Abstract
RNA-mediated pathogenesis is a recently developed disease model that proposes that certain types of mutant genes produce toxic transcripts that inhibit the activities of specific proteins. This pathogenesis model was proposed first for the neuromuscular disease myotonic dystrophy (DM), which is associated with the expansion of structurally related (CTG)n and (CCTG)n microsatellites in two unrelated genes. At the RNA level, these expansions form stable hairpins that alter the pre-mRNA splicing activities of two antagonistic factor families, the MBNL and CELF proteins. It is unclear which altered activity is primarily responsible for disease pathogenesis and whether other factors and biochemical pathways are involved. Here, we show that overexpression of Mbnl1 in vivo mediated by transduction of skeletal muscle with a recombinant adeno-associated viral vector rescues disease-associated muscle hyperexcitability, or myotonia, in the HSALR poly(CUG) mouse model for DM. Myotonia reversal occurs concurrently with restoration of the normal adult-splicing patterns of four pre-mRNAs that are misspliced during postnatal development in DM muscle. Our results support the hypothesis that the loss of MBNL1 activity is a primary pathogenic event in the development of RNA missplicing and myotonia in DM and provide a rationale for therapeutic strategies designed either to overexpress MBNL1 or inhibit MBNL1 interactions with CUG and CCUG repeat expansions.
Keywords: microsatellite, muscle, pathogenesis, splicing, transgenic
During mammalian embryonic and postnatal periods, tissues are initially formed and then extensively remodeled to meet the evolving demands of the developing organism. This postnatal remodeling process requires a dynamic series of biochemical events at both the transcriptional and posttranscriptional levels. For vertebrate skeletal muscle, myogenic precursor cells derived from somites migrate during embryogenesis and, ultimately, fuse to form primary and secondary myofibers (1). After birth, fetal muscle must be modified to perform adult functions, and this transition (e.g., a suckling versus a highly mobile adult mouse) requires the sequential replacement of fetal protein isoforms with their adult counterparts. For some muscle genes, this process involves skipping of fetal exons and/or the inclusion of adult exons during pre-mRNA splicing. Remarkably, this normal splicing transition is altered specifically in the neuromuscular disease myotonic dystrophy (DM) (2–4).
Adult-onset DM is a multisystemic degenerative disease (4). Characteristic features of this disorder include skeletal muscle myotonia (muscle hyperexcitability), weakness/wasting, heart conduction defects, particulate subcapsular cataracts, and insulin insensitivity. The genetic basis of DM is unusual because it is associated with different unstable microsatellites in two unrelated genes. DM type 1 (DM1) is caused by the expansion of a (CTG)n repeat in the 3′ UTR of the DMPK(dystrophia myotonica protein kinase) gene (5). Normal DMPKmicrosatellite lengths range from 5–37, whereas the DM1-affected population carries expansions from >50 to ≈3,000 CTG repeats. In contrast, DM type 2 (DM2) results from a (CCTG)n expansion (within a more complex [TG]n[TCTG]n[CCTG]n repeat motif) in the first intron of ZNF9(zinc finger 9) (6). These DM2 expansions can be massive with a disease range of 75 to >11,000 CCTG repeats.
Early pathogenesis models for DM1 speculated that the CTG repeat expansion resulted in DMPKlocus-specific effects either by causing haploinsufficiency of DMPKalone or in combination with the tightly linked genes, DMWDand SIX5(2–4). The discovery of the DM2 mutation in the unlinked ZNF9gene provided a strong argument against these models. Alternatively, several lines of evidence support an RNA-mediated pathogenesis mechanism that proposes that CUG and CCUG repeats in the pathogenic range fold into RNA hairpins that are not exported from the nucleus but instead accumulate within nuclear, or ribonuclear, foci (7–9). Several studies have confirmed the existence of these dsRNA structures, including a recent crystal structure of an 18-bp RNA containing six CUG repeats that stacks end-on-end to form pseudocontinuous A form helices (10–14). The RNA pathogenesis model also has been directly tested by the generation of transgenic mice carrying a human skeletal α-actin (HSA) gene modified by the insertion of either 5 (HSA short repeat, HSASR) or 250 (HSA long repeat, HSALR) CTG repeats in the HSA 3′ UTR (15). The HSASR mice carrying a repeat in the normal range are indistinguishable from control FVB/n (the background strain for HSALR) mice. In contrast, HSALR mice develop severe myotonia and dystrophic muscle features characteristic of DM disease. Importantly, lines that fail to express the HSALR transgene, presumably due to integration at a silenced site, do not develop myotonia. Thus, transcription of this CTG expansion is required for the development of myotonia and muscle pathology.
Why are (CUG)n and (CCUG)n expansions pathogenic? Current evidence indicates that these mutant RNAs interfere with the activities of two protein families that act antagonistically during alternative splicing, the muscleblind-like (MBNL) and CUGBP1/ETR-3-like factors (CELF) (8, 16–19). During development, the CELF proteins promote the inclusion of specific fetal exons in embryonic and neonatal tissues, whereas postnatal activation of MBNL leads to fetal exon skipping and expression of adult protein isoforms. The MBNL proteins, which have a high affinity for (CUG)n and (CCUG)n expansion RNAs, are sequestered in DM skeletal muscle and neuronal ribonuclear foci (8, 20–24). Consequently, MBNL splicing activity is compromised, resulting in the retention of fetal isoforms in the adult (18, 19). This MBNL loss-of-function hypothesis for DM pathogenesis was tested by generating an isoform-specific knockout of mouse Mbnl1(Mbnl1ΔE3/ΔE3) (18). These Mbnl1ΔE3/ΔE3 mice develop characteristic features associated with this disease, including myotonia and histopathological changes to skeletal muscle (central nuclei, fiber size heterogeneity), subcapsular cataracts, and improper splicing of those exons that are incorrectly spliced in DM tissues. These observations suggest that loss of alternative splicing regulation results directly from the loss of MBNL1 function and unopposed CELF activity.
Although MBNL1 sequestration appears to play a prominent role in DM pathogenesis, additional factors have been implicated in the disease process. For example, the DM1 and DM2 expansions also affect CELF activity because CUGBP1 steady-state levels and splicing activity increase in DM1 cells (25–27). Although it is unclear how (CUG)n and (CCUG)n expansion RNAs up-regulate CELF protein levels, transgenic mice overexpressing CUGBP1 develop DM-associated missplicing patterns (28). In addition to MBNL, another factor implicated in the regulation of alternative splicing, hnRNP H, also accumulates in ribonuclear foci (29). Finally, CUG and CCUG repeat expansion RNAs might sequester transcription factors (30). In this study, we address the role of Mbnl1 sequestration in vivo by using adeno-associated virus (AAV)-mediated transduction to overexpress this protein in HSALR skeletal muscle. Our results demonstrate that elevated expression of Mbnl1 alone is sufficient to rescue the myotonia and aberrant splicing of specific gene transcripts characteristic of DM skeletal muscle.
Results
Selection of the Mbnl Isoform for Overexpression in Skeletal Muscle.
There are three MBNLgenes in humans and mice (8, 31). Cotransfection analysis of HEK293 and HeLa cells with minigene splicing reporters and human MBNL expression plasmids demonstrates that all three MBNL proteins (MBNL1, MBNL2, and MBNL3) promote fetal exon exclusion to a similar extent (19). Nevertheless, we chose Mbnl1 for AAV-mediated overexpression in HSALR skeletal muscle primarily because transgenic HSALR and Mbnl1ΔE3/ΔE3 knockout models for DM both develop myotonia, distinctive morphological changes to muscle structure, and remarkably similar adult missplicing patterns (15, 18, 24). Because loss of Mbnl1 function and expression of a CUG repeat expansion results in a similar muscle phenotype, this study was designed to test the prediction that overexpression of a single Mbnl1 isoform would reverse RNA missplicing and histopathology in HSALR muscle. We speculated that overexpression of any Mbnl1 isoform with a high affinity for CUG repeat expansions might saturate dsCUG-binding sites and release the endogenous Mbnl pool. This speculation is supported by a recent fluorescence recovery after photobleaching study on DM1 fibroblasts that reported relatively rapid exchange rates between MBNL proteins within ribonuclear foci and the surrounding nucleoplasmic pool (32).
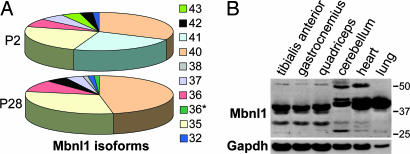
Another question focused on which Mbnl1 isoform should be overexpressed in HSALR skeletal muscle. The mouse Mbnl1gene encodes at least 14 isoforms that use two different initiation codons (18). Isoforms that initiate in exon 3 contain four copies of a zinc-knuckle-like motif CCCH (4XC3H proteins) that is required for RNA binding (8, 18, 23). The isoforms that use the exon 4 initiation codon contain only two C3H motifs (2XC3H), similar to the Drosophila muscleblind proteins (8). To identify candidates for AAV-mediated muscle expression, the predominant Mbnl1 isoforms expressed in adult mouse skeletal muscle were identified. Because loss of 4XC3H Mbnl1 isoforms alone is sufficient to cause the multisystemic DM-like phenotype in Mbnl1ΔE3/ΔE3 mice, coding sequences for these isoforms were PCR-amplified from mouse postnatal day (P) 2 hindlimb and P28 quadriceps cDNAs by using primers positioned in Mbnl1exons 3 and 13. This RNA splicing transition from fetal exon inclusion to exclusion occurs before P20 (24). For each time point, the complete DNA sequences of 72–81 Mbnl1 cDNA clones were determined. At P2, the major Mbnl1 4XC3H isoforms are 41, 40, and 35 kDa, whereas, by P28, the 41-kDa isoform was no longer expressed (Fig. 1A). Seven additional isoforms (43, 42, 38, 37, 36, 36*, and 32) also were expressed at significantly lower levels. Immunoblot analysis by using the A2764 polyclonal antibody directed against the Mbnl1-specific carboxyl-terminal peptide confirmed that the major adult protein in three different skeletal muscles (tibialis anterior, gastrocnemius, and quadriceps) and heart was 40 kDa, whereas the major isoforms in cerebellum and lung were slightly larger (Fig. 1B).
Fig. 1.
Expression of Mbnl1 isoforms in skeletal muscle. (A) Distribution of Mbnl1 isoforms in neonatal (P2) and P28 muscle. Ten hindlimb muscle isoforms were identified by RT-PCR followed by cDNA sequencing. Isoforms (color coded) are as follows (in kDa): 43, lime green; 42, black; 41, turquoise; 40, orange; 38, purple; 37, light blue; 36, red; a 36 spliced variant (36*, dark green); 35, yellow; and 32, blue. (B) The major Mbnl1 protein in adult skeletal muscle is 40 kDa. Immunoblot analysis was performed on three skeletal muscles (tibialis anterior, gastrocnemius, and quadriceps), cerebellum, heart, and lung by using either anti-Mbnl1 or anti-Gapdh (loading control).
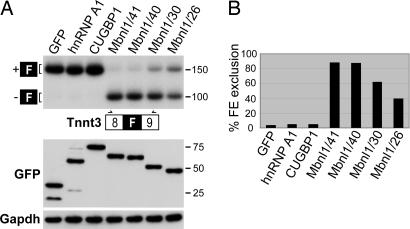
We pursued the possibility of using the Mbnl1 41-kDa isoform (Mbnl1/41) for AAV-mediated transduction for several reasons. First, MBNL1/41 has a high affinity for a CUG repeat expansion RNA [(CUG)54] with a Kd of 2.5 nM (data not shown). Second, this isoform was absent in adults, so the relative levels of endogenous versus transduced Mbnl1 could be simultaneously ascertained by immunoblotting with an anti-Mbnl1 antibody. Third, we wanted to determine whether Mbnl1/41 would function as a splicing factor in adult muscle. To address this latter question, we first used a HEK293T cotransfection assay to compare the splicing activities of various Mbnl1 isoforms and unrelated RNA-binding proteins. For the minigene reporter, fast skeletal muscle troponin T (Tnnt3) was selected because the splicing of this fetal exon is very sensitive to Mbnl1 levels in vivo (18). For protein expression, GFP fusion expression plasmids were used so that transfection efficiency could be monitored readily. Although overexpression of other RNA-binding proteins, including the splicing factors hnRNP A1 and CUGBP1, failed to alter the F exon-splicing pattern, all Mbnl1 isoforms tested significantly enhanced F exon skipping (Fig. 2). The Mbnl1 40- and 41-kDa isoforms showed the highest Tnnt3 F exon-skipping activity. Importantly, there was no difference in the splicing activities of these 4XC3H isoforms in this assay with ≈85% Tnnt3 fetal exon exclusion in cells expressing either GFP-Mbnl1/40 or GFP-Mbnl1/41.
Fig. 2.
Splicing assay for Mbnl1 isoforms. (A Upper) RNA splicing in HEK293T cells transfected with a Tnnt3 minigene reporter and protein expression plasmids containing GFP, GFP-hnRNP A1 and GFP-CUGBP1, and GFP-Mbnl1 4XC3H (Mbnl1/40 and Mbnl1/41) and 2XC3H (Mbnl1/30 and Mbnl1/26) isoforms. (A Lower) An immunoblot of GFP fusion protein expression showing equivalent expression levels for GFP and GFP-fusion proteins after transfection. Protein loading control is Gapdh. PCR primers (arrows) are located in Tnnt3 exons 8 and 9 (open boxes) bordering the alternatively spliced fetal (F) exon (filled box). (B) Phosphorimager quantification of percent fetal exon (FE) exclusion from the PCR data shown in A.
Reversal of Myotonia in a Poly(CUG) Model for DM.
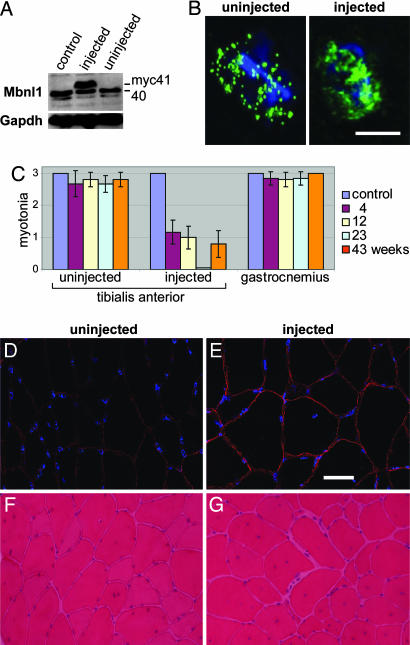
For Mbnl1 overexpression, we selected the tibialis anterior (TA) muscle because it is relatively small, readily accessible without surgery, and efficiently transduced by AAV (33). Four-week-old HSALR mice were injected with 1 × 1011 vector genomes in the right TA with AAV2/1 (AAV2 ITR in an AAV1 capsid) modified to express the myc-tagged Mbnl1 41-kDa isoform (AAV2/1-mycMbnl1/41). Relative levels of endogenous 40 kDa versus exogenous mycMbnl1/41 expression were assessed by immunoblotting. At 23 weeks after injection, the mycMbnl1/41 protein was present in injected, but not in uninjected, TA muscles (Fig. 3A). Compared with the uninjected TA, the level of the endogenous 40-kDa protein in the injected muscle was reduced ≈20% when normalized to Gapdh. Despite this reduction, there was an overall 2-fold increase in Mbnl1 protein due to AAV2/1-mycMbnl1/41 expression. Because of the high expression level of the HSALR transgene, numerous discrete ribonuclear foci containing Mbnl1, which colocalizes with (CUG)250 RNA (20), were detectable in transgenic myonuclei (Fig. 3B). In contrast, Mbnl1 was more diffusely distributed in the nucleus after AAV-mediated overexpression, suggesting that a subpopulation of this splicing factor was no longer sequestered in these foci.
Fig. 3.
Rescue of myotonia after Mbnl1 overexpression. (A) Mbnl1 is overexpressed after AAV2/1-mycMbnl1/41 transduction of TA muscle. TA muscles (23 weeks after injection) were dissected from either uninjected (control) or injected HSALR mice (both injected and uninjected muscles are shown) and total protein immunoblotted with anti-MBNL1 antibody. Gapdh is the protein loading control. (B) Distribution of Mbnl1 protein in transverse sections of skeletal muscle. Shown are max-value projections of deconvolved images obtained under identical exposure settings. HSALR sections from uninjected (Left) and injected (Right) TA muscles were stained by using the anti-Mbnl1 antibody. (Scale bar: 5 μm.) (C) Myotonia was assessed by electromyography on HSALR mice injected in the right TA. The uninjected left TA and gastrocnemius muscles also were tested. The electromyography scale is as follows: 0, no myotonia; 1, occasional myotonic discharge in <50% of needle insertions; 2, myotonic discharge with >50% of insertions; 3, myotonic discharge with nearly all insertions. Uninjected control and injected HSALR mice were tested at 4 (purple), 12 (yellow), 23 (turquoise), and 43 (orange) weeks after injection. (D and E) Restoration of Clcn1 protein levels in myofiber membranes after Mbnl1 overexpression. Clcn1 protein levels were detected in uninjected (D) and injected (E) transverse muscle sections at 23 weeks after injection by using an anti-Clcn1 polyclonal antibody (red). DNA distribution is shown by DAPI staining (blue). (Scale bar: 10 μm.) (F and G) Muscle histology (H&E staining) of muscle sections from uninjected (F) and injected (G) TA at 43 weeks after injection.
Electrical myotonia is a prominent pathological feature of both human DM and mouse HSALR skeletal muscle. Electromyography revealed a striking reduction of myotonia specifically in the HSALR-injected TA by 4 weeks after injection and a complete absence of myotonic discharges at 23 weeks (Fig. 3C). By 43 weeks after injection, muscle hyperexcitability again was detectable in some mice, whereas the myotonia present in both uninjected HSALR TA and gastrocnemius muscles was unaffected at all time points assayed. Moreover, control TA injections with AAV expressing only GFP driven by the CBA promoter (AAV2/1-GFP) showed no effect in this model (data not shown).
Current evidence suggests that myotonia in DM results from missplicing of the major skeletal muscle chloride channel CLCN1/ClC-1 in adults, which results in loss of functional membrane-associated chloride channels. Although Clcn1 protein levels were reduced markedly in HSALR muscle compared with control FVB muscle levels, Clcn1 was restored to near wild-type levels in injected TA muscles at 23 weeks after injection (Fig. 3 D and E). Mouse HSALR muscles show DM-relevant morphological abnormalities, including centralized nuclei, split fibers, and fiber size heterogeneity in the absence of significant muscle necrosis (15). In contrast to the myotonia, these abnormalities still were present in injected TA muscles at all time points examined (Fig. 3 F and G).
Severity of Myotonia Correlates with Altered Splicing of Specific Developmentally Regulated Exons.
Recent studies have shown that the splicing of specific pre-mRNAs is affected in human DM and HSALR and Mbnl1ΔE3/ΔE3 mouse models. Because myotonia was eliminated by 23 weeks after injection of AAV2/1-mycMbnl1/41, we characterized the alternative splicing of these exons in FVB/n, HSALR, and AAV2/1-mycMbnl1/41-injected mice.
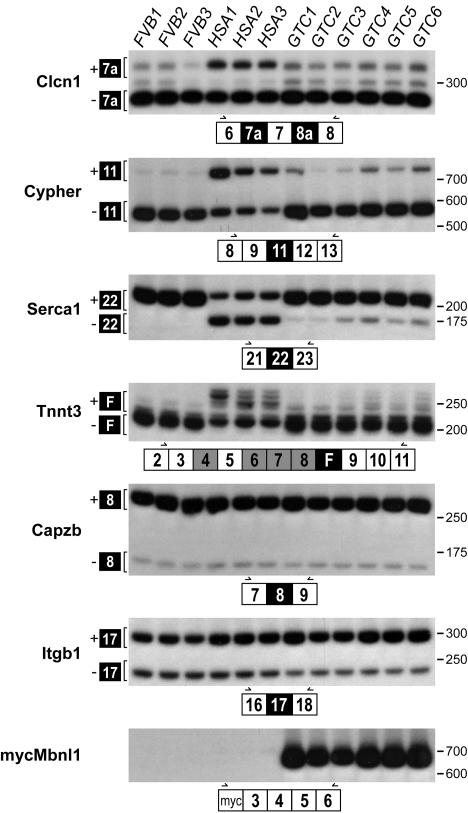
Although myotonia in humans is associated with mutations in the skeletal muscle sodium (SCN5A) and chloride (CLCN1/ClC-1) channels, several studies have concluded that DM-associated myotonia is due to CLCN1 missplicing (34, 35). In normal neonatal and adult DM muscle, CLCN1 intron 2 and exons 7a and 8a are more frequently included during splicing. These sequences contain in-frame termination codons that may reduce CLCN1 mRNA levels, via the nonsense-mediated decay pathway, or result in the synthesis of dominant-negative truncated CLCN1 proteins (36). Whereas in wild-type FVB mice exons 6, 7, and 8 are spliced directly together, exon 7a is included in HSALR muscle (Fig. 4). Overexpression of Mbnl1 reversed this splicing defect by promoting Clcn1 exon 7a exclusion.
Fig. 4.
Mbnl1 overexpression promotes adult splicing patterns. RT-PCR splicing assays of uninjected FVB/n and HSALR (three mice each) or AAV2/1-mycMbnl1/41 (gene therapy group C, GTC) injected TA at 23 weeks after injection (six mice). Developmentally regulated exons (filled boxes) are either dysregulated in DM (Clcn1, Ldb3/Cypher, Serca1, and Tnnt3) or not affected by the DM expansion mutations (Capzb and Itgb1). Primer positions (arrows) are illustrated below each autoradiograph within constitutively spliced exons (open boxes). The overexpressed mycMbnl1/41 is detectable only in the GTC mice (Bottom).
Another pre-mRNA that is misspliced in DM is Ldb3/Cypher/Zasp, which encodes a striated muscle PDZ-LIM domain protein that localizes to the Z line and interacts with α-actinin 2. Although Cypher expression is not essential for sarcomerogenesis or Z line function, Cypher proteins are important for normal muscle function because Cypher knockout mice die perinatally because of severe congenital myopathy, and Cypher is misspliced in DM1 and DM2 muscle (24, 37). Interestingly, human CYPHER/ZASP mutations recently have been linked to a novel autosomal dominant muscular dystrophy (38). During development, the embryonic Cypher isoform (Cypher1S) is replaced postnatally by Cypher 3S by skipping of Cypher exon 11. As expected, normal FVB adults showed exon 11 skipping, whereas HSALR mice recapitulated the wild-type neonatal pattern with equivalent levels of exon 11 inclusion and exclusion. At 23 weeks after injection, the adult Cypher splicing pattern was restored in AAV2/1-mycMbnl1/41-injected HSALR TA muscles, with the majority of mRNAs excluding exon 11.
Intracellular skeletal muscle calcium homeostasis is regulated by the sarcoplasmic reticulum (SR) proteins, ryanodine receptor 1 (RyR1) and SR/endoplasmic reticulum Ca2+ ATPase (Serca). Whereas RyR1 releases Ca2+ from the SR, Serca is the skeletal muscle SR Ca2+ reuptake pump. A recent study demonstrated that Serca1 exon 22 is included in adult muscle but excluded in neonatal, as well as adult DM1 and mouse HSALR, muscle (39). In agreement, exon 22 was predominantly included in FVB adult Serca1 mRNA, whereas the neonatal pattern of ∼50/50 exon 22 inclusion/exclusion was observed in HSALR adult TA. Overexpression of Mbnl1 after AAV2/1-mycMbnl1/41 injection led to a significant increase in exon 22 inclusion and near-normal adult Serca1 splicing.
Fast skeletal muscle troponin T (TNNT3) is the subunit of the troponin complex that binds to tropomyosin. Although TNNT3 contains several alternatively spliced exons (4, 6, 7, 8, F, 16, and 17), only F exon splicing is altered in DM (18). The requirement for F exon inclusion in fetal muscle is unclear. In HSALR adult muscle, the F exon is included together with different combinations of alternatively spliced exons 4–8, which yields multiple cDNAs containing the F exon, whereas the F exon is excluded from normal FVB TA adult muscle. In agreement with the hypothesis that Mbnl1 is the primary regulator of this exon, nearly all F exon inclusion was eliminated in injected TA muscle. In contrast, alternative splicing of Capzb exon 8 and Itgb1 exon 17, two pre-mRNAs whose splicing is not affected by the DM expansion mutation, were unaffected in HSALR and AAV-transduced mice compared with FVB controls.
Discussion
Our results demonstrate that AAV-mediated overexpression of a single Mbnl1 isoform reverses the muscle hyperexcitability and aberrant splicing of those exons misspliced in HSALR muscle. An initial concern with our experimental strategy was that HSALR myonuclei express a CUG repeat expansion at a higher level than detectable in human DM muscle because transgene expression is driven by the human α-actin, rather than the considerably less active DMPK, promoter. Moreover, Mbnl1 levels in skeletal myonuclei increased only 2-fold after AAV-mediated transduction. Surprisingly, this level of Mbnl1 overexpression led to a significant reduction in myotonia as early 4 weeks after injection of AAV2/1-mycMbnl1/41. Muscle hyperexcitability was absent by 23 weeks after injection with the concurrent reversal of the fetal splicing patterns for several pre-mRNAs (Clcn1, Cypher, Serca1, and Tnnt3) that are misspliced in HSALR adult muscle. Absence of myotonia correlated with a slightly higher level of myc-positive myonuclei and myofibers at 23 weeks as detected by immunohistochemistry (Fig. 5, which is published as supporting information on the PNAS web site). We conclude that the myotonia and RNA-splicing defects in this poly(CUG) mouse model for DM are coupled and are the direct result of functional inactivation of Mbnl1.
The return of myotonia detected at 43 weeks after AAV-mediated transduction might result from the enhanced regeneration typical of HSALR muscle coupled with inefficient transduction of satellite cells by AAV2/1-mycMbnl1/41. Muscle turnover also may be promoted by expression of the nontolerized myc-tagged Mbnl1/41 protein, leading to an elevated immune response and a decrease in the number of myofibers expressing mycMbnl1. In support of this interpretation, a previous study on the mdx mouse model for Duchenne muscular dystrophy, as well as CVBA3′ minidystrophin mdx mice, demonstrated loss of AAV-mediated transgene expression likely due to an elevated immune response (40). Testing this possibility by overexpressing the untagged Mbnl1 40-kDa adult isoform in HSALR skeletal muscle should be performed. A study also should address the possibility that overexpression of the Mbnl1 41-kDa isoform, which is expressed in neonatal but not adult skeletal muscle, is incompatible with long-term rescue of adult myotonia.
Whereas Mbnl1 overexpression rescued myotonia and RNA missplicing, normal myofiber structure was not restored to AAV-transduced HSALR muscle. Of course, HSALR muscle histopathology may not be reversible. However, AAV-mediated expression of δ-sarcoglycan (δ-SG) restores normal muscle weight, specific twitch and titanic force, and normal muscle histology within 14 weeks after virus injection of the δ-SG-deficient Bio14.6 hamster model for limb girdle muscular dystrophy 2F (33). DM muscle is characterized by an unusual combination of numerous centralized nuclei, ringed fibers, and type 1 fiber atrophy without the high degree of necrosis, fibrosis, and regenerative activity typical of other muscular dystrophies, including Duchenne muscular dystrophy. Our results support previous observations that DM-relevant histological changes probably do not result from prolonged myotonia. Centralized nuclei are uncommon in myotonia congenita, and adr mice, which lack Clcn1 expression, develop severe myotonia without a significant increase in centralized myonuclei and fiber size variability (41). Moreover, double mutant adr-mdx mice show a less severe dystrophic phenotype than mdx mice.
We favor the possibility that the level of Mbnl1 overexpression achieved in this study was insufficient to saturate dsCUG-binding sites in HSALR muscle and, thus, release endogenous Mbnl, and/or other sequestered, proteins required to restore normal myofiber structure. Because the entire repertoire of Mbnl pre-mRNA splicing targets is unknown, missplicing of uncharacterized target exons may be responsible for the histopathological defects characteristic of DM muscle. Alternatively, (CUG)n expansion RNA may impair other biochemical pathways. Indeed, MBNL2 recently has been implicated in mRNA-directed protein localization of integrin α3 to adhesion complexes (42).
Materials and Methods
Transgenic Mice.
The HSALR 20b line was used for this study. Intergenerational stability of the (CTG)250 expansion was monitored by PCR with AmpliTaq Gold (Applied Biosystems, Foster City, CA) as described in ref. 15.
AAV2/1-mycMbnl1/41 Virus Preparation and Injection Protocol.
A myc tag was added to the human MBNL1/41 ORF (ORF, AF401998) by using sequential PCR rounds with the same reverse primer (MSS1580) and the following four forward primers: 1, MSS1576; 2, MSS1577; 3, MSS1578; 4, MSS1579 (see Data Set 1, which is published as supporting information on the PNAS web site). Amplification for each round was performed for 25 cycles (98°C for 20 sec, 60°C for 30 sec, and 72°C for 90 sec). The PCR product was subcloned into pCR4-TOPO (Invitrogen, Carlsbad, CA) to create mycMBNL1/41. Human and mouse protein sequences vary by 2 aa in exon 5. Therefore, human cDNA was converted into a cDNA that encodes the mouse protein sequence by digestion of the mouse Mbnl1/41 ORF with HincII and HindIII followed by ligation into HincII/HindIII-digested mycMBNL1/41.
The mycMbnl1/41 cDNA was subcloned into SpeI/ClaI-cut pTR-UF12Δ (IRES-GFP deleted) to create pTR-UF12Δ-mycMbnl1/41. The plasmids pTR-UF12Δ-mycMbnl1/41 and pXYZ1 were used for virus preparation in HEK293 cells by using a two plasmid cotransfection protocol (43). The AAV-mycMbnl1/41 vector was purified further by using a HiTrap Q column (GE Healthcare, Piscataway, NJ) (44), and the concentration (1.24 × 1013 vector genomes/ml) was determined by using a standardized real-time qPCR titration technique (45). The right TA muscles of 4- to 5-week-old HSALR 20b mice were injected with one vector dose (1 × 1011 vector genomes in 30 μl of PBS) by using a 29-gauge needle.
Analysis of Mbnl1 Isoforms in Adult Muscle.
To determine the major Mbnl1 isoforms expressed in C57BL6/J skeletal muscle, total RNA was isolated from P2 hindlimb muscle and P28 quadriceps by using TRI Reagent (Sigma, St. Louis, MO). cDNA was generated by using 5 μg of RNA by random hexamer-primed reverse transcription by using SuperScript II (Invitrogen) followed by digestion with 0.2 units/μl RNase H (Invitrogen) at 37°C for 15 min. Mbnl1 isoforms were PCR-amplified through 35 cycles (95°C for 30 sec, 50°C for 30 sec, and 72°C for 90 sec) by using forward (MSS2161) and reverse (MSS2162) primers, and the product was cloned into pCR4-TOPO. Multiple clones from P2 and P28 were randomly picked, and full-length cDNA coding sequences were obtained for both P2 (n = 72) and P28 (n = 81).
For immunological detection of Mbnl1, 9-month-old HSALR tissues (tibialis anterior, gastrocnemius, quadriceps, cerebellum, heart, and lung) were homogenized in 50 mM Tris·Cl, pH = 6.8/1 mM EDTA/2% SDS/0.5 mM phenylmethylsulfonyl fluoride/5 μg/ml pepstatin A/1 μg/ml chymostatin/1 mM aminocaproic acid/1 mM p-aminobenzamidine/1 μg/ml leupeptin/2 μg/ml aprotinin. Homogenates were centrifuged at 16,000 × g for 10 min, and supernatant proteins were fractionated on 12.5% SDS-polyacrylamide gels (50 μg protein per lane), electroblotted to nitrocellulose, immunoblotted, and visualized by ECL. Antibodies for immunoblotting included the rabbit anti-Mbnl1 polyclonal A2764 (1:1,000) and the anti-GAPDH monoclonal 6C5 (1:10,000; Novus Biologicals, Littleton, CO). The same procedure was performed for immunological detection of mycMbnl1/41 in AAV-transduced TA muscles.
RNA Splicing.
For splicing analysis in cell culture, HEK293T cells were plated in six-well plates in DMEM (Invitrogen), supplemented with 10% FBS (Invitrogen) and 1% penicillin-streptomycin (Invitrogen). The next day, the cells were cotransfected with 1 μg of the Tnnt3 minigene and 10 ng of protein expression plasmids by using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. Protein and RNA were harvested 48 h after transfection. For analysis of GFP-fusion protein expression after transfection, proteins (20 μg per lane) were fractionated and detected by immunoblotting with anti-GFP (1:1,000; Roche, Indianapolis, IN) and anti-GAPDH mAb 6C5 (1:10,000). For RNA-splicing analysis, first-strand cDNA was generated from total RNA by reverse transcription with 5 μg of total RNA and SuperScript II RNase H-RT (Invitrogen). Subsequent PCR (28 cycles at 95°C for 30 sec, 55°C for 30 sec, and 72°C for 30 sec) was performed by using 20% of the reverse transcription reaction as a template. Each PCR was spiked with 5 μCi (1 Ci = 37 GBq) of [α32P]-dCTP (PerkinElmer, Wellesley, MA). Tnnt3 minigene mRNA was analyzed by using exon 8 forward (MSS1956) and exon 9 reverse (MSS1938) primers. PCR products were resolved on 10% nondenaturing polyacrylamide gels followed by autoradiography with Biomax MS film (Eastman Kodak, Rochester, NY). Splicing shifts were measured by using a Pharox FX plus Molecular Imager (Bio-Rad, Hercules, CA).
Splicing patterns of rAAV-injected and uninjected muscles were monitored by radioactive RT-PCR as described in ref. 18. Primers used were as follows: Serca1, exon 21 forward (MSS2761) and exon 23 reverse (MSS2762); Cypher, exon 8 forward (MSS2763) and exon 13 reverse (MSS2764); Tnnt3, exon 2 and 3 overlapping forward (MSS1677) and exon 11 reverse (MSS1678); Clcn1, exon 6 forward (MSS2788) and exon 8 reverse (MSS1653); Capzb, exon 7 forward (MSS2765) and exon 9 reverse (MSS2766); Itgb1, exon 16 forward (MSS2767) and exon 18 reverse (MSS2768); myc-Mbnl1. forward primers in myc (MSS1579) and reverse primers in Mbnl1 exon 6 (MSS1656).
Electromyography.
Before electromyography, mice were anesthetized by using i.p. 100 mg/kg ketamine, 10 mg/kg xylazine, and 3 mg/kg acepromazine or 250 mg/kg 2,2,2 tribromoethanol. Electromyography on tibialis anterior and gastrocnemius muscles was performed as described in ref. 18 by using 30 gauge concentric needle electrodes and a minimum of 10 needle insertions for each muscle. For each time point (4, 12, 23, and 43 weeks after injection), at least 6 mice were evaluated, and both uninjected and injected TA, as well as gastrocnemius, muscles were tested.
Histology and Immunohistochemistry.
Frozen sections (10 μm) of TA muscle were either prepared for routine H&E staining or immunostained by using antibodies against the ClC-1 C terminus (Alpha Diagnostic, San Antonio, TX) or the Mbnl1 C-terminal peptide (rabbit polyclonal antibody A2764) as described in ref. 34 with the following modifications. For Mbnl1, sections were fixed in 4% paraformaldehyde for 15 min at room temperature and permeabilized with 2% acetone in PBS for 5 min. For ClC-1, sections were unfixed. Primary antibody concentrations used were 1:10,000 for Mbnl1 and 1:50 for ClC-1. Secondary antibodies were goat anti-rabbit Alexa Fluor 488 and 546 (Invitrogen) at 1:400. Stacks of Z plane images were deconvolved by using a blind point spread function (46). For comparison of injected and uninjected muscle, sections were stained on the same slide, and images were obtained and processed under the same exposure and threshold settings (see Supporting Materials and Methods, which is published as supporting information on the PNAS web site).
Supplementary Material
Acknowledgments
We thank Regina Shaw of the Interdisciplinary Center for Biotechnology Research Genomics Core for sequence analysis and Karen Johnstone and Chris Futtner (University of Florida) for gifts of FVB/n mice. This study was supported by a grant from the Muscular Dystrophy Association and National Institutes of Health Grants AR46799 and NS48843 (to M.S.S.).
Abbreviations
- AAV
adeno-associated virus
- CELF
CUGBP1/ETR3-like factors
- DM
myotonic dystrophy
- DMPK
dystrophia myotonica protein kinase
- HSA
human skeletal α-actin
- MBNL
muscleblind-like
- Pn
postnatal day n
- TA
tibialis anterior.
Footnotes
Conflict of interest statement: M.S.S. is associated with a gene therapy company that has optioned a license for AAV-mediated correction of myotonic dystrophy.
References
- 1.Charge S. B., Rudnicki M. A. Physiol. Rev. 2004;84:209–238. doi: 10.1152/physrev.00019.2003. [DOI] [PubMed] [Google Scholar]
- 2.Ranum L. P., Day J. W. Trends Genet. 2004;20:506–512. doi: 10.1016/j.tig.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 3.Nykamp K. R., Swanson M. S. Prog. Mol. Subcell. Biol. 2004;35:57–77. doi: 10.1007/978-3-540-74266-1_3. [DOI] [PubMed] [Google Scholar]
- 4.Thornton C. A., Swanson M. S., Cooper T. A. In: Genetic Instabilities and Hereditary Neurological Diseases. Ashizawa T., Wells R.D., editors. San Diego, CA: Elsevier-Academic; 2006. [Google Scholar]
- 5.Brook J. D., McCurrach M. E., Harley H. G., Buckler A. J., Church D., Aburatani H., Hunter K., Stanton V. P., Thirion J. P., Hudson T., et al. Cell. 1992;68:799–808. doi: 10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- 6.Liquori C. L., Ricker K., Moseley M. L., Jacobsen J. F., Kress W., Naylor S. L., Day J. W., Ranum L. P. Science. 2001;293:864–867. doi: 10.1126/science.1062125. [DOI] [PubMed] [Google Scholar]
- 7.Taneja K. L., McCurrach M., Schalling M., Housman D., Singer R. H. J. Cell Biol. 1995;128:995–1002. doi: 10.1083/jcb.128.6.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller J. W., Urbinati C. R., Teng-Umnuay P., Stenberg M. G., Byrne B. J., Thornton C. A., Swanson M. S. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis B. M., McCurrach M. E., Taneja K. L., Singer R. H., Housman D. E. Proc. Natl. Acad. Sci. USA. 1997;94:7388–7393. doi: 10.1073/pnas.94.14.7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Napierala M., Krzyzosiak W. J. J. Biol. Chem. 1997;272:31079–31085. doi: 10.1074/jbc.272.49.31079. [DOI] [PubMed] [Google Scholar]
- 11.Michalowski S., Miller J. W., Urbinati C. R., Paliouras M., Swanson M. S., Griffith J. Nucleic Acids Res. 1999;27:3534–3542. doi: 10.1093/nar/27.17.3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tian B., White R. J., Xia T., Welle S., Turner D. H., Mathews M. B., Thornton C. A. RNA. 2000;6:79–87. doi: 10.1017/s1355838200991544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sobczak K., de Mezer M., Michlewski G., Krol J., Krzyzosiak W. J. Nucleic Acids Res. 2003;31:5469–5482. doi: 10.1093/nar/gkg766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mooers B. H. M., Logue J. S., Berglund J. A. Proc. Natl. Acad. Sci. USA. 2005;102:16626–16631. doi: 10.1073/pnas.0505873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mankodi A., Logigian E., Callahan L., McClain C., White R., Henderson D., Krym M., Thornton C. A. Science. 2000;289:1769–1773. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- 16.Timchenko L. T., Miller J. W., Timchenko N. A., DeVore D. R., Datar K. V., Lin L., Roberts R., Caskey C. T., Swanson M. S. Nucleic Acids Res. 1996;24:4407–4414. doi: 10.1093/nar/24.22.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Philips A. V., Timchenko L. T., Cooper T. A. Science. 1998;280:737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- 18.Kanadia R. N., Johnstone K. A., Mankodi A., Lungu C., Thornton C. A., Esson D., Timmers A. M., Hauswirth W. W., Swanson M. S. Science. 2003;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- 19.Ho T. H., Charlet B. N., Poulos M. G., Singh G., Swanson M. S., Cooper T. A. EMBO J. 2004;23:3103–3112. doi: 10.1038/sj.emboj.7600300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mankodi A., Urbinati C. R., Yuan Q. P., Moxley R. T., Sansone V., Krym M., Henderson D., Schalling M., Swanson M. S., Thornton C. A. Hum. Mol. Genet. 2001;10:2165–2170. doi: 10.1093/hmg/10.19.2165. [DOI] [PubMed] [Google Scholar]
- 21.Mankodi A., Teng-Umnuay P., Krym M., Henderson D., Swanson M., Thornton C. A. Ann. Neurol. 2003;54:760–768. doi: 10.1002/ana.10763. [DOI] [PubMed] [Google Scholar]
- 22.Jiang H., Mankodi A., Swanson M. S., Moxley R. T., Thornton C. A. Hum. Mol. Genet. 2004;13:3079–3088. doi: 10.1093/hmg/ddh327. [DOI] [PubMed] [Google Scholar]
- 23.Kino Y., Mori D., Oma Y., Takeshita Y., Sasagawa N., Ishiura S. Hum. Mol. Genet. 2004;13:495–507. doi: 10.1093/hmg/ddh056. [DOI] [PubMed] [Google Scholar]
- 24.Lin X., Miller J. W., Mankodi A., Kanadia R. N., Yuan Y., Moxley R. T., Swanson M. S., Thornton C. A. Hum. Mol. Genet. 2006;15:2087–2097. doi: 10.1093/hmg/ddl132. [DOI] [PubMed] [Google Scholar]
- 25.Savkur R. S., Philips A. V., Cooper T. A. Nat. Genet. 2001;29:40–47. doi: 10.1038/ng704. [DOI] [PubMed] [Google Scholar]
- 26.Timchenko N. A., Cai Z. J., Welm A. L., Reddy S., Ashizawa T., Timchenko L. T. J. Biol. Chem. 2001;276:7820–7826. doi: 10.1074/jbc.M005960200. [DOI] [PubMed] [Google Scholar]
- 27.Dansithong W., Paul S., Comai L., Reddy S. J. Biol. Chem. 2005;280:5773–5780. doi: 10.1074/jbc.M410781200. [DOI] [PubMed] [Google Scholar]
- 28.Ho T. H., Bundman D., Armstrong D. L., Cooper T. A. Hum. Mol. Genet. 2005;14:1539–1547. doi: 10.1093/hmg/ddi162. [DOI] [PubMed] [Google Scholar]
- 29.Kim D. H., Langlois M. A., Lee K. B., Riggs A. D., Puymirat J., Rossi J. J. Nucleic Acids Res. 2005;33:3866–3874. doi: 10.1093/nar/gki698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ebralidze A., Wang Y., Petkova V., Ebralidse K., Junghans R. P. Science. 2004;303:383–387. doi: 10.1126/science.1088679. [DOI] [PubMed] [Google Scholar]
- 31.Fardaei M., Rogers M. T., Thorpe H. M., Larkin K., Hamshere M. G., Harper P. S., Brook J. D. Hum. Mol. Genet. 2002;11:805–814. doi: 10.1093/hmg/11.7.805. [DOI] [PubMed] [Google Scholar]
- 32.Ho T. H., Savkur R. S., Poulos M. G., Mancini M. A., Swanson M. S., Cooper T. A. J. Cell Sci. 2005;118:2923–2933. doi: 10.1242/jcs.02404. [DOI] [PubMed] [Google Scholar]
- 33.Xiao X., Li J., Tsao Y. P., Dressman D., Hoffman E. P., Watchko J. F. J. Virol. 2000;74:1436–1442. doi: 10.1128/jvi.74.3.1436-1442.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mankodi A., Takahashi M. P., Jiang H., Beck C. L., Bowers W. J., Moxley R. T., Cannon S. C., Thornton C. A. Mol. Cell. 2002;10:35–44. doi: 10.1016/s1097-2765(02)00563-4. [DOI] [PubMed] [Google Scholar]
- 35.Charlet B. N., Savkur R. S., Singh G., Philips A. V., Grice E. A., Cooper T. A. Mol. Cell. 2002;10:45–53. doi: 10.1016/s1097-2765(02)00572-5. [DOI] [PubMed] [Google Scholar]
- 36.Berg J., Jiang H., Thornton C. A., Cannon S. C. Neurology. 2004;63:2371–2375. doi: 10.1212/01.wnl.0000148482.40683.88. [DOI] [PubMed] [Google Scholar]
- 37.Zhou Q., Chu P. H., Huang C., Cheng C. F., Martone M. E., Knoll G., Shelton G. D., Evans S., Chen J. J. Cell Biol. 2001;155:605–612. doi: 10.1083/jcb.200107092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Selcen D., Engel A. G. Ann. Neurol. 2005;57:269–276. doi: 10.1002/ana.20376. [DOI] [PubMed] [Google Scholar]
- 39.Kimura T., Nakamori M., Lueck J. D., Pouliquin P., Aoike F., Fujimura H., Dirksen R. T., Takahashi M. P., Dulhunty A. F., Sakoda S. Hum. Mol. Genet. 2005;14:2189–2200. doi: 10.1093/hmg/ddi223. [DOI] [PubMed] [Google Scholar]
- 40.Yuasa K., Sakamoto M., Miyagoe-Suzuki Y., Tanouchi A., Yamamoto H., Li J., Chamberlain J. S., Xiao X., Takeda S. Gene Ther. 2002;9:1576–1588. doi: 10.1038/sj.gt.3301829. [DOI] [PubMed] [Google Scholar]
- 41.Kramer R., Lochmuller H., Abicht A., Rudel R., Brinkmeier H. Neuromuscul. Disord. 1998;8:542–550. doi: 10.1016/s0960-8966(98)00078-9. [DOI] [PubMed] [Google Scholar]
- 42.Adereth Y., Dammai V., Kose N., Li R., Hsu T. Nat. Cell Biol. 2005;7:1140–1147. doi: 10.1038/ncb1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harris J. D., Beattie S. G., Dickson J. G. Methods Mol. Med. 2003;76:255–267. doi: 10.1385/1-59259-304-6:255. [DOI] [PubMed] [Google Scholar]
- 44.Zolotukhin S., Potter M., Zolotukhin I., Sakai Y., Loiler S., Fraites T. J., Jr., Chiodo V. A., Phillipsberg T., Muzyczka N., Hauswirth W. W., et al. Methods. 2002;28:158–167. doi: 10.1016/s1046-2023(02)00220-7. [DOI] [PubMed] [Google Scholar]
- 45.Veldwijk M. R., Topaly J., Laufs S., Hengge U. R., Wenz F., Zeller W. J., Fruehauf S. Mol. Ther. 2002;6:272–278. doi: 10.1006/mthe.2002.0659. [DOI] [PubMed] [Google Scholar]
- 46.Holmes T. J., O’Connor N. J. J. Microsc. 2000;200:114–127. doi: 10.1046/j.1365-2818.2000.00751.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.