Abstract
The green lineage is reportedly 1,500 million years old, evolving shortly after the endosymbiosis event that gave rise to early photosynthetic eukaryotes. In this study, we unveil the complete genome sequence of an ancient member of this lineage, the unicellular green alga Ostreococcus tauri (Prasinophyceae). This cosmopolitan marine primary producer is the world’s smallest free-living eukaryote known to date. Features likely reflecting optimization of environmentally relevant pathways, including resource acquisition, unusual photosynthesis apparatus, and genes potentially involved in C4 photosynthesis, were observed, as was downsizing of many gene families. Overall, the 12.56-Mb nuclear genome has an extremely high gene density, in part because of extensive reduction of intergenic regions and other forms of compaction such as gene fusion. However, the genome is structurally complex. It exhibits previously unobserved levels of heterogeneity for a eukaryote. Two chromosomes differ structurally from the other eighteen. Both have a significantly biased G+C content, and, remarkably, they contain the majority of transposable elements. Many chromosome 2 genes also have unique codon usage and splicing, but phylogenetic analysis and composition do not support alien gene origin. In contrast, most chromosome 19 genes show no similarity to green lineage genes and a large number of them are specialized in cell surface processes. Taken together, the complete genome sequence, unusual features, and downsized gene families, make O. tauri an ideal model system for research on eukaryotic genome evolution, including chromosome specialization and green lineage ancestry.
Keywords: genome heterogeneity, genome sequence, green alga, Prasinophyceae, gene prediction
The smallest free-living eukaryote known so far is Ostreococcus tauri (1). This tiny unicellular green alga belongs to the Prasinophyceae, one of the most ancient groups (2) within the lineage giving rise to the green plants currently dominating terrestrial photosynthesis (the green lineage) (3, 4). Consequently, since its discovery, there has been great interest in O. tauri, which, because of its apparent overall simplicity, a naked, nonflagellated cell possessing a single mitochondrion and chloroplast, in addition to its small size and ease in culturing, renders it an excellent model organism (5). Furthermore, it has been hypothesized, based on its small cellular and genome sizes (2, 6), that it may reveal the “bare limits” of life as a free-living photosynthetic eukaryote, presumably having disposed of redundancies and presenting a simple organization and very little noncoding sequence.
Since its identification in 1994, Ostreococcus has been recognized as a common member of the natural marine phytoplankton assemblage. It is cosmopolitan in distribution, having been found from coastal to oligotrophic waters, including the English Channel, the Mediterranean and Sargasso Seas, and the North Atlantic, Indian, and Pacific Oceans (7–12). Eukaryotes within the picosize fraction (<2- to 3-μm diameter) have been shown to contribute significantly to marine primary production (9, 13). Ostreococcus itself is notable for its rapid growth rates and potential grazer susceptibility (9, 14). Furthermore, dramatic blooms of this organism have been recorded off the coasts of Long Island (15) and California (11). At the same time, attention has focused on the tremendous diversity of picoeukaryotes (16, 17), which holds true for Ostreococcus as well. Recently, Ostreococcus strains isolated from surface waters were shown to represent genetically and physiologically distinct ecotypes, with light-regulated growth optima different from those isolated from the deep chlorophyll maximum (18). These findings are similar to the niche adaptations documented in different ecotypes of the abundant marine cyanobacteria Prochlorococcus (19, 20).
Overall, marine picophytoplankton play a significant role in primary productivity and food webs, especially in oligotrophic environments where they account for up to 90% of the autotrophic biomass (9, 13, 21, 22). Several recent studies have undertaken a genome sequencing approach to understand the ocean ecology of phytoplankton. To date, these studies have focused on the bacterial component of the plankton, particularly on the picocyanobacteria Prochlorococcus (20) and Synechococcus (23), for which 9 complete genome sequences are already publicly available and >13 others on the way. Much less is known about eukaryotic phytoplankton, because only one, the diatom Thalassiosira pseudonana, has a complete genome sequenced (24). Picoeukaryotes are especially interesting in the context of marine primary production, given the combination of their broad environmental distribution and the fact that their surface area to volume ratio, a critical factor in resource acquisition and success in oligotrophic environments (25), is similar to that of prokaryotic counterparts generally considered superior in uptake and transport of nutrients.
In this article, we describe the complete genome sequence of O. tauri OTH95, a strain isolated in the Thau lagoon (France) in which this species makes recurrent, quasimonospecific blooms in summer (1). This genome is particularly significant in that it represents a complete genome sequence of a member of the Prasinophyceae, which diverged at the base of the green lineage (2). It is also the complete genome sequence of a picoeukaryote thought to be of ecological importance to primary production. Analysis of the O. tauri genome and comparison with other genomes available to date, including algal, plant, and fungal genomes, allowed delineation of both specific gene features and identification of unique aspects of this genome.
Results and Discussion
Global Genome Structure.
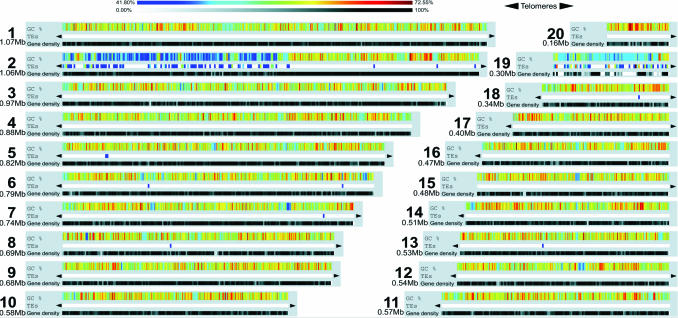
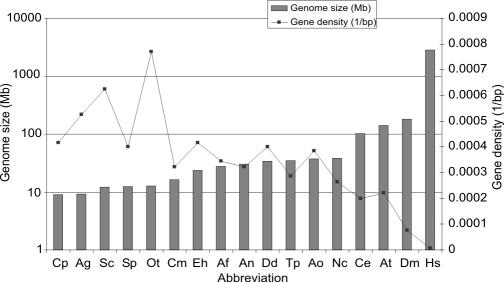
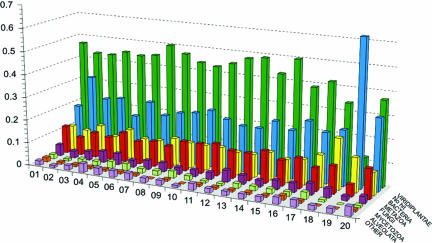
Whole genome shotgun sequencing and an oriented walking strategy were used to sequence the genome of O. tauri strain OTH95 (Tables 2 and 3, which are published as supporting information on the PNAS web site). A genome size of 12.56 Mb distributed in 20 superscaffolds corresponding to 20 chromosomes was determined by means of sequence assembly (Fig. 1; and Figs. 4 and 5, which are published as supporting information on the PNAS web site), fully consistent with pulsed-field gel electrophoresis results indicating a total size of 12.5 to 13 Mb (Fig. 4 and Supporting Text, which are published as supporting information on the PNAS web site). This genome size is similar to that of the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe, despite their larger cell size, but smaller than any other oxyphototrophic eukaryote known so far, including the red alga Cyanidioschyzon merolae (26) (Fig. 2 and Table 1). The G+C content of O. tauri is more akin to that of C. merolae than to that of plants, fungi, or even T. pseudonana (Table 1). As shown in Fig. 2 and Table 1, 8,166 protein-coding genes were predicted in the nuclear genome, making O. tauri the most gene dense free-living eukaryote known to date. Only the chromosomes of the nucleomorphs within chlorachniophyte and cryptophyte algae are more gene-dense bodies (27), which are internally contained and not capable of independent propagation. We found that 6,265 genes are supported by homology with known genes in public databases (e-value <10−5), of which the majority (46%) were most similar to plant orthologs (Fig. 3). Very few repeated sequences have been found in this genome, except for a long internal duplication of 146,028 kb on chromosome 19. Because the duplicated sequence is >99% identical, it is probably of recent origin.
Fig. 1.
General characteristics of the 20 O. tauri chromosomes. TEs, transposon frequency. Size is indicated to the left of each chromosome (Mb). Colored bars indicate the percentage G+C content (upper bar) and of transposons (lower bar).
Fig. 2.
Genome size and gene density for various eukaryote genomes. Cp, Cryptosporidium parvum; Ag, Ashbya gossypii, Sp, Schizosaccharomyces pombe; Sc, Saccharomyces cerevisiae; Ot, Ostreococcus tauri; Cm, Cyanydioschyzon merolae; Eh, Entamoeba hemolytica; Af, Aspergillus fumigatu; An, Aspergillus niger; Dd, Dictyostelium discoidum; Tp, Thalassiosira pseudonana; Ao, Aspergillus oryzae; Nc, Neurospora crassa; Ce, Caenorhabditis elegans; At, Arabidopsis thaliana; Dm, Drosophila melanogaster; Hs, Homo sapiens.
Table 1.
General features of the O. tauri genome
| Feature | O. tauri | T. pseudonana | C. merolae | Arabidopsis thaliana | Ashbya gossypii | S. cerevisiae | S. pombe | Cryptosporidium parvum |
|---|---|---|---|---|---|---|---|---|
| Size, Mbp | 12.56 | 34.50 | 16.52 | 140.12 | 9.20 | 12.07 | 12.46 | 9.10 |
| No. of chromosomes | 20 | 24 | 20 | 5 | 7 | 16 | 3 | 8 |
| G+C content, % | 58.0 (59.0*) | 47.0 | 55.0 | 36.0 | 52.0 | 38.3 | 36.0 | 30.0 |
| Gene number | 8,166 | 11,242 | 5,331 | 26,207 | 4,718 | 6,563 | 4,824 | 3,807 |
| Gene density, kb per gene | 1.3 | 3.5 | 3.1 | 4.5 | 1.9 | 1.6 | 2.5 | 2.4 |
| Mean gene size, bp per gene† | 1,257 | 992 | 1,552 | 2,232 | N.A. | N.A. | 1,426 | 1,795 |
| Mean inter-ORF distance | 197 | N.A. | 1,543 | 2,213 | 341 | N.A. | 952 | 566 |
| Genes with introns, % | 39 | N.A. | 0.5 | 79 | 5 | 5 | 43 | 5 |
| Mean length of introns, bp | 103 (187*) | N.A. | 248 | 164 | N.A. | N.A. | 81 | N.A. |
| Coding sequences, % | 81.6 | N.A. | 44.9 | 33.0 | 79.5 | N.A. | 57.5 | 75.3 |
| No. of ribosomal RNA units | 4 | N.A. | 3 | 700–800 | 50 | 100–150 | 200–400 | 5 |
Data for the yeast S. cerevisiae compiled from refs. 24, 26, 47–49 and from Saccharomyces Genome Database at www.yeastgenome.org; N.A., not available.
*Data that exclude chromosomes 2 and 19.
†Data that exclude introns.
Fig. 3.
Taxon distribution of best hits for genes from each of the O. tauri chromosomes. Green, viridiplantae; blue, no hit; yellow, bacteria; red, metazoa; pink, fungi; gray, mycetozoa; orange, alveolates; purple, others. Annotation of genes on the low G+C part of chromosome 2 is difficult, and the percentage of genes having no hit on chromosome 2 can be slightly overestimated. See Genome Heterogeneity for details.
Genome Heterogeneity.
In view of what is currently known about eukaryotic nuclear genomes, one of the most striking features of the O. tauri genome is its heterogeneity, a feature which is not only unusual but also perplexing from an evolutionary perspective. Two chromosomes (2 and 19) are different from the other 18, in terms of organization for chromosome 2 and function for chromosome 19 (Fig. 1; and Fig. 6, which is published as supporting information on the PNAS web site). Both of these chromosomes have lower G+C content than the 59% G+C of the other 18 chromosomes (Fig. 1). Chromosome 2 is composed primarily of two blocks, one with a G+C content similar to that of the other chromosomes and the other with a markedly lower G+C content (52%). The average G+C content of the entire chromosome 2 amounts to 55%. Likewise, the G+C content of chromosome 19 (54%) is similar to the atypical region of chromosome 2. Taken together, these two aberrant chromosomes contain 77% of the 417 transposable elements (TEs), or relics thereof, which are identified in the genome (57% in chromosome 2 and 20% in chromosome 19) (Fig. 1 and Table 4, which is published as supporting information on the PNAS web site). Other chromosomes therefore contain very few or no TEs. TEs have a G+C content similar to the rest of the genome and cannot explain the global lower G+C content observed in these two chromosomal regions. Moreover, almost all of the known TE types can be found in the O. tauri genome: fifteen class I TE families [i.e., 3 TY1/Copia-like LTR-retrotransposons and 12 terminal-repeat retrotransposons in miniature (TRIMs)], nine transposon families, [4 Mariner-like elements, 2 P instability factors (PIFs), 1 homology and transposition (hAT), 1 foldback, and 1 unclassified (28)], and three miniature inverted repeat transposable element (MITE) families were identified. Only long interspersed nuclear elements (LINEs), short interspersed nuclear elements (SINEs), and helitrons were not detected. In the case of O. tauri, the distribution bias could have two origins: either the species originated from an allopolyploidization event between two donor parents with a genome contrasting for their TE content or there is a strong insertion bias for the TEs on both chromosomes 2 and 19. For most of the TE families, several partial copies or relics can be found throughout the 20 chromosomes (Table 4), indicating their ancient origin in the genome, therefore not supporting the first hypothesis. Nevertheless, further analyses are needed to conclude on this matter.
Chromosome 2 has additional unique features aside from differences in G+C content and the occurrence of many transposons. In particular, codon usage for genes in the low G+C region of this chromosome is different from that of all other chromosomes (Table 5, which is published as supporting information on the PNAS web site). Many of the genes in this low G+C region also contain multiple small introns with specific features (Fig. 7 a and b, which is published as supporting information on the PNAS web site). These two differences make gene modeling more complicated for this region, although at least 61 predicted peptides were supported by ESTs (see Table 6, which is published as supporting information on the PNAS web site). Chromosome 2 small introns differ in many respects from the other introns, such as their size (40–65 bp), composition (they are AT rich and richer by ≈10% than the neighboring exons), and splice sites and branch points that are less conserved than for other introns (Fig. 7b). Interestingly, phylogenetic analysis (see Materials and Methods) shows that 43% of the genes on this chromosome, including the small intron-containing genes, have green lineage ancestry (Fig. 3). Of those, 44% cluster specifically (with bootstrap values >70%) with genes of Chlamydomonas reinhardtii (data not shown but available on request). Together with the fact that the genes encoded in this region are essential housekeeping genes not duplicated elsewhere in the genome, this observation argues against an alien (horizontal transfer) origin for the low G+C region of chromosome 2. Thus, the origin of the chromosome 2 peculiarities remains elusive. One possibility is that it represents a sexual chromosome. It has been shown before that such chromosomes possess distinctive features for avoiding recombination and are characterized by an unusual richness in transposable elements (29). Meiosis has not been observed in culture, and no equivalent of a mating-type locus has been found akin to that in C. reinhardtii. Nevertheless, the presence of most of the core meiotic genes homologous to those identified in other organisms found in O. tauri (Table 7, which is published as supporting information on the PNAS web site) is at least a strong indication that O. tauri may be a sexual organism (30). Indeed other marine algae known to undergo sexual reproduction commonly suppress this capability in culture (31).
With respect to chromosome 19, phylogenetic analysis shows that only 18% of the peptide-encoding genes are related to the green lineage, a significantly lower percentage than that for the 19 other chromosomes. Others resemble proteins from various origins, mainly bacterial, although generally poorly conserved (Fig. 3; and Table 8, which is published as supporting information on the PNAS web site). Interestingly, most (84%) of the ones having a documented function belong to a few functional categories, primarily encoding surface membrane proteins or proteins involved in the building of glycoconjugates (Table 8). Based on these features, we hypothesize that chromosome 19 is of a different origin than the rest of the genome. This putatively alien material could have yielded some selective advantages in cell surface processes, potentially related, for example, to defense against pathogens or other environmental interactions.
Genome Compaction.
A second remarkable feature of the O. tauri genome is the intense degree of genome compaction, which appears to be the result of several processes. Shortening of intergenic regions is clearly a major factor. The average intergenic size is only 196 bp, which is shorter than that of other eukaryotes having a similar genome size (Table 1). Two other important factors are gene fusion, for which several cases are observed (Fig. 8, which is published as supporting information on the PNAS web site), and reduction of the size of gene families. For example, the gene complement involved in cell division control is one of the most complete across eukaryotes, although there is only one copy of each gene (32). Although this type of reduction is often the case in O. tauri, there are some exceptions. For example, the full set of partially redundant enzymes required for polysaccharide metabolism in land plants is present. Here, the maintenance of 27 genes, including multicopy genes, related to synthesis and breakdown of only two types of chemical linkages in the chloroplast, seems excessive for building the semicrystalline starch granule of O. tauri (33). Indeed, apicomplexa parasites or even red algae require only 10 genes to build and degrade simple polymers in their cytoplasm (34). O. tauri appears to be quite similar to other unicellular organisms in terms of numbers of transcription factors, with no further reduction than what has commonly been reported. Approximately 2.5–3.8% of predicted proteins of unicellular organisms fall within the category for transcription factors (Table 9, which is published as supporting information on the PNAS web site). This finding is in contrast to multicellular organisms, for which 12–15% of the predicted proteins generally fall within the transcription factor category (see e.g., Table 9).
With respect to pigment biosynthesis and photosynthesis, many genes involved in these pathways are found in multiple copies in other photosynthetic eukaryotes. In O. tauri, they also form multigene families, but the copy number is generally lower (e.g., Table 10, which is published as supporting information on the PNAS web site; and see also ref. 35). As expected, O. tauri maintains all essential enzymes for carbon fixation (Table 10), and, based on available data for other algae and land plants, homologs are generally present at half the copy number (35). Double sets of several carbon metabolism-related genes, including phosphoglycerate kinase, ribulose-bisphosphate carboxylase, and triosephosphate isomerase, can be found in the O. tauri genome. Based on both best hit and subsequent phylogenetic analyses these “doubles” each appear to have different origins (bacterial versus eukaryotic).
O. tauri Metabolic Pathways.
O. tauri displays some other characteristics unusual for land plants and algae. For instance, the typical genes encoding the major light-harvesting complex proteins associated with photosystem II (LHCII) are lacking. Instead, paralogs encoding prasinophyte-specific chlorophyll-binding proteins are present, making a special antenna as previously observed in Mantoniella squamata (35). Interestingly, O. tauri also possesses a small set of five lhcA genes, encoding an LHCI antenna. Combined with the absence of major LHCII protein-encoding genes, this finding supports the hypothesis that the LHCI antenna type is more ancestral than is LHCII (35). Unique features are also seen in the carbon assimilation machinery. Only one carbonic anhydrase (CA), most similar to bacterial β-CA, was identified. No carbon-concentrating mechanism (CCM) genes (36) comparable with those of C. reinhardtii or common to organisms that actively or passively enhance inorganic carbon influx were found. However, genes putatively encoding all of the enzymes required for C4 photosynthesis were identified (see Table 10). Whereas C4 photosynthesis has yet to be unequivocally shown in unicellular organisms (24, 25, 37, 38), C4 in the absence of Kranz anatomy is now well documented, especially in Hydrilla verticillata, a facultative C4 aquatic monocot (39). Unlike T. pseudonana, which appears to lack plastid-localized NADP-dependent malic enzymes (NADP-ME), O. tauri has two NADP-ME orthologs most similar to H. verticillata (40) with at least one apparently targeted to the chloroplast based on ChloroP and TargetP predictions. O. tauri also has phosphoenolpyruvate (PEP) carboxylase, NADP+ malate dehydrogenase, and pyruvate-orthophosphate dikinase (Table 10), with predicted chloroplast targeting transit peptides in the latter two. C4 photosynthesis is thought to have evolved multiple times from C3 ancestors. Although timing is uncertain, it is currently thought to have first evolved 24–35 million years ago in relation to environmental pressures (e.g., declining atmospheric CO2) (36, 38). Interestingly, only one member of the Chlorophyta, the macroalga Udotea, has been shown to perform C4 photosynthesis. Udotea utilizes PEP carboxykinase (PEPCK) (NADP-ME being absent) (41), a C4 photosynthesis form variant to that suggested here, although not yet confirmed experimentally, for O. tauri. Despite its energetic cost, if O. tauri is capable of C4 photosynthesis, it could constitute a critical ecological advantage in the CO2-limiting conditions of phytoplankton blooms, especially in circumstances where competitors have lower CCM efficiencies (or no CCM at all).
Resource acquisition is critical to survival in the frequently limiting marine environment, and here O. tauri seems to have developed competitive strategies currently thought uncommon amongst eukaryotic algae. Nitrogen is typically a major limiting nutrient of marine phytoplankton growth. O. tauri is known to grow on nitrate, ammonium, and urea (9), and complete sets of genes allowing transport and assimilation of these substrates have been identified (Fig. 9 and Table 11, which are published as supporting information on the PNAS web site). Interestingly, four genes encoding ammonium transporters were identified, two being green lineage-related and the other two prokaryote-like. Eukaryotic algae are generally considered ineffective competitors for ammonium; however, the high number of ammonium transporters in O. tauri (unlike e.g., T. pseudonana) indicates it may be a strong competitor for this resource. All other genes related to nitrogen acquisition and assimilation are found in a single copy, including those for nitrate, again in contrast to T. pseudonana. It is notable that eight of the genes involved in nitrate uptake and assimilation are found next to each other on chromosome 10 (Fig. 9A), as well as four genes for urea assimilation genes on chromosome 15 (Fig. 9B). A comparable clustering of nitrate assimilation genes was also observed in C. reinhardtii (41) but grouping fewer genes. This organization is reminiscent of prokaryotes, especially cyanobacteria (20), and indicates a possible selective pressure for optimization of nitrate and urea uptake and assimilation, although experimental evidence for the regulation of expression of these genes is currently lacking. The nitrite reductase (NIR) apoenzyme has a unique structure, with two additional redox domains at the C terminus of canonical ferredoxin-NIR, rubredoxin and cytochrome b5 reductase (Fig. 8). This structure should allow this enzyme to use NAD(P)H directly as reducing substrate, which may also contribute to optimization of the pathway. Within this cluster, Snt encodes a protein with weak similarity to sulfate transporters. Nonetheless, its specific position in the cluster suggests that Snt probably encodes a molybdate transporter, a gene predicted to exist but so far unidentified in any species. Taken together with the possibility that O. tauri may be capable of C4 photosynthesis and the relatively high surface area to volume ratio of this tiny phytoplankter, these various ways to optimize nitrogen assimilation could yield a major competitive advantage over other unicellular phytoplankton. This adaptation would be particularly important to its relative success under environmental scenarios, such as intense bloom conditions, where limitation of multiple resources can be encountered.
Finally, O. tauri displays a few traits seemingly more characteristic of land plants than green algae. These traits include the absence of genes encoding the three subunits of the light-independent protochlorophyllide reductase. Thus, like angiosperms, chlorophyll can only be synthesized during the day, owing to the light-dependent protochlorophyllide oxido-reductase gene, present in two copies in the genome. In contrast, the large number of kinase-encoding and calcium-binding domains (Table 12, which is published as supporting information on the PNAS web site) suggests that, as in Arabidopsis and Chlamydomonas, phosphorelay-based calcium-dependent signal transduction systems are commonly used. However, tyrosine kinases appear to be more highly represented in O. tauri than in plants, as is also the case in Chlamydomonas.
In conclusion, the genome structure of O. tauri generally follows predictions of compaction and streamlining that might be driven by its specific lifestyle and ecology. However, the heterogeneity we reveal here concerning two chromosomes raises the challenge of elucidating its origin, which could either be a reminiscence of this alga’s ancient nature or on the contrary more recent adaptations to its environmental niche. It also raises the question of whether this type of heterogeneity is in fact not unique to O. tauri, but rather a common feature of some eukaryotes, given that current understanding of eukaryotic genomes relies on a genome database so far dominated by “higher organisms”. Understanding features specific to success in the marine environment as well as of evolutionary processes within the green lineage relies on new hypotheses and further experimentation for which this complete genome sequence provides a powerful resource. The exceptional features unveiled in the genome of this ubiquitous, ancient, autonomous unicell highlight the fundamental level at which we might reconsider current paradigms.
Materials and Methods
BAC Library.
Genomic DNA was prepared by embedding O. tauri cells in agarose strings, subsequently lysed with proteinase K and partially digested by HindIII. DNA fragments were separated according to size by using pulsed-field gel electrophoresis and electroeluted from the gel. DNA fragments were then ligated to pINDIGO BAC5–HindIII cloning ready (Epicentre Technologies) at a molar ratio insert/vector of 10/1. The ligation product was mixed with EC100 electrocompetent cells (Epicentre Technologies) and electroporated. After 20 h at 37°C on LB chloramphenicol (12.5 μg/ml) plates, recombinant colonies were picked into 384-well microtitre plates containing 60 μl of 2YT medium plus 5% glycerol and 12.5 μg/ml chloramphenicol, grown for 18 h at 37°C, duplicated and stored at −80°C. Two BAC libraries having inserts of ≈50 kb and 130 kb, were prepared, representing a 7-fold coverage of the genome. Clones of both libraries were spotted on high-density filters for further hybridizations, and their ends were sequenced.
Shotgun Libraries.
Purified DNA was broken by sonication, and, after filling ends, DNA fragments ranging from 1 to 5 kb were separated in an agarose gel. Blunt-end fragments were inserted into pBluescript II KS (Stratagene) digested with EcoRV and dephosphorylated. About 60,000 clones were isolated from four independent O. tauri shotgun libraries. Plasmid DNA from recombinant Escherichia coli strains was extracted according to the TempliPhi method (GE Healthcare), and inserts were sequenced on both strands by using universal forward and reverse M13 primers and the ET DYEnamic terminator kit (GE Healthcare). Sequences were obtained with MegaBace 1000 automated sequencers (GE Healthcare). Data were analyzed, and contigs were assembled by using Phred-Phrap (42) and Consed software packages. Gaps were filled through primer-directed sequencing by using custom made primers.
cDNA Library.
Two cDNA libraries were generated from cultures grown under different conditions to improve the representation of the expressed sequences. Exponentially growing cells sampled at various stages of the cell cycle of cultures synchronized by light/dark cycles were mixed with a stationary stage culture. Poly(A) mRNAs from the different cultures were isolated and then mixed together. One cDNA library was created in the λ ZAP vector (Stratagene) and the second in the Gateway system according to the manufacturer’s instructions (Invitrogen). The average insert size analyzed on agarose gels was ≈1.5 kb for both libraries. Sequences were obtained by using the forward primer, and single reads were assembled in contigs by using Phred-Phrap (42).
Genome Annotation.
The genomic sequence of O. tauri was annotated by using the EuGène (43) gene finding system with SpliceMachine (44) signal sensor components trained specifically on O. tauri datasets. A set of 152 GT donor and 152 AG acceptor sites was constructed to optimize the SpliceMachine context representations and to train the splice site sensors that were used to recognize O. tauri splice sites. We found GT donor sites to be highly conserved, which resulted in a highly accurate donor site signal sensor. For acceptor sites, the AG consensus pattern was less conserved, whereas the branch point motif was again highly conserved. SpliceMachine was able to extract this branch point pattern and to use it to recognize AG acceptor sets, again resulting in a highly accurate acceptor site sensor. The content sensor used by EuGène to recognize coding sequences is an interpolated Markov model that was computed from 145 O. tauri ORFs and 167 intron sequences (used as background). Training EuGène requires the estimation of scaling parameters from known O. tauri genes within their genomic context. As such, 17 genomic O. tauri sequences that each contained abutting genes were constructed and used to train EuGène.
Peptides for two deviant chromosomes, numbers 2 and 19, were modeled by using EuGène and SpliceMachine trained specifically on low GC chromosome 2 special genes. A set of 253 GT donor and 253 AG acceptor sites was constructed to optimize the SpliceMachine context representations and to train the sensors used to recognize the splice sites on these two deviant chromosomes. In contrast to the splice sites of the normal O. tauri genes, these GT–AG splice sites were less conserved, resulting in less accurate splice site sensors. However, splice site recognition accuracy was boosted by incorporating intron length constraints (introns in these genes are shorter than in so-called normal genes, with lengths typically between 40 and 60 bp, compared with 170–190 bp for the 18 other chromosomes) at the level of gene recognition. The interpolated Markov model used by EuGène to recognize the special coding sequences was computed from 43 O. tauri ORFs and 209 intron sequences (used as background). Ten genes within their genomic context were used to optimize the scaling parameters within EuGène.
The data sources used to complement the ab initio part of EuGène were composed of O. tauri expressed sequence tags (ESTs), proteins, and genomic sequences. ESTs sequenced over the course of the project were aligned on the genome and used as the most reliable source of extrinsic information. For BlastX, the Swissprot protein dataset (v. 42), C. merolae proteins (26), publicly available C. reinhardtii proteins, and predicted proteins from Sargasso Sea environmental sequences (45) were used in a decreasing order of priority to avoid error propagation, because the latter dataset is the least reliable.
The functional annotation resulted from the synthesis of InterPro and Gene Ontogeny (GO) assignments based on domain occurrences in the predicted proteins by using the InterPro scripts, BlastP against the clusters of eukaryotic orthologous groups (KOG) database, and a top four of BlastP hits (e-value <10−5) against the nonredundant UniProt database. Throughout this process, genes and pathways of particular importance were curated manually by specialists and integrated into the genome annotation. The resulting database is publicly available at http://bioinformatics.psb.ugent.be/genomes/ostreococcus_tauri/in a format that includes browse and query options.
Phylogenetic Analyses.
Homologous genes of O. tauri were searched for in public databases by using BlastP. All top hits were retrieved (up to a significant rise in e-value), and the amino acid sequences were aligned by using ClustalW. Alignment columns containing gaps were removed when a gap was present in >10% of the sequences. To reduce the chance of including misaligned amino acids, all positions in the alignment left or right from the gap were also removed until a column in the sequence alignment was found where the residues were conserved in all genes included in our analyses. Column conservation was determined as follows: For every pair of residues in the column, the BLOSUM62 value was retrieved. If at least half of the pairs had a BLOSUM62 value = 0, the column was considered as conserved.
Neighbor-joining trees were constructed by using TreeCon (46), based on Poisson- and Kimura-corrected distances. Bootstrap analyses with 500 replicates were performed to test the significance of the nodes. Genes were only ascribed to a certain taxon if supported at a bootstrap level >70%.
Supplementary Material
Acknowledgments
This paper is dedicated to André Picard, who passed away in November 2004. He made a major contribution to the field of cell biology applied to marine models. We thank B. Khadaroo and C. Schwartz for technical help in preparation of cDNA libraries, X. Sabau for macroarrays, C. Courties and P. Lagoda for discussions, and F. Dierick and E. Bonnet for bioinformatics help. We also thank I. Grigoriev, J. Grimwood, and B. Palenik for sharing information that helped confirm our assembly. This work was supported by the Génopole Languedoc-Roussillon and the French research ministry and by Région Bretagne (Phostreo) Grant 1043-266-2003 (to F.P. and A.Z.W.). A.Z.W. acknowledges support from a Gordon and Betty Moore Foundation investigator grant. S. Robbens thanks the Institute for the Promotion of Innovation by Science and Technology in Flanders. The work presented here was conducted within the framework of the “Marine Genomics Europe” European Network of Excellence (2004–2008) (GOCE-CT-2004-505403).
Abbreviation
- TE
transposable element.
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The genome data have been submitted to the European Molecular Biology Laboratory, www.embl.org [accession nos. CR954201 (Chrom 1), CR954202 (Chrom 2), CR954203 (Chrom 3), CR954204 (Chrom 4), CR954205 (Chrom 5), CR954206 (Chrom 6), CR954207 (Chrom 7), CR954208 (Chrom 8), CR954209 (Chrom 9), CR954210 (Chrom10), CR954211 (Chrom 11), CR954212 (Chrom 12), CR954213 (Chrom 13), CR954214 (Chrom 14), CR954215 (Chrom 15), CR954216 (Chrom 16), CR954217 (Chrom 17), CR954218 (Chrom 18), CR954219 (Chrom 19), and CR954220 (Chrom 20)].
See Commentary on page 11433.
References
- 1.Courties C., Vaquer A., Troussellier M., Lautier J., Chrétiennot-Dinet M. J., Neveux J., Machado C., Claustre H. Nature. 1994;370:255. [Google Scholar]
- 2.Courties C., Perasso R., Chrétiennot-Dinet M.-J., Gouy M., Guillou L., Troussellier M. J. Phycol. 1998;34:844–849. [Google Scholar]
- 3.Baldauf S. L. Science. 2003;300:1703–1706. doi: 10.1126/science.1085544. [DOI] [PubMed] [Google Scholar]
- 4.Yoon H. S., Hackett J. D., Ciniglia C., Pinto G., Bhattacharya D. Mol. Biol. Evol. 2004;21:809–818. doi: 10.1093/molbev/msh075. [DOI] [PubMed] [Google Scholar]
- 5.Chrétiennot-Dinet M.-J., Courties C., Vaquer A., Neveux J., Claustre H., Lautier J., Machado M. C. Phycologia. 1995;34:285–292. [Google Scholar]
- 6.Derelle E., Ferraz C., Lagoda P., Eychenié S., Cooke R., Regad F., Sabau X., Courties C., Delseny M., Demaille J., et al. J. Phycol. 2002;38:1150–1156. [Google Scholar]
- 7.Díez B., Pedrós-Alió C., Massana R. Appl. Environ. Microbiol. 2001;67:2932–2941. doi: 10.1128/AEM.67.7.2932-2941.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guillou L., Eikrem W., Chrétiennot-Dinet M.-J., Le Gall F., Massana R., Romari K., Pedrós-Alió C., Vaulot D. Protist. 2004;155:193–214. doi: 10.1078/143446104774199592. [DOI] [PubMed] [Google Scholar]
- 9.Worden A. Z., Nolan J. K., Palenik B. Limnol. Oceanogr. 2004;49:168–179. [Google Scholar]
- 10.Zhu F., Massana R., Not F., Marie D., Vaulot D. FEMS Microbiol. Ecol. 2005;52:79–92. doi: 10.1016/j.femsec.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 11.Countway P. D., Caron D. A. Appl. Environ. Microbiol. 2006;72:2496–2506. doi: 10.1128/AEM.72.4.2496-2506.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Worden A. Z. Aquat. Microb. Ecol. 2006;43:165–175. [Google Scholar]
- 13.Li W. K. W. Limnol. Oceanogr. 1994;39:169–175. [Google Scholar]
- 14.Fouilland E., Descolas-Gros C., Courties C., Collos Y., Vaquer A., Gasc A. Microb. Ecol. 2004;48:103–110. doi: 10.1007/s00248-003-2035-2. [DOI] [PubMed] [Google Scholar]
- 15.O’Kelly C. J., Sieracki M. E., Their E. C., Hobson I. C. J. Phycol. 2003;39:850–854. [Google Scholar]
- 16.López-García P., Rodríguez-Valera F., Pedrós-Alió C., Moreira D. Nature. 2001;409:603–607. doi: 10.1038/35054537. [DOI] [PubMed] [Google Scholar]
- 17.Moon-van der Staay S. Y., De Watcher R., Vaulot D. Nature. 2001;409:607–610. doi: 10.1038/35054541. [DOI] [PubMed] [Google Scholar]
- 18.Rodríguez F., Derelle E., Guillou L., Le Gall F., Vaulot D., Moreau H. Environ. Microbiol. 2005;7:853–859. doi: 10.1111/j.1462-2920.2005.00758.x. [DOI] [PubMed] [Google Scholar]
- 19.Moore L. R., Rocap G., Chisholm S. W. Nature. 1998;393:464–467. doi: 10.1038/30965. [DOI] [PubMed] [Google Scholar]
- 20.Rocap G., Larimer F. W., Lamerdin J., Malfatti S., Chain P., Ahlgren N. A., Arellano A., Coleman M., Hauser L., Hess W. R., et al. Nature. 2003;424:1042–1047. doi: 10.1038/nature01947. [DOI] [PubMed] [Google Scholar]
- 21.Campbell L., Nolla H. A., Vaulot D. Limnol. Oceanogr. 1994;39:954–961. [Google Scholar]
- 22.Rocap G., Distel D. L., Waterbury J. B., Chisholm S. W. Appl. Environ. Microbiol. 2002;68:1180–1191. doi: 10.1128/AEM.68.3.1180-1191.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palenik B., Brahamsha B., Larimer F. W., Land M., Hauser L., Chain P., Lamerdin J., Regala W., Allen E. E., McCarren J., et al. Nature. 2003;424:1037–1042. doi: 10.1038/nature01943. [DOI] [PubMed] [Google Scholar]
- 24.Armbrust E. V., Berges J. A., Bowler C., Green B. R., Martinez D., Putnam N. H., Zhou S., Allen A. E., Apt K. E., Bechner M., et al. Science. 2004;306:79–86. doi: 10.1126/science.1101156. [DOI] [PubMed] [Google Scholar]
- 25.Raven J. A., Kübler J. E. J. Phycol. 2002;38:11–16. [Google Scholar]
- 26.Matsuzaki M., Misumi O., Shin-i T., Maruyama S., Takahara M., Miyagishima S.-y., Mori T., Nishida K., Yagisawa F., Nishida K., et al. Nature. 2004;428:653–657. doi: 10.1038/nature02398. [DOI] [PubMed] [Google Scholar]
- 27.Gilson P. R. Genome Biol. 2001;2:1022.1–1002.5. doi: 10.1186/gb-2001-2-8-reviews1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feschotte C., Wessler S. R. Proc. Natl. Acad. Sci. USA. 2002;99:280–285. doi: 10.1073/pnas.022626699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fraser J. A., Heitman J. Mol. Microbiol. 2004;51:299–306. doi: 10.1046/j.1365-2958.2003.03874.x. [DOI] [PubMed] [Google Scholar]
- 30.Ramesh M. A., Malik S.-B., Logsdon J. M., Jr. Curr. Biol. 2005;15:185–191. doi: 10.1016/j.cub.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 31.Chepurnov V. A., Mann D. G., Sabbe K., Vyverman W. Int. Rev. Cytol. 2004;237:91–154. doi: 10.1016/S0074-7696(04)37003-8. [DOI] [PubMed] [Google Scholar]
- 32.Robbens S., Khadaroo B., Camasses A., Derelle E., Ferraz C., Inzé D., Van de Peer Y., Moreau H. Mol. Biol. Evol. 2005;22:589–597. doi: 10.1093/molbev/msi044. [DOI] [PubMed] [Google Scholar]
- 33.Ral J.-P., Derelle E., Ferraz C., Wattebled F., Farinas B., Corellou F., Buléon A., Slomianny M.-C., Delvalle D., d’Hulst C., et al. Plant Physiol. 2004;136:3333–3340. doi: 10.1104/pp.104.044131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coppin A., Varré J.-S., Lienard L., Dauvillée D., Guérardel Y., Soyer-Gobillard M.-O., Buléon A., Ball S., Tomavo S. J. Mol. Evol. 2005;60:257–267. doi: 10.1007/s00239-004-0185-6. [DOI] [PubMed] [Google Scholar]
- 35.Six C., Worden A. Z., Rodríguez F., Moreau H., Partensky F. Mol. Biol. Evol. 2005;22:2217–2230. doi: 10.1093/molbev/msi220. [DOI] [PubMed] [Google Scholar]
- 36.Giordano M., Beardall J., Raven J. A. Annu. Rev. Plant Biol. 2005;56:99–131. doi: 10.1146/annurev.arplant.56.032604.144052. [DOI] [PubMed] [Google Scholar]
- 37.Reinfelder J. R., Milligan A. J., Morel F. M. M. Plant Physiol. 2004;135:2106–2111. doi: 10.1104/pp.104.041319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sage R. F. New Phytol. 2004;161:341–370. doi: 10.1111/j.1469-8137.2004.00974.x. [DOI] [PubMed] [Google Scholar]
- 39.Rao S. K., Magnin N. C., Reiskind J. B., Bowes G. Plant Physiol. 2002;130:876–886. doi: 10.1104/pp.008045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bowes G., Rao S. K., Estavillo G. M., Reiskind J. B. Funct. Plant Biol. 2002;29:379–392. doi: 10.1071/PP01219. [DOI] [PubMed] [Google Scholar]
- 41.Quesada A., Galván A., Schnell R. A., Lefebvre P. A., Fernández E. Mol. Gen. Genet. 1993;240:387–394. doi: 10.1007/BF00280390. [DOI] [PubMed] [Google Scholar]
- 42.Ewing B., Hillier L., Wendl M. C., Green P. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 43.Schiex T., Moisan A., Rouzé P. Lect. Notes Comput. Sci. 2001;2066:111, 125. [Google Scholar]
- 44.Degroeve S., Saeys Y., De Baets B., Rouzé P., Van de Peer Y. Bioinformatics. 2005;21:1332–1338. doi: 10.1093/bioinformatics/bti166. [DOI] [PubMed] [Google Scholar]
- 45.Venter J. C., Remington K., Heidelberg J. F., Halpern A. L., Rusch D., Eisen J. A., Wu D., Paulsen I., Nelson K. E., Nelson W., et al. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 46.Van de Peer Y., De Wachter R. Comput. Appl. Biosci. 1997;13:227–230. doi: 10.1093/bioinformatics/13.3.227. [DOI] [PubMed] [Google Scholar]
- 47.Abrahamsen M. S., Templeton T. J., Enomoto S., Abrahante J. E., Zhu G., Lancto C. A., Deng M., Liu C., Widmer G., Tzipori S., et al. Science. 2004;304:441–445. doi: 10.1126/science.1094786. [DOI] [PubMed] [Google Scholar]
- 48.Haas B. J., Wortman J. R., Ronning C. M., Hannick L. I., Smith R. K., Jr., Maiti R., Chan A. P., Yu C., Farzad M., Wu D., et al. BMC Biol. 2005;3:7. doi: 10.1186/1741-7007-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dietrich F. S., Voegeli S., Brachat S., Lerch A., Gates K., Steiner S., Mohr C., Pöhlmann R., Luedi P., Choi S., et al. Science. 2004;304:304–307. doi: 10.1126/science.1095781. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.