Abstract
Lim1, Ssdp1, and Ldb1 proteins are components of the Ldb1-associated transcriptional complex, which is important in the head-organizing activity during early mouse development. Depletion of each individual protein alone causes a headless phenotype. To explore in more detail the modular architecture of the complex, we have generated two different gene-trapped mouse lines that express truncated forms of Ssdp1. Embryos derived from the gene-trapped line that encodes a truncated Ssdp1 lacking the proline-rich sequence exhibit a lethal abnormal head-development phenotype, resembling mouse embryos deficient for Lim1, Ssdp1, or Otx2 genes. Embryos derived from the second gene-trapped line, in which most of the proline-rich domain of Ssdp1 is retained, did not show abnormalities in head development. Our data demonstrate that components of the Ldb1-dependent module can be subdivided further into discrete functional domains and that the proline-rich stretch of Ssdp1 is critical for embryonic head development. Furthermore, phylogenetic comparisons revealed that in Caenorhabditis elegans, a similar proline-rich sequence is absent in Ssdp but present in Ldb1. We conclude that although the overall architecture of the Ldb1-dependent module has been preserved, the genetic specification of its individual components has diversified during evolution, without compromising the function of the module.
Keywords: gene trap, Ldb1, module
Classical evolutionary biology has traditionally considered body parts as semiautonomous units that are built according to distinct structural plans and undergo relatively independent evolution. This characteristic feature of all organisms is named modularity, and it is likely to be important for adaptive phenotypic evolution, allowing optimization of different traits individually by natural selection (1). The dissociability of such modules in evolution is believed to be facilitated by a corresponding modular organization of interacting proteins controlling their development, such that changes that occur within one module have less chance of impacting negatively on others (2). The module is defined as a group of proteins that carry out a semiautonomous function (3, 4). Accordingly, protein interactions can be classified either as intramodule hubs (simultaneous interactions) or intermodule hubs (interactions at different times). The intramodule hubs belong to a single module, whereas intermodule hubs interact with multiple modules (3). Head development is a perfect example of modularity: genetic perturbation of the head organizer in mice results in embryos that are missing anterior structures but in which the trunk and tail regions develop normally (5). In the present work, we suggest that the Lbd1 (LIM domain-binding protein 1, also known as Clim2 and NL1)-associated transcriptional complex serves as an intramodule core of head organizer, and we start to investigate its modular architecture.
The ubiquitous nuclear Ldb1 interacts with multiple proteins, and its interaction domains are highly conserved through evolution (6, 7). The LIM-interaction domain is located in the C-terminal part of Ldb1, and it is sufficient for binding LIM domains of diverse LIM-homeodomain (LIM-HD) and LIM-only (LMO) proteins with high affinity and specificity. The fundamental and universal nature of this interaction is clear from evolutionary comparisons. The Ldb1 ortholog in Caenorhabditis elegans is also able to bind LIM domains (8), and Chip, the Drosophila homolog of Ldb1, can interact with fly and mammalian LIM proteins (9).
The second important domain of Ldb1 is located in the middle of the protein, and it is responsible for binding to single-stranded DNA-binding proteins of the Ssdp family (10). This protein interaction is also evolutionarily ancient, and it has been found in vertebrates, Drosophila, and plants. The Arabidopsis proteins LEUNIG and SEUSS, which share local domain similarity with Ssdp and Ldb/Chip proteins, interact specifically with each other (11). Ssdp1 was shown to bind single-stranded pyrimidine-rich sequences, and it is composed of a well conserved FORWARD/LUFS domain at the N-terminal end and a proline-rich sequence in the central region (11–13). The FORWARD domain, comprising three α-helices, is responsible for Ssdp interactions with Ldb1 (10). The C-terminal end of Ssdp1 was shown to possess the transcriptional activity independent of the interaction with Ldb1 (14).
Finally, the N-terminal part of Ldb1 contains a dimerization domain that is necessary to bridge two LIM-HD proteins and maintain the stoichiometry of the complex (15). These three types of interaction (LIM binding, Ssdp binding, and dimerization) allow Ldb1 to form diverse complexes with different protein partners and, therefore, to serve as a basis for multiple functions of Ldb/Chip in development. A good example is the Chip-mediated partnership of LIM-HD and HD proteins during Drosophila development: composition of Chip-associated transcriptional complexes is different in developing wing, neural tissues, tarsus 4, tarsus 5, and pretarsus (16).
There are several critical molecules important during vertebrate head development, including Ldb1 and Lim1, the Ldb1-interacting LIM-HD protein. According to the role of Ldb1 as a central core for multiple complexes in many developing structures, Ldb1−/− embryos do not survive beyond embryonic day 9.5 because of pleiotropic abnormalities, including forebrain truncations (17). Lim1 is peripheral to Ldb1, and it is expected to show more local effects. Lim1 was shown to be an essential regulator of the head organizer (5). Lim1−/− embryos lacked anterior head structures, but the remaining body developed normally (5). In transcriptional assays, activating synergism of Ldb1 and Lim1 requires both the LIM-interaction domain and the dimerization domain of Ldb1, indicating the importance of Ldb1 as a dimer (15, 18, 19).
A similar headless phenotype was also observed in knockouts of homeobox protein Otx2 (20–22), which directly associates with Lim1 (23). The C-terminal region of Otx2 binds to the Lim1 homeodomain (HD), whereas both HD and C-terminal regions of Otx2 are responsible for interaction with the hepatocyte nuclear factor 3β (HNF-3β) fork head domain (23). In addition, a member of the Otx family interacts specifically with Ldb1 (24). HNF-3β and Lim1 were shown to work synergistically to regulate organizer activity of the anterior visceral endoderm (25). It is conceivable that different HD proteins can either contribute to the specification or facilitate DNA binding in the Ldb1-dependent module.
In the present work, we have obtained evidence that the proline-rich domain of Ssdp1 performs a critical function in the Ldb1-associated complex. We have shown that deletion of the proline-rich domain has a deleterious effect in anterior patterning, leading to head truncation, and that the absence of the proline-rich domain is likely to change the protein-interaction interface of the Ldb1-dependent transcription complex. It has been demonstrated that proline-rich sequences are important in transient protein–protein interactions, and many such elements possess trans-activation abilities (26). Therefore, the deletion of the proline-rich domain of Ssdp1 may lead to a failure to interact with proteins involved in head development. Furthermore, we suggest that the Ldb1-dependent transcriptional module serves as an intramodular core of the head organizer, and we provide evolutionary data that support this idea.
Results and Discussion
Phylogenetic Comparison of Ldb1 and Ssdp Proteins.
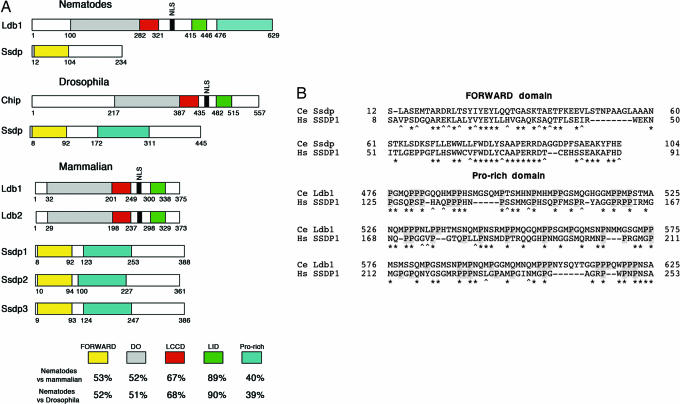
Recent studies revealed that intramodule hubs evolve more slowly than intermodule hubs (3), reinforcing the concept that modules must preserve their functions over vast stretches of evolutionary time, leading to greater constraint on intramodule hubs. Consistent with this idea, Ldb proteins demonstrate a remarkably high level of structural conservation. Two very similar mammalian proteins, Ldb1 and Ldb2, the Drosophila homolog Chip, and the C. elegans homolog Ldb1 (GenBank accession no. NP_509844) all have the same composition of highly conserved domains (Fig. 1A).
Fig. 1.
Phylogenetic comparison of Ldb1 and Ssdp proteins. (A) Domain architecture of the invertebrate and vertebrate Ssdp and Ldb proteins. At the bottom, homology is shown as percent similarity between corresponding domains. DD, dimerization domain; LCCD, Ldb1-Chip conservative domain; LID, LIM-interaction domain; Pro-rich, proline-rich sequence. Amino acid numbers in diagrams of nematode and mammalian proteins are shown for C. elegans and human, respectively. (B) Alignment of the proline-rich regions of Ssdp1 and Ldb1. Ce, C. elegans; Hs, Homo sapiens. Prolines are on a gray background. Identical amino acids are indicated by asterisks, and conserved amino acid substitutions are indicated by carets. Spaces were introduced to optimize the alignment, and they are indicated by dashes. Amino acid numbers are shown to the left and to the right of the corresponding sequences.
Based on the similarity of the N-terminal FORWARD domain (13), invertebrate Ssdp homologs were previously described in Drosophila (CG7187) (27) and in C. elegans (2N612; GenBank accession no. NM_064415). The reason for conservation of the FORWARD domain of Ssdp proteins is clear because it is responsible for binding the partner domain within Ldb proteins (10). However, we paid attention to the remaining part of the mammalian Ssdp proteins, which is rich in proline and glycine residues.
Although the FORWARD domain is more conserved than the proline-rich tail, all vertebrate orthologs and paralogs of Ssdp proteins display a high level of sequence identity with mouse and human Ssdp1 over the entire coding sequences. In contrast, the single Ssdp ortholog in C. elegans is short and contains only the N-terminal FORWARD domain (Fig. 1A). We used the proline-rich stretch of mammalian Ssdp1 (segment from Pro-150 to Thr-289) as a template in BLAST and Position-Specific Iterated (PSI)-BLAST searches with standard parameters in worm databases, and, surprisingly, we identified a sequence of significant similarity in C. elegans Ldb1. The level of homology of this region of C. elegans Ldb1 protein and human Ssdp1 (38% similarity and 31% identity) is lower but comparable with that between the FORWARD domains of human and nematode Ssdp proteins (55% similarity and 43% identity) (Fig. 1B). Moreover, alignment shows strong conservation of proline residues.
In Drosophila Ssdp, a sequence corresponding to the vertebrate Ssdp1 proline-rich domain is also present (54% similarity and 49% identity) (Fig. 1A); but in contrast to C. elegans, many of its proline residues have not been conserved. Thus, our phylogenetic comparison led us to investigate the importance of the Ssdp1 proline-rich sequence in the function of the Ldb1-associated transcriptional complex in vertebrates.
Generation and Characterization of Mutant Mice.
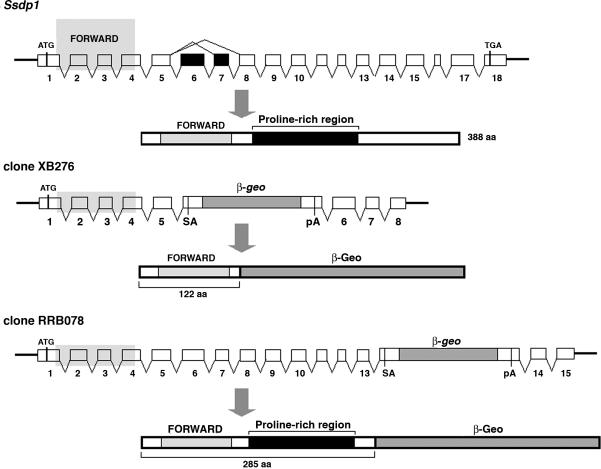
To obtain mutants for the Ssdp1 locus, we used a library of insertional mutations produced in ES cells by BayGenomics (http://baygenomics.ucsf.edu). A survey of this gene-trap database revealed clones XB276 and RRB078 with the β-geo insertion within the 5th and 13th introns of the Ssdp1 allele, respectively (Fig. 2). Because of these insertions, the first 122 (clone XB276) and 285 (clone RRB078) amino acids of Ssdp1 were fused to the β-galactosidase-neomycin protein (β-Geo) (Fig. 2). As a result, the proline-rich stretch is eliminated from the 122-aa truncated form of Ssdp1 (Ssdp1122). However, the 285-aa form (Ssdp1285) truncates only the C-terminal 103 residues, and thus it retains most of the proline-rich stretch. Therefore, these two alleles will allow us to define the role of the Ssdp1 proline-rich sequence within the Ldb1-dependent complex.
Fig. 2.
Generation and phenotype of Ssdp1 mutant mice. The wild-type Ssdp1 allele and gene-trapped clones with the β-geo insertion within the 5th and 13th introns of Ssdp1 are shown. On the schematic representation of the wild-type Ssdp1 protein, the two conserved regions (FORWARD domain and proline-rich stretch) are indicated, as are the truncated protein products formed in the mutant mice.
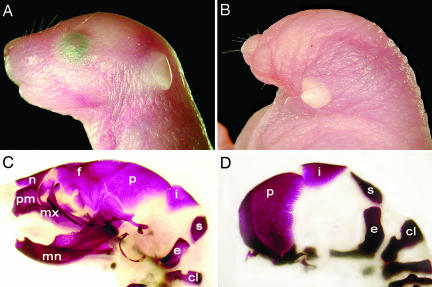
The gene-trapped clones were injected into blastocysts to produce chimeras, and the F1 heterozygotes derived from both lines were phenotypically indistinguishable from wild type, indicating that disruption of one allele of Ssdp1 does not cause dominant-negative effects. Western blot analysis confirmed that heterozygotes express chimeric proteins composed of N-terminal Ssdp1 polypeptides fused to β-Geo (not shown). Embryos derived from both lines were analyzed at different stages of gestation. Ssdp1122−/− embryos exhibited a lethal phenotype associated with abnormal head development (Fig. 3). This headless phenotype is associated with the deletion of the fore- and midbrain and phenocopies the reported Ssdp1-null mice (28). In contrast, Ssdp1285−/− embryos did not show abnormalities in head development. These results demonstrate that the proline-rich domain of Ssdp1 has a critical role in embryonic head development.
Fig. 3.
Lateral view of embryonic day 18.5 normal (Ssdp1285−/−) and mutant (Ssdp1122−/−) mice. (A and C) Control Ssdp1285−/− embryo. (B and D) Mutant embryo (Ssdp1122−/−) displays a severe anterior truncation. Bone staining (red) is shown in C and D. Ssdp1122−/− mouse lacks bones anterior to the parietal bone (p). c1, c1 vertebra; e, exoccipital bone; f, frontal bone; i, interparietal bone; mn, mandible; mx, maxillary bone; n, nasal bone; p, parietal bone; pm, premaxillary bone; s, supraoccipital bone.
Evolution of the Ldb1-Associated Transcriptional Complex.
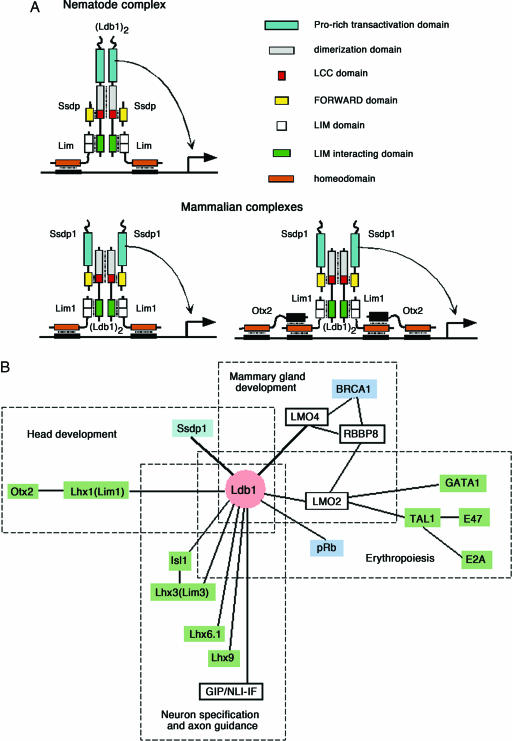
Together with the finding that nematodes lack the proline-rich domain found in Drosophila and the mouse, our experimental data allow us to see evolution of the Ldb1-associated transcriptional complex in a new light (Fig. 4A). Because the fly and the nematode are much more closely related to each other than either is to the mouse, we propose that the proline-rich domain was ancestrally associated with Ssdp, that the Ldb1 in the nematode lineage gained a proline-rich domain, and that Ssdp subsequently lost the functionally equivalent region. Although we cannot categorically rule out the alternative scenario that an ancient Ssdp protein gained a new functional proline-rich domain before invertebrate and vertebrate separation and that Ldb1 lost the consequently redundant element, the functional data confirm our model of the evolutionary change in the nematode lineage. It has been shown that Ldb1 is actively expressed during C. elegans development, that it is required for some neuronal function, and that knockdown of its expression by RNA interference leads to uncoordinated locomotion (8). However, Ssdp is expressed at a low level, and its RNA interference shows no obvious phenotype (29). Thus, both Ldb1 and Ssdp1 are equally important for development in vertebrates, whereas in C. elegans Ssdp lacking the proline-rich domain is dispensable.
Fig. 4.
Model of the Ldb1-associated complex. (A) Hypothetical structure of the Ldb1-associated complex as an intramodule hub. In C. elegans, Ldb1 performs dual functions: bridging together LIM-HD proteins and trans-activation. In vertebrates, the trans-activator function is performed by Ssdp1. LIM-HD proteins serve as a DNA-binding part of the complex. Otx2 activates transcription of its target genes synergistically with Lim1 (Lhx1) and Ldb1 and binds directly to the Lim1 homeodomain with its C-terminal region (23). Therefore, in addition to the hexameric Ssdp1-containing complex, we predict an octameric complex in which both HD proteins, Otx2 and Lim1, contribute to DNA binding. These two complexes are similar in their structure to (Lim3)2·(Ldb1)2 and (Lim1)2·(Isl1)2·(Ldb1)2 complexes described for neuron specification and axon guidance, respectively (33). HNF-3β could serve as a negative regulator for an Otx2-containing Ldb1-associated complex: HNF-3β can repress Otx2-directed gene expression by binding to both the HD and C-terminal regions of Otx2 (23) and excluding Otx2 from the complex. Similarly, LMOs could serve as negative regulators for all Ldb1-associated complexes: they can occupy LIM-interacting domains of Ldb1 and prevent binding to DNA by excluding LIM-HD proteins from the complex. (B) Multiple interactions of the Ldb1 protein during vertebrate development. The diagram of protein–protein interactions with Ldb1 participation was prepared by using BIND, iHOP, and STRING databases. The diagram shows only known interactions in the mammalian system, and it is not exhaustive. Ldb1 serves as a core for different oligoprotein modules linking DNA-binding (shown in green) and trans-activating (shown in blue) proteins. Both DNA-binding and trans-activating parts of the complexes can be attached to Ldb1 either directly or through LMO proteins Lmo2 and Lmo4 (6).
The Ldb1-Associated Transcriptional Complex Is the Intramodule Hub.
Our data can be put into the frame of functional modules, as originally described by Hartwell et al. (4). A functional module was defined as a discrete entity whose function is separable from the functions of other modules. The Ldb1-associated module behaves as the intramodule hub in the head organizer based on the properties of the protein products that constitute this complex. This notion is supported by the individual knockouts of Otx2, Lim1, Ldb1, and Ssdp1 genes: mutant embryos display a similar headless phenotype (5, 17, 20–22, 28).
Based on the accumulated data, we propose a model that describes the role of the Ldb1-associated module during head development. This intramodule hub consists of the Ldb1·Ssdp1·Lim1 core hexamer, which in turn works with Otx2 and HNF-3β through a specific association with Lim1 (Fig. 4). In addition, there are likely to be other, as yet unknown, proteins that interact with the proline-rich stretch of Ssdp1 and the activation domain of Lim1. Further biochemical and genetic studies will shed more light on the structure and function of this complex. Furthermore, the individual components of the module may belong to different modules during development. In Drosophila, Chip can form complexes with different LIM-HD and LMO partners affecting the development of legs or wings (16). The LIM-HD proteins behave as versatile factors in development based on recent studies in Drosophila. It was proposed that different developmental outcomes of LIM-HD protein function could be explained by the dosage and precise identity of cofactors available locally (16). We suggest that the core of the Ldb-dependent intramodule hub (Ldb·Ssdp·LIM domain proteins) stays the same but that it would employ different components (that constitute intermodule hubs) in various tissues and/or at times of development according to required functions.
It is interesting to mention that point mutations within the LIM domain of Lim1 result in a phenocopy of Lim1-null mice (30). These results could be explained in light of our proposed model (Fig. 4A). The LIM domain is critical for a specific association with Ldb1, thus keeping Lim1 as an integral component of the module. The disruption of the Lim1–Ldb1 interaction causes a failure to create a functional Ldb1-associated module, leading to misregulation of downstream target genes involved in head development. Furthermore, our model predicts that mutations within critical domains of individual components that constitute the module would cause head defects in developing mouse embryos. These mutations would therefore change the ability of the Ldb1-associated module to interact with other modules, possibly preventing intermodule hubs. Together with our finding that deletion of the Ssdp1 proline-rich sequence disrupts the activity of the Ldb1-associated complex, these results demonstrate that components of this complex can be subdivided further into discrete functional domains.
The success of the Ldb-dependent module in controlling different aspects of development depends on the core components of the module. Because of this stringent requirement, genes that encode these proteins are resistant to dramatic mutational changes over a long evolutionary time, thus leading to a greater constraint on the intramodule hub. It appears that proteins that constitute the Ldb-dependent module have changed very little during evolution. According to our model, the Ldb·Ssdp·LIM domain interactions constitute a core of the module. The Ssdp proteins do not possess nuclear localization signals; however, the association with the Ldb proteins allows them to enter the nucleus (10). There are two paralogs of Ldb, three paralogs for Ssdp proteins, and several LIM-HD and LMO proteins in vertebrates. They display different expression patterns during development, and therefore, the ability to create a functional Ldb-associated module will depend on the rate and availability of individual components within any given tissue/organ and time of development (Fig. 4B).
In conclusion, we provide evidence that the structure and function of the Ldb-dependent module is governed by the general design principle, which was shaped by the constraints of evolution.
Materials and Methods
Protein sequence databases were searched by using standard protein–protein BLAST and the PSI-BLAST with standard parameters at the National Center for Biotechnology Information BLAST Server (www.ncbi.nlm.hih.gov/BLAST). Pairwise and multiple alignments were performed by using the MacVector 7.2 program (Oxford Molecular Group) and ClustalW algorithm (31). Functional associations among proteins were retrieved by using STRING (http://string.embl.de), Biomolecular Interaction Network Database (BIND) (http://bind.ca), and Database of Interacting Protein (DIP) (http://dip.doe-mbi.ucla.edu) sites and the Information Hyperlinked over proteins (iHOP) database (www.pdg.cnb.uam.es/UniPub/iHOP). Mouse ES cell lines XB276 and RRB078 with the β-geo insertion within the 5th and 13th introns of the Ssdp1 allele were identified in the BayGenomics gene-trap database (http://baygenomics.ucsf.edu). These ES clones were injected into C57BL/6 blastocysts to generate chimeric mice, which were bred to establish Ssdp1 mutant mice. To genotype animals at weaning, dot blots of DNA prepared from tail biopsies were probed with the β-galactosidase probe, or PCR was performed with the β-galactosidase-specific primers. Skeletal preparations were carried out by standard methods (32).
Acknowledgments
We thank Hunter Fraser, Gunter Wagner, Richard Behringer, and Heiner Westphal for a critical reading of the manuscript; the Animal Genomics Services at Yale for the blastocyst injections; and Timothy Brend and Timothy Nottoli for comments and editorial assistance. This work was supported in part by National Institutes of Health Centers of Biomedical Research Excellence Grant 5 P20 RR17702-04.
Abbreviations
- HD
homeodomain
- HNF-3β
hepatocyte nuclear factor 3β
- Lbd1
LIM-binding domain 1
- LIM
Lim domain
- LIM-HD
LIM-homeodomain
- LMO
LIM-only
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Needham J. Biol. Rev. 1933;8:180–233. [Google Scholar]
- 2.Schlosser G., Wagner G. P., editors. Modularity in Development and Evolution. Chicago: Univ. of Chicago Press; 2004. [Google Scholar]
- 3.Fraser H. B. Nat. Genet. 2005;37:351–352. doi: 10.1038/ng1530. [DOI] [PubMed] [Google Scholar]
- 4.Hartwell L. H., Hopfield J. J., Leibler S., Murray A. W. Nature. 1999;402:C47–C52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- 5.Shawlot W., Behringer R. R. Nature. 1995;374:425–430. doi: 10.1038/374425a0. [DOI] [PubMed] [Google Scholar]
- 6.Matthews J. M., Visvader J. E. EMBO Rep. 2003;4:1132–1137. doi: 10.1038/sj.embor.7400030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hobert O., Westphal H. Trends Genet. 2000;16:75–83. doi: 10.1016/s0168-9525(99)01883-1. [DOI] [PubMed] [Google Scholar]
- 8.Cassata G., Rohrig S., Kuhn F., Hauri H. P., Baumeister R., Burglin T. R. Dev. Biol. 2000;226:45–56. doi: 10.1006/dbio.2000.9846. [DOI] [PubMed] [Google Scholar]
- 9.Torigoi E., Bennani-Baiti I. M., Rosen C., Gonzalez K., Morcillo P., Ptashne M., Dorsett D. Proc. Natl. Acad. Sci. USA. 2000;97:2686–2691. doi: 10.1073/pnas.050586397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Meyel D. J., Thomas J. B., Agulnick A. D. Development (Cambridge, U.K.) 2003;130:1915–1925. doi: 10.1242/dev.00389. [DOI] [PubMed] [Google Scholar]
- 11.Franks R. G., Wang C., Levin J. Z., Liu Z. Development (Cambridge, U.K.) 2002;129:253–263. doi: 10.1242/dev.129.1.253. [DOI] [PubMed] [Google Scholar]
- 12.Bayarsaihan D., Soto R. J., Lukens L. N. Biochem. J. 1998;331:447–452. doi: 10.1042/bj3310447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bayarsaihan D. Biochim. Biophys. Acta. 2002;1599:152–155. doi: 10.1016/s1570-9639(02)00376-x. [DOI] [PubMed] [Google Scholar]
- 14.Wu L. Biochem. Biophys. Res. Commun. 2006;339:977–984. doi: 10.1016/j.bbrc.2005.11.098. [DOI] [PubMed] [Google Scholar]
- 15.Jurata L. W., Pfaff S. L., Gill G. N. J. Biol. Chem. 1998;273:3152–3157. doi: 10.1074/jbc.273.6.3152. [DOI] [PubMed] [Google Scholar]
- 16.Pueyo J. I., Couso J. P. Development (Cambridge, U.K.) 2004;131:3107–3120. doi: 10.1242/dev.01161. [DOI] [PubMed] [Google Scholar]
- 17.Mukhopadhyay M., Teufel A., Yamashita T., Agulnick A. D., Chen L., Downs K. M., Schindler A., Grinberg A., Huang S. P., Dorward D., Westphal H. Development (Cambridge, U.K.) 2003;130:495–505. doi: 10.1242/dev.00225. [DOI] [PubMed] [Google Scholar]
- 18.Jurata L. W., Gill G. N. Mol. Cell. Biol. 1997;17:5688–5698. doi: 10.1128/mcb.17.10.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Breen J. J., Agulnick A. D., Westphal H., Dawid I. B. J. Biol. Chem. 1998;273:4712–4717. doi: 10.1074/jbc.273.8.4712. [DOI] [PubMed] [Google Scholar]
- 20.Acampora D., Mazan S., Lallemand Y., Avantaggiato V., Maury M., Simeone A., Brulet P. Development (Cambridge, U.K.) 1995;121:3279–3290. doi: 10.1242/dev.121.10.3279. [DOI] [PubMed] [Google Scholar]
- 21.Ang S. L., Jin O., Rhinn M., Daigle N., Stevenson L., Rossant J. Development (Cambridge, U.K.) 1996;122:243–252. doi: 10.1242/dev.122.1.243. [DOI] [PubMed] [Google Scholar]
- 22.Matsuo I., Kuratani S., Kimura C., Takeda N., Aizawa S. Genes Dev. 1995;9:2646–2658. doi: 10.1101/gad.9.21.2646. [DOI] [PubMed] [Google Scholar]
- 23.Nakano T., Murata T., Matsuo I., Aizawa S. Biochem. Biophys. Res. Commun. 2000;267:64–70. doi: 10.1006/bbrc.1999.1872. [DOI] [PubMed] [Google Scholar]
- 24.Bach I., Carriere C., Ostendorff H. P., Andersen B., Rosenfeld M. G. Genes Dev. 1997;11:1370–1380. doi: 10.1101/gad.11.11.1370. [DOI] [PubMed] [Google Scholar]
- 25.Perea-Gomez A., Shawlot W., Sasaki H., Behringer R. R., Ang S. Development (Cambridge, U.K.) 1999;126:4499–4511. doi: 10.1242/dev.126.20.4499. [DOI] [PubMed] [Google Scholar]
- 26.Kay B. K., Williamson M. P., Sudol M. FASEB J. 2000;14:231–241. [PubMed] [Google Scholar]
- 27.Chen L., Segal D., Hukriede N. A., Podtelejnikov A. V., Bayarsaihan D., Kennison J. A., Ogryzko V. V., Dawid I. B., Westphal H. Proc. Natl. Acad. Sci. USA. 2002;99:14320–14325. doi: 10.1073/pnas.212532399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishioka N., Nagano S., Nakayama R., Kiyonari H., Ijiri T., Taniguchi K., Shawlot W., Hayashizaki Y., Westphal H., Behringer R. R., et al. Development (Cambridge, U.K.) 2005;132:2535–2546. doi: 10.1242/dev.01844. [DOI] [PubMed] [Google Scholar]
- 29.Kamath R. S., Fraser A. G., Dong Y., Poulin G., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., et al. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 30.Cheah S. S., Kwan K. M., Behringer R. R. Genesis. 2000;27:12–21. doi: 10.1002/1526-968x(200005)27:1<12::aid-gene30>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 31.Thompson J. D., Higgins D. G., Gibson T. J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hogan B., Beddington R., Constantini F., Lacy E. Manipulating the Mouse Embryo: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1994. [Google Scholar]
- 33.Thaler J. P., Lee S. K., Jurata L. W., Gill G. N., Pfaff S. L. Cell. 2002;110:237–249. doi: 10.1016/s0092-8674(02)00823-1. [DOI] [PubMed] [Google Scholar]