer-specific targeting in the visual system and displays phenotypes indistinguishable from those associated with mutations in LAR and N-cadherin. (A–F) Cross-sectional views of the lamina. R1–R6 axons express LacZ under the control of the Rh1 promoter (green); all R cells are counterstained with the R cell-specific antibody mAb24B10 (red). (A) WT R1–R6 axons assemble into fascicles, denoted cartridges (circled), containing 6 R cell axons. R7 and R8 sit outside of each cartridge. (B–D) Liprin-α somatic mosaic animals in which photoreceptor axons are homozygous mutant. Individual cartridges are of unequal size and contain variable numbers of R1–R6 termini. (E) Lar2127. (F) N-cadherinΔ14. The phenotypes observed in E and F are indistinguishable from those seen in B–D. (G–N) Horizontal section of the medulla in eye-specific mosaic adult flies. (G–K) R7 axons express lacZ under the control of the Rh3 promoter (green); all R cell axons are counterstained with mAb24B10 (red). Note that not all R7 cells express Rh3lacZ; a subset express the R4 opsin and, thus, are labeled only with mAb24B10 (red). (G and H) WT. R7 axons invariably stop in a layer more proximal than R8; hash marks denote each layer. (I) Liprin-αE. (J) Lar2127. (K) N-cadherinΔ14. In these three mutant backgrounds, R7 axons sometimes stop in the R8 recipient layer instead of the R7 recipient layer (arrowheads), leaving gaps in the array of otherwise regular R7 termini. (L–N) R8 axons are labeled with lacZ expressed under the control of the Rh5 promoter (green); all R cell axons are stained with mAb24B10 (red). Note that not all R8 axons express Rh5lacZ; some normally express Rh6 and, thus, are labeled only with mAB24B10 (red). (L and M) WT. (N) Liprin-αE. Layer-specific targeting of R8 is unaffected by the loss of Liprin-α. (O–Q) R1–R6 axons are labeled with lacZ under the control of the Rh1 promoter; all R cell axons are stained with mAb24B10 (red). (O and P) WT. (Q) Liprin-αE. The ganglion-specific targeting of R1–R6 axons to the lamina occurs normally in Liprin-α mutants. (Scale bars: A–F, 5 μm; G–N, 30 μm.)
To identify additional phenotypes in Liprin-α mutants, we used markers that specifically label either R7 or R8 to examine targeting of these cells in eye-specific mosaics. In WT animals, R7 and R8 innervated distinct layers in the outer medulla (Fig. 1 G and H). In Liprin-α mutants, R7 axons frequently stopped at abnormally distal positions within the R8 recipient layer (Fig. 1I). These phenotypes were qualitatively indistinguishable from those associated with mutations in N-cadherin and LAR (Fig. 1 J and K). We note, however, that mutations in Liprin-α and LAR cause R7 targeting phenotypes that were somewhat lower in expressivity than those associated with N-cadherin mutants. None of these three mutants affected the ganglion-specific targeting of R1–R6 axons to the lamina nor did they affect the layer-specific targeting of R8 axons within the medulla (Fig. 1 L–Q).
These defects did not reflect errors in cell fate specification early in eye development, nor were they the result of defects in the guidance of R cell axons to the target field. In particular, in Liprin-α eye-specific mosaics, R cells proliferated normally during the third instar larval stage and displayed normal morphological differentiation during pupal and adult stages (Fig. 2A and B; data not shown). Moreover, Liprin-α mutant R cell axons selected appropriate ganglion-specific targets in the lamina and the medulla and induced appropriate differentiation of the neurons in the lamina target field, as assessed by using the antibodies directed against the pan-neural protein Elav, and the nuclear L5 marker BSH (Fig. 2 C and D). These axons also elaborated topographically appropriate maps in each region (Fig. 2 E and F), and glial cell differentiation in these areas was largely normal (Fig. 2 G and H).
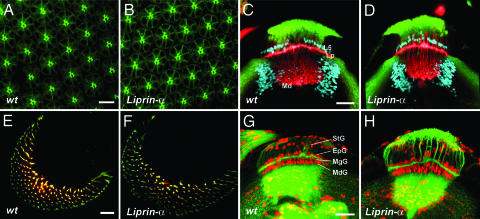
Fig. 2.
R cell fate determination, axon guidance into the brain, and brain development are normal in Liprin-α mutants. (A and B) Cross-sections of the retina visualized during mid-pupal development, labeling R cells with the R cell-specific antibody mAb24B10 (green). (A) WT. (B) Liprin-α1. R cell shape and rhabdomere morphology are unaffected by the loss of Liprin-α. (C and D) Horizontal views of the brain during the third instar larval stage. R cell axons are visualized with mAb24B10 (red); neuronal nuclei are marked with antibodies directed against Elav (green); L5 lamina neurons and some medullar neurons are labeled with antibodies directed against the Brain-Specific Homeobox (blue). Lp, lamina plexus; Md, medulla. (C) WT. (D) Liprin-α1. (E and F) En face views of R cell axons entering the lamina; R cell axons are labeled with mAb24B10 (red); R2-R5 axons express tau-lacZ under the control of the rough promoter (green). (G and H) Horizontal views of the brain during the third instar larval stage. Neuronal processes are labeled with anti-horseradish peroxidase-FITC (green); glial nuclei are stained with an antibody directed against Repo (red). StG, satellite glia; MgG, marginal glia; EpG, epithelial glia; MdG, medulla glia. (G) WT. (H) Liprin-α1. (Scale bar: A and B, 10 μm; C–F, 20 μm; G and H, 30 μm.)
Taken together, these studies demonstrate that Liprin-α function is required in R cell axons for normal target selection. The extensive phenotypic similarities observed between Liprin-α, LAR, and N-cadherin mutant R cells suggest that these three genes control a common process in these developing axons.
Liprin-α Is Expressed in R Cell Axons and Their Targets.
To determine when and where Liprin-α is expressed, we stained developing optic lobes with antibodies directed against Liprin-α at multiple developmental stages (Fig. 7 A–D, which is published as supporting information on the PNAS web site). During the third larval stage, Liprin-α was broadly expressed in retinal precursors, differentiating R cells, lamina neurons, and most other cells in the visual system (Fig. 7A). During early pupal development, when R7 axons make their initial extensions into the medulla, strong Liprin-α staining was observed in the medulla and lamina neuropil (Fig. 7B). This staining remained strong within the lamina plexus during mid-pupal development, when R1–R6 axons select targets (Fig. 7 C and D). This expression of Liprin-α substantially overlapped with the expression of LAR and N-cadherin (Fig. 7 E–H). These localization studies demonstrate that Liprin-α, LAR, and N-cadherin are expressed in the right time and place to function together in regulating axon extension.
Liprin-α Acts Before Synapse Formation in R Cell Axons.
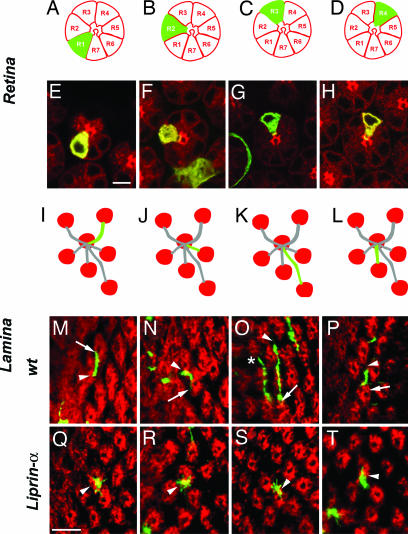
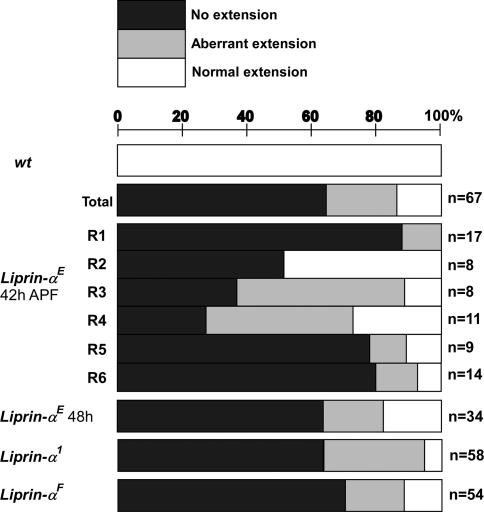
Because R1–R6 axon extension takes place in a single, morphologically defined step (unlike the two-step targeting process described for R7 axons) and has been shown to depend on N-cadherin-mediated interactions between pre- and postsynaptic cells, we focused our developmental analysis of Liprin-α function on R1–R6 cells. To determine whether the defects we observed in Liprin-α mutants reflected cell-autonomous requirements in these cells, we generated single-cell mutant clones by using mosaic analysis with a repressible cell marker (MARCM) method (12, 26). In this experiment, single-mutant R cells in an otherwise WT animal were visualized during mid-pupal development. WT R1–R6 axons from the same ommatidium extended into the brain as part of a fascicle. Upon reaching the lamina plexus, each axon defasciculated, and extended across the surface of the lamina to innervate a single target. Each R cell subtype made a characteristic projection that grew in a stereotyped direction and was invariant in shape and length (Fig. 3 A–P). Synapses assemble between R cell axons and their targets after this final projection has formed (16). The behavior of Liprin-α mutant axons was indistinguishable from wild type along their trajectories into the lamina plexus, with each axon remaining tightly associated with the axon bundle of its WT neighbors from the same ommatidium. However, once within the lamina plexus, unlike WT R cells, Liprin-α mutant R cells typically displayed specific defects in axon extension toward their targets (Fig. 3 Q–T). We observed two types of defects (Fig. 4). In particular, 64% (n = 67) of Liprin-αE mutant axons completely failed to extend away from the ommatidial bundle, whereas 21% (n = 67) made weak, morphologically abnormal extensions; the remaining axons extended normally. All R cell subtypes were equally affected, and each of our Liprin-α alleles caused quantitatively comparable phenotypic effects. This latter observation is particularly striking in the case of Liprin-αF, where only a small portion of the C terminus is removed. Because this mutant protein is expressed at normal levels (data not shown) and because this portion of vertebrate Liprin-α mediates binding to many proteins including LAR, one or more of these interactions are likely to be critical to Liprin-α function R cells. The defects we observed did not reflect simple developmental delays in axon extension because both our analysis of R1–R6 targeting in adult animals (Fig. 1) and single-cell MARCM at a slightly later developmental stage (Fig. 4) revealed comparably expressive phenotypes. These results demonstrate that Liprin-α activity is required cell-autonomously in R cell axons, acting before synapse formation, to allow axons to extend to their targets. These defects are qualitatively indistinguishable from, and quantitatively similar to, those associated with single-cell LAR and N-cadherin mutant clones (refs. 12 and 18; data not shown). Thus Liprin-α function is required cell-autonomously in developing R cells and is necessary for R cell axons to extend to appropriate targets. These results demonstrate that the phenotypic similarities between Liprin-α, LAR, and N-cadherin mutants extend to the single-cell level.
Fig. 3.
Liprin-α is required cell autonomously in R1–R6 axons for target selection in the lamina. Single-mutant R cells were generated by mitotic recombination and visualized at 42 h after puparium formation to capture axonal extension. R cell clones were labeled with GFP (green), and cartridges were visualized by staining all R cells with mAb24B10 (red). (A–D) Schematic cross-sectional views of the retina. Each R cell type is identifiable by its characteristic morphology and position. (E–H) Cross-sectional images of the retina. (I–L) Schematic representation of the projection of each R cell subtype. Each R cell axon makes a projection that is invariant in morphology, and which projects in an orientation that can be predicted from the position of its cell body within the ommatidium. Each R cell extends from one cartridge (arrowhead) to a neighboring cartridge (arrow). (M–P) WT. (Q–T) Liprin-αE. Most Liprin-α mutant R cell growth cones fail to extend across the surface of the lamina. (A, E, I, M, and Q) R1. (B, F, J, N, and R) R2. (C, G, K, O, and S) R3. The asterisk in o denotes an R4 axon from a neighboring ommatidium (which extends normally). (D, H, L, P, and T) R4. (Scale bars: 5 μm.)
Fig. 4.
Liprin-α function is required in all R cell subtypes. Each axonal projection was categorized as absent (no extension, black bars), present but abnormal in morphology (aberrant, gray bars), or normal (white bars). n, number of fibers of each type examined; the lower three graphs (Liprin-αE, Liprin-α1, and Liprin-αF) combine data across all R cell subtypes. These data demonstrate that Liprin-α function is critical to R cell axon extension and that this requirement is not restricted to particular R cell subtypes.
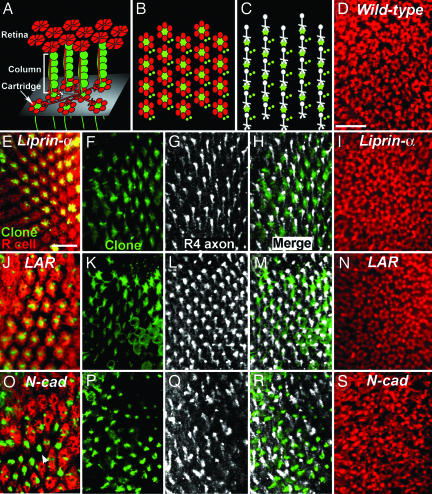
Liprin-α and LAR Are Not Required in Lamina Neurons for R Cell Target Selection.
R1–R6 cell axons extend through an environment composed of the processes of lamina neurons, glia, and other R cells. Each R cell axon selects a single group of five lamina neurons, designated a column, as its target (Fig. 5A). Once they reach their target, R cell axons and lamina neuron processes form a highly organized fascicle, called a cartridge, in which R cell axons surround, and are flanked by, the processes of the lamina neurons (Fig. 5B). N-cadherin is required both in R cells and lamina neurons for R cell axon extension: Columns that lack all N-cadherin activity typically fail to receive innervation (12). Because R cells mutant for LAR and Liprin-α displayed phenotypes similar to those associated with N-cadherin mutations and because these genes were expressed both pre- and postsynaptically, we examined whether Liprin-α and LAR are required in lamina neurons for R cell axon extension. Using the MARCM method, we generated large clones of mutant lamina neurons innervated by heterozygous R cell axons. Within each clone, a variable number of lamina neurons in each column were made homozygous mutant: Most columns contained mixtures of mutant and WT cells; some columns were completely mutant. Using R cell-specific markers, we then scored whether these lamina neuron clones caused defects in R cell target selection (Fig. 5). In animals in which R cell targeting occurs normally, cross-sectional views of the lamina revealed a highly regular pattern of cartridges comprising intermingled R cell axons and lamina neuron processes (Fig. 5 A and B). In 13 of 13 patches in which lamina neurons were made homozygous for Liprin-α1, including >300 columns containing mutant cells, we invariably observed normal spacing and structure of each cartridge (Fig. 5E). Similar results were obtained in 11 of 11 patches, including >200 columns, in which groups of lamina neurons were made homozygous for LARomb451 (Fig. 5J). By contrast, in six of six patches (>100 columns) made homozygous for N-cadherinΔ14, we observed that the pattern of cartridges was highly disrupted (Fig. 5O). To extend this analysis to the single-cell level, we incorporated a reporter construct that specifically labels R4 axons into our MARCM scheme and examined the projections of single WT R4 axons into and away from these mutant target columns. R4 axons that target normally form regular arrays of parallel extensions within the lamina plexus (Fig. 5C). In Liprin-α and LAR clones, the projections of R4 are invariably unaffected by the presence of mutant target cells (Fig. 5 F–H and K–M, respectively). By contrast, in N-cadherin mutant target clones, the array of R4 axons appears disrupted, with many axons failing to extend and others targeting inappropriately (Fig. 5 P–R). To confirm that these apparent differences in cellular requirement do not reflect differences in expressivity between Liprin-α, LAR, and N-cadherin, we used the “ELF” system to generate target clones in which most target neurons are rendered homozygous mutant (27) and visualized the lamina of adult flies by using an R cell-specific marker (Fig. 5 D, I, N, and S). Using this method, in animals in which target neurons are homozygous for a control chromosome, R cell targeting errors are observed much <1% of the time (Fig. 5D; n = 11 lobes, ≈480 cartridges, corresponding to 2,880 R cell-targeting events). In animals in which target neurons are homozygous for Liprin-α or LAR, R cell targeting events are similarly infrequent (Fig. 5 I and N; n = 15 lobes, 580 cartridges, 3,480 R cell targeting events for Liprin-α; n = 8 lobes, 440 cartridges, and 2,640 R cell targeting events for LAR). By contrast, in animals in which target neurons are made homozygous for N-cadherin, the cartridge array is severely disrupted, reflecting frequent errors in R cell targeting and preventing estimation of the number of cartridges scored (Fig. 5S; n = 9 lobes). Together, these results demonstrate that Liprin-α and LAR, unlike N-cadherin, are not required in postsynaptic neurons for R cell target selection.
Fig. 5.
Liprin-α and LAR, unlike N-cadherin, are not required in lamina neurons for normal R cell target selection. (A) Schematic view of the retina and lamina. R cells (red) extend axons out of the retina in fascicles before choosing targets in the lamina. Each fascicle is associated with a column of five lamina neurons (green) and contains all eight R cells from the same ommatidium. Within the lamina plexus (gray plane), R1–R6 axons extend laterally across the surface, choosing targets arranged in invariant relative positions. The R4 axon is highlighted (white). Upon reaching their target, each R cell axon joins a new fascicle, the cartridge, comprising R cell axon termini surrounding lamina neuron processes. (B) Schematic cross-sectional views of lamina cartridges. R1–R6 (red) surround, and are flanked by, lamina neuron processes (green). (C) Schematic cross-sectional view illustrating R4 axon projections (white) within a group of lamina neuron processes (green). R4 axons extend downward from one column (white dot), terminating in a growth cone adjacent to a second column (green). (D, I, N, and S) Single cross-sections of the adult lamina in which target neurons are homozygous for a control chromosome, and R cell axons termini are labeled red. (E–I) Liprin-α1. (J–N) LARomb451. (O–S) N-cadherinΔ14. (E, J, and O) Single cross-sections of the lamina plexus. Large clones of mutant lamina neurons labeled with GFP (green) were generated by using the MARCM system; photoreceptor axons were stained by using mAb24B10 (red). In this section plane, lamina neuron processes are visible as small profiles, whereas clusters of R cell axon termini label each cartridge. In Liprin-α and LAR mutant target clones, as in wild type, the overall spacing of cartridges is regular, and each cartridge contains R cell axon termini closely associated with lamina neuron processes. In N-cadherin clones, the spacing of cartridges is disrupted, and many lamina neuron processes lack associated R cell axons (arrowhead). (F–H, K–M, and P–R) Cross-sections of the lamina plexus within large target cell clones in which single R4 axons were labeled with mδ-lacZ. (F, K, and P) Target clone (green). (G, L, and Q) mδ-lacZ (white). (H, M, and R) Merge. In Liprin-α and LAR mutant clones, extended R4 axons form parallel rows of fibers both inside and outside the clone. In N-cadherin mutant clones, R4 axons frequently fail to extend or extend to inappropriate targets. (Scale bars: 10 μm.)
Discussion
Liprin-α is required for R cell axons to reach their postsynaptic targets. In the lamina, Liprin-α function is cell-autonomous to each R1–R6 cell subtype and is required before synapse formation. This phenotype is substantially identical to phenotypes described for N-cadherin and LAR in these cells (11–15, 17–18). Expression studies reveal that these three genes are expressed in largely overlapping patterns. These extensive similarities suggest that these genes act at the same step in the target selection process in R1–R6 axons. However, further somatic mosaic analysis revealed a critical distinction amongst the functions of these genes. That is, whereas N-cadherin is required both pre- and postsynaptically, Liprin-α and LAR are required only in R1–R6 cell axons, not their targets. Because work in other systems has demonstrated that Liprin-α, LAR, and N-cadherin form a complex (28) and that LAR can regulate the critical cadherin effector, β-catenin (28, 29), we speculate that homophillic, N-cadherin-mediated adhesive interactions might be differentially regulated between pre- and postsynaptic cells.
Defining a Previously Undescribed Function for Liprin-α.
Previous studies of Liprin-α have demonstrated that it functions as a key regulator of active zone structure and synaptic function (3–5, 7). Indeed, Liprin-α mutations cause significant defects in the size, structure, and physiology of synaptic boutons and defects in active zone size and the localization of synaptic vesicle components (5, 7). Intriguingly, in these studies, axonal innervation of the postsynaptic target was completely normal. Our work has demonstrated that this observation is not true in the developing visual system: Photoreceptors lacking Liprin-α function frequently fail to reach their appropriate postsynaptic targets. Because ultrastructural analysis of the development of this system reveals that synapses do not form until well after photoreceptor axons have reached their terminal target (16), our studies have defined a previously undescribed function for Liprin-α in target selection.
What does Liprin-α do in this context? Our studies demonstrating that R1–R6 cells mutant for Liprin-α, LAR, or N-cadherin display identical axonal phenotypes, both in adult animals and during development, argue that these genes act in the same process during target selection. These genes also are required for the layer-specific targeting of R7 axons (Fig. 2; refs. 11, 14, and 18). In this context, our results are consistent with all three genes acting together during the second step of R7 layer-specific targeting; during the first step of the R7-targeting process, N-cadherin acts independently of LAR (and presumably of Liprin-α) (13). Extensive evidence in other systems suggests biochemical and regulatory interactions between LAR and N-cadherin and between Liprin-α and LAR. Recent studies have proposed that LAR, N-cadherin, and Liprin-α are cotransported to the postsynaptic densities of excitatory synapses in adult brains and that LAR phosphatase activity regulates membrane insertion of this complex in dendrites (28). In addition, LAR associates directly with β-catenin and can influence its phosphorylation in vitro (28, 29). Moreover, protein tyrosine phosphatase activity can modulate cadherin-dependent neurite outgrowth in culture (20). Taken together, we speculate that Liprin-α and LAR act as regulators of N-cadherin-mediated adhesion in R1–R6 cell axons.
A critical, very early step in R1–R6 target selection is a homophillic, N-cadherin-mediated interaction between R cells and their presumptive targets that occurs before the ultimate choice of synaptic partner (12). One possibility is that Liprin-α acts before N-cadherin during the target selection process to control the trafficking of molecules necessary for R cell axons to stably contact their targets. Such a view would be conceptually consistent with the previously described role for Liprin-α as a regulator of axonal trafficking (30). Indeed, N-cadherin itself or one of its effectors would be likely candidates. However, inconsistent with this notion, we have been unable to detect gross changes in the levels or localization of N-cadherin, β-catenin, or LAR in Liprin-α mutant R cell growth cones (K-M.C., S.P., and T.R.C., data not shown). The alternative model is that Liprin-α acts after N-cadherin, recruiting additional components to the presynaptic terminal that are involved in initiating active zone assembly and maintaining contact between pre- and postsynaptic cells. Here, the formation of N-cadherin-mediated adhesive interactions between R cell axons and their targets would alter the activity of Liprin-α at the future synapse, affecting the trafficking of synaptic vesicle components in the region. Such a notion also is consistent with the observed biochemical interactions in mammalian cells between Liprin-α and other presynaptic components, as well as genetic studies demonstrating that Liprin-α is required for active zone assembly and recruitment of synaptic vesicle components (5, 7, 30, 31). Broadly speaking, a role for Liprin-α downstream of adhesion molecules involved in target selection raises the possibility that, in many contexts, Liprin-α may directly link the process of choosing a synaptic partner to synapse assembly.
Liprin-α and LAR as Asymmetric Regulators of R Cell Target Selection.
Cadherin function has been studied extensively in the context of symmetric interactions between epithelial cells, and models derived from these studies have been applied to interactions between neurons. In this context, our work raises the possibility that cadherin function might be asymmetrically regulated between axons and dendrites. In particular, our experiments demonstrate that the mutant phenotypes associated with the loss of Liprin-α, LAR, or N-cadherin from R1–R6 cell axons are indistinguishable. However, although N-cadherin also is required postsynaptically, Liprin-α and LAR are not, demonstrating that the relative contributions of each component differ in R1–R6 cells and their targets. These results suggest that the molecular mechanisms that stabilize connections between R cell axons and their targets differ pre- and postsynaptically. Given that N-cadherin is a critical component on both sides of this interaction, and that LAR, in other contexts, has been shown to influence N-cadherin adhesivity, we speculate that these differences may be reflected in how cadherin-mediated adhesion complexes are used or regulated in axons and dendrites.
Materials and Methods
Genetics.
Eye-specific mosaic flies were generated by using ey3.5FLP; FRT40cycEAR95/GlaBc in which the FLP recombinase was expressed under control of a retina-specific eyeless promotor fragment, and twinspots were eliminated by the recessive cell lethal cycE mutation (19, 27). MARCM analyses on R1–R6 and lamina target neurons were performed as described by using the elavGAL4 transgene to drive expression of mCD8GFP (12). In particular, both R cell and target clone mosaics were generated by using a heat-shock inducible FLP recombinase, following a protocol in which animals were heat shocked (at 37°C for 30 min) either during the early third larval stage (to generate R cell clones), or during the second instar stage (to generate target cell clones). Target clones also were generated by using the “ELF” system as described in ref. 27. The lethal noncomplementation screen to identify additional Liprin alleles was performed by using ethylmethane sulfonate treatment under standard conditions (32).
Immunohistochemistry.
Fly brains were dissected, fixed in 2% paraformaldehyde, and stained as described in ref. 18. Rabbit anti-Liprin-α was used at 1:125 dilution (7). Mouse mAb24B10 [Developmental Studies Hybridoma Bank (DSHB) at the University of Iowa, Iowa City, IA] was used at 1:50 to stain photoreceptor neurons. Goat anti-HRP FITC (Jackson ImmunoResearch) was used at 1:100 to stain neuronal processes. Rat anti-elav antibody (DSHB) was used at 1:100 to stain neuronal nuclei. Mouse anti-repo antibody (DSHB) was used at 1:100 to stain differentiated glial cells. Guinea pig anti-Bsh antibody was used at 1:500 to stain L5 during pupal development. Mouse IgG2a anti-LacZ (Promega) and rabbit anti-GFP (Molecular Probes) were used at 1:100. The secondary antibodies, goat anti-mouse, rat, or rabbit IgG coupled to Alexa488, Cy3, Alexa594, or Cy5 (Molecular Probes), were used at 1:100. Images were collected on a Leica TCS SP2 AOBS, deconvolved by using Huygens Pro (Scientific Volume Imaging) and visualized by using Imaris (Bitplane).
Supplementary Material
Acknowledgments
We thank K. Hofmeyer and J. E. Treisman for communicating their results to us before publication; R. Heisinger (Baylor College of Medicine, Houston, TX) for providing us with critical antibody stocks; and D. Van Vactor (Harvard Medical School, Boston, MA), B. J. Dickson (Research Institute of Molecular Pathology, Vienna, Austria), T. Suzuki (Max Planck Institute of Neurobiology, Martinsreid, Germany), A. H. Brand (Cambridge University, Cambridge, U.K.), C. A. Brennan (Emory University, Atlanta, GA), M. F. Wernet (Stanford University, Stanford, CA), and the Bloomington Stock Center (Bloomington, IN) for giving us Drosophila stocks necessary to complete this work. We received valuable technical advice from H. Lee, and we thank J. D. Mast for his initial characterization of the pseudopupils in our behavioral mutants. Finally, we thank members of the T.R.C. laboratory for helpful discussions regarding this work. This work was supported in part by National Institutes of Health Grant R01 EY015231-01A1 (to T.R.C.). T.R.C. is a Sloan Fellow, a Searle Scholar, and a recipient of a Burroughs–Wellcome Career Development Award.
Abbreviation
- MARCM
mosaic analysis with a repressible cell marker.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Serra-Pages C., Kedersha N. L., Fazikas L., Medley Q., Debant A., Streuli M. EMBO J. 1995;14:2827–2838. doi: 10.1002/j.1460-2075.1995.tb07282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Serra-Pages C., Medley Q. G., Tang M., Hart A., Streuli M. J. Biol. Chem. 1998;273:15611–15620. doi: 10.1074/jbc.273.25.15611. [DOI] [PubMed] [Google Scholar]
- 3.Wyszynski M., Kim E., Dunah A. W., Passafaro M., Valtschanoff J. G., Serra-Pages C., Streuli M., Weinberg R. J., Sheng M. Neuron. 2002;34:39–52. doi: 10.1016/s0896-6273(02)00640-2. [DOI] [PubMed] [Google Scholar]
- 4.Ko J., Kim S., Valtschanoff J. G., Shin H., Lee J. R., Sheng M., Premont R. T., Weinberg R. J., Kim E. J. Neurosci. 2003;23:1667–1677. doi: 10.1523/JNEUROSCI.23-05-01667.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhen M., Jin Y. Nature. 1999;401:371–375. doi: 10.1038/43886. [DOI] [PubMed] [Google Scholar]
- 6.Ackley B. D., Harrington R. J., Hudson M. L., Williams L., Kenyon C. J., Chisholm A. D., Jin Y. J. Neurosci. 2005;25:7517–7528. doi: 10.1523/JNEUROSCI.2010-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaufmann N., DeProto J., Ranjan R., Wan H., Van Vactor D. Neuron. 2002;34:27–38. doi: 10.1016/s0896-6273(02)00643-8. [DOI] [PubMed] [Google Scholar]
- 8.Krueger N. X., Van Vactor D., Wan H. I., Gelbart W. M., Goodman C. S., Saito H. Cell. 1996;84:611–622. doi: 10.1016/s0092-8674(00)81036-3. [DOI] [PubMed] [Google Scholar]
- 9.Hummel T., Zipursky S. L. Neuron. 2004;42:77–88. doi: 10.1016/s0896-6273(04)00158-8. [DOI] [PubMed] [Google Scholar]
- 10.Iwai Y., Usui T., Hirano S., Steward R., Takeichi M., Uemura T. Neuron. 1997;19:77–89. doi: 10.1016/s0896-6273(00)80349-9. [DOI] [PubMed] [Google Scholar]
- 11.Lee C. H., Herman T., Clandinin T. R., Lee R., Zipursky S. L. Neuron. 2001;30:437–450. doi: 10.1016/s0896-6273(01)00291-4. [DOI] [PubMed] [Google Scholar]
- 12.Prakash S., Caldwell J. C., Eberl D. F., Clandinin T. R. Nat. Neurosci. 2005;8:443–450. doi: 10.1038/nn1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ting C. Y., Yonekura S., Chung P., Hsu S. N., Robertson H. M., Chiba A., Lee C. H. Development (Cambridge, U.K.) 2005;132:953–963. doi: 10.1242/dev.01661. [DOI] [PubMed] [Google Scholar]
- 14.Iwai Y., Hirota Y., Ozaki K., Okano H., Takeichi M., Uemura T. Mol. Cell. Neurosci. 2002;19:375–388. doi: 10.1006/mcne.2001.1081. [DOI] [PubMed] [Google Scholar]
- 15.Nern A., Nguyen L-V. T., Herman T., Prakash S., Clandinin T. R., Zipursky S. L. Proc. Natl. Acad. Sci. USA. 2005;102:12944–12949. doi: 10.1073/pnas.0502888102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meinertzhagen I. A., Hanson T. E. In: The Development of Drosophila melanogaster. Bate M., Martinez-Arias A., editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 1363–1491. [Google Scholar]
- 17.Maurel-Zaffran C., Suzuki T., Gahmon G., Treisman J. E., Dickson B. J. Neuron. 2001;32:225–235. doi: 10.1016/s0896-6273(01)00471-8. [DOI] [PubMed] [Google Scholar]
- 18.Clandinin T. R., Lee C. H., Herman T., Lee R. C., Yang A. Y., Ovasapyan S., Zipursky S. L. Neuron. 2001;32:237–248. doi: 10.1016/s0896-6273(01)00474-3. [DOI] [PubMed] [Google Scholar]
- 19.Burden-Gulley S. M., Brady-Kalnay S. M. J. Cell Biol. 1999;144:1323–1336. doi: 10.1083/jcb.144.6.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu G., Arregui C., Lilien J., Balsamo J. J. Biol. Chem. 2002;277:49989–49997. doi: 10.1074/jbc.M206454200. [DOI] [PubMed] [Google Scholar]
- 21.Balsamo J., Leung T., Ernst H., Zanin M. K., Hoffman S., Lilien J. J. Cell Biol. 1996;134:801–813. doi: 10.1083/jcb.134.3.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brady-Kalnay S. M., Mourton T., Nixon J. P., Pietz G. E., Kinch M., Chen H., Brackenbury R., Rimm D. L., Del Vecchio R. L., Tonks N. K. J. Cell Biol. 1998;141:287–296. doi: 10.1083/jcb.141.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balsamo J., Arregui C., Leung T., Lilien J. J. Cell Biol. 1998;143:523–532. doi: 10.1083/jcb.143.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kypta R. M., Su H., Reichardt L. F. J. Cell Biol. 1996;134:1519–1529. doi: 10.1083/jcb.134.6.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brady-Kalnay S. M., Rimm D. L., Tonks N. K. J. Cell Biol. 1995;130:977–986. doi: 10.1083/jcb.130.4.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee T., Luo L. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 27.Chotard C., Leung W., Salecker I. Neuron. 2005;48:237–251. doi: 10.1016/j.neuron.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 28.Dunah A. W., Hueske E., Wyszynski M., Hoogenraad C. C., Jaworski J., Pak D. T., Simonetta A., Liu G., Sheng M. Nat. Neurosci. 2005;8:458–467. doi: 10.1038/nn1416. [DOI] [PubMed] [Google Scholar]
- 29.Muller T., Choidas A., Reichmann E., Ullrich A. J. Biol. Chem. 1999;274:10173–10183. doi: 10.1074/jbc.274.15.10173. [DOI] [PubMed] [Google Scholar]
- 30.Miller K. E., DeProto J., Kaufmann N., Patel B. N., Duckworth A., Van Vactor D. Curr. Biol. 2005;15:684–689. doi: 10.1016/j.cub.2005.02.061. [DOI] [PubMed] [Google Scholar]
- 31.Ko J., Na M., Kim S., Lee J. R., Kim E. J. Biol. Chem. 2003;278:42377–42385. doi: 10.1074/jbc.M307561200. [DOI] [PubMed] [Google Scholar]
- 32.Ashburner M. Drosophila, A Laboratory Handbook. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.