Abstract
We have previously shown that ≈5% of the genes encoded by the genome of Mycobacterium tuberculosis are specifically required for the growth or survival of this bacterium during infection. This corresponds to hundreds of genes, most of which have no identifiable function. As a unique approach to characterize these genes, we developed a method to rapidly delineate functional pathways by identifying mutations that modify each other’s phenotype, i.e., “genetic interactions”. Using this method, we have defined a complex set of interactions between virulence genes in this pathogen, and find that the products of unlinked genes associate to form multisubunit transporters that are required for bacterial survival in the host. These findings implicate a previously undescribed family of transport systems in the pathogenesis of tuberculosis, and identify genes that are likely to function in the metabolism of their substrates. This method can be readily applied to other organisms at either the single pathway level, as described here, or at the system level to define quantitative genetic interaction networks.
Keywords: epistasis, mce, transport, tuberculosis
Functional assignments cannot be made for a large fraction of genes in even the most thoroughly studied organisms (1). It has been even more difficult to characterize the genomes of pathogens, because a large fraction of the encoded functions are specialized for survival in experimentally intractable environments. Genes of unknown function are generally classified through the identification of physical (2–4) regulatory (5), or phylogenetic (6) associations that allow genes to be associated with pathways of known function. A more comprehensive approach is the definition of genetic interactions. In this type of analysis, the phenotypes of single and double mutants are compared to find mutations that alter the phenotypic consequence of inactivating a gene of interest. This general phenomenon is termed “epistasis,” and encompasses several types of interaction (7). “Negative” (i.e., synthetic or synergistic) interactions are defined as mutations that produce a larger than additive phenotypic effect when present together, and “positive” (i.e., buffering or antagonistic) interactions occur between mutations produce a less than additive effect. “Suppressors” are a distinct subcategory of positive interactions, which alleviate the consequence of gene loss.
Although the interpretation of these interactions is not always straightforward, for null alleles, negative interactions are often found between genes of separate pathways that perform redundant functions. Positive interactions, excluding suppressors, occur largely between genes of the same pathway that depend on each other for their function. Suppressor mutations can act by inducing a compensatory pathway or by relieving a toxic effect of the primary mutation. Thus, genetic interactions can be used to define the individual steps of a pathway, identify parallel pathways that contribute to similar biological processes, and even provide clues to the molecular basis of a mutant phenotype.
Large scale maps of genetic interactions have previously only been generated in a small number of easily manipulated model organisms by determining the relative in vitro growth rates of thousands of individually constructed single and double mutant strains. With notable exceptions (8, 9), these cumbersome screens provide largely qualitative data and define only the most dramatic interactions. We developed a microarray-based technique, called transposon site hybridization (TraSH), which can be used to compare the growth rates of individual transposon mutants in large pools and is easily applied to less tractable pathogenic organisms (10). The mutant libraries used in these studies were made by using a minitransposon that encodes outward-facing T7 RNA polymerase promoters. The labeled RNA probe generated from these promoters is complementary to the chromosomal DNA flanking each insertion in the mutant pool, and effectively marks the approximate position of each transposon. The relative composition of any two pools can be rapidly determined by competitively hybridizing the corresponding probes to a microarray. Because mutant pools can be analyzed after being subjected to any number of selective conditions, including passage through animal hosts, this approach can be used to study virulence genes that are only essential during infection.
We have used this method to identify genes that interact with the “mce loci” of M. tuberculosis, which have been implicated as important determinants of virulence. These loci consist of four homologous 8-13 gene operons, mce1–mce4, that appear to have arisen by the duplication of a single ancestral locus (11). The first mce gene to be discovered was found to promote the uptake of bacteria into nonphagocytic cells (12, 13) and was thus designated to function in mycobacterial cell entry (“mce”). However, mutations in these loci have been reported to have varying effects, either increasing or decreasing virulence (14–16), leaving their role during infection unclear. A systematic genetic analysis of the mce loci has yet to be performed, and no biochemical function for their products has been proposed.
The identification of unlinked genes that genetically interact with the mce loci has provided numerous functional insights. Most significantly, we report that the product of one interacting gene represents an ATPase that associates with mce-encoded transmembrane proteins to form multisubunit transport systems resembling ATP-binding cassette (ABC) transporters. Mutagenesis of this essential subunit indicates that Mce-mediated transport is critical for tuberculosis pathogenesis, and other genetic interactions suggest that these transporters may function in the translocation of lipid substrates. In addition to defining a mechanism by which this pathogen interacts with the host, this work demonstrates the general utility of this approach for the characterization of virulence genes for which little functional information is available.
Results
Generation of a Genetic Interaction Map for the mce Loci.
Previous TraSH analyses indicated that mutations in the mce1 and mce4 operons produced distinct in vivo growth defects (15). To characterize these loci further, we used TraSH to identify genes that positively interact with them and are, therefore, likely to function in the same pathway (Fig. 1). Whole genome data are provided in Tables 1 and 2, which are published as supporting information on the PNAS web site. Interacting genes were defined by mutations that reduced the in vivo growth rate of wild-type bacteria, but had a significantly less severe effect in mce-deficient backgrounds (see Experimental Procedures). Thirty-five and 31 positively interacting mutations were identified for the mce1 and mce4 loci, respectively, approximately one-third of which appear to act as suppressors (Tables 3 and 4, which are published as supporting information on the PNAS web site). The total number of interactions corresponds to ≈1% of the genome, which is similar to large-scale interaction screens in Saccharomyces cerevisiae (8, 9, 17).
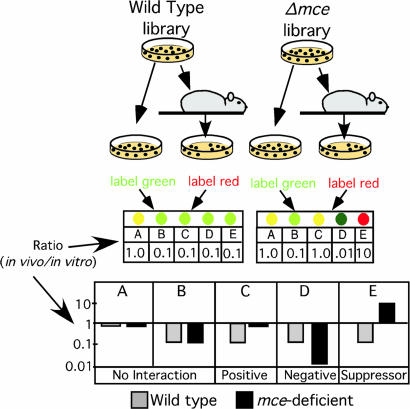
Fig. 1.
Strategy for identifying genes that interact with the mce1 and mce4 loci. Saturated transposon libraries were generated in wild-type or mce-deficient strains and the resulting libraries were used to infect groups of mice. After a period of infection, the surviving bacteria were recovered from spleen homogenates by plating. The unselected pools were replated in parallel to serve as a control. Each of these libraries was collected and “TraSH probe” was generated, which was complementary to the chromosomal DNA flanking each insertion in a pool. In vitro- and in vivo-selected pools were labeled with different fluorophores and compared by competitive hybridization to a microarray. (Lower) Idealized data. Genes (A–E) that are specifically required for in vivo growth will produce microarray ratios (in vivo/in vitro) of less than 1. Transposon mutations that cause a quantitatively similar phenotype in both genetic backgrounds (i.e., no interaction) will produce the same microarray ratios in both pools (gene B). Positively interacting transposon mutations will produce numerically larger ratios in the mutant background (gene C), and negatively interacting mutations will produce numerically smaller ratios (gene D). Suppressor mutations will produce ratios greater than 1 in the mutant background (gene E).
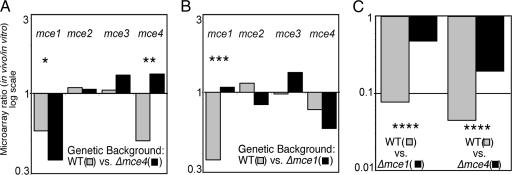
The mce1 and mce4 mutations used in these studies deleted a portion of each locus that was expected to abrogate function (15), but left a number of the genes in each operon intact. Predictably, genes adjacent to the mce1 and mce4 deletions, which are likely to act in concert with the deleted genes, were found to interact with the corresponding mutation (rv3502c of the mce4 locus, and lprK of the mce1 locus). Although all of the genes of a locus did not meet the stringent criteria that we set, they behaved similarly. Therefore, to increase the statistical power of this analysis, each mce locus was analyzed in composite by averaging the ratios for each intact gene. This strategy verified that, although insertions in the mce1 and mce4 loci produce attenuation in a wild-type background, these mutations have virtually no effect if the corresponding locus contains a second inactivating mutation (“mce4” in Fig. 2A and “mce1” in Fig. 2B). Mutations in the mce2 and mce3 loci had no significant effect in any genetic background.
Fig. 2.
Positive and negative interactions identified by using TraSH. (A and B) Microarray ratios from each gene of the indicated mce loci were averaged. Only genes that remain intact in the Δmce1 and Δmce4 strains were included in this analysis. Ratios from the wild-type (gray bars) or the indicated mce-deficient backgrounds (black bars) are plotted on a log scale. (C) Ratios for mceG (i.e., rv0655) mutants are plotted in a similar manner. Asterisks indicate statistical significance by t test (∗, P = 0.029; ∗∗, P = 0.008; ∗∗∗, P = 0.044) or ANOVA (∗∗∗∗).
In addition, this analysis identified a negative interaction between mutations in the mce1 and mce4 loci (“mce1” in Fig. 2A). This interaction was statistically significant only in the Δmce4 background, although a similar trend is noted in both experiments. The decreased significance of this interaction in the Δmce1 strain is likely a result of the relatively slow replication of this mutant during the period of in vivo selection. In general, some replication is required to discriminate between authentic positive interactions and unrelated mutations whose phenotypes are buffered simply due to the lack of growth. In this case, the statistical significance of some mce1 interactions was likely reduced and as a result, this screen may have been less sensitive than the mce4 experiment.
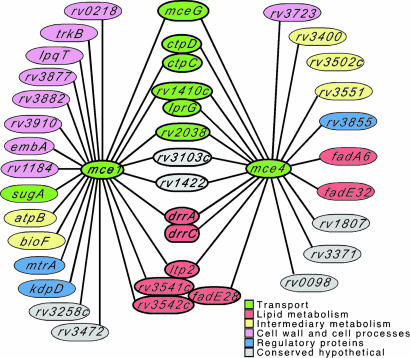
The negative interaction between mce1 and mce4 mutations suggested that these loci perform similar or even partially redundant functions. This model was supported by the large proportion of genes that interacted positively with both loci, as depicted in Fig. 3. Greater than 25% of the genes that interacted with each mce mutation were also found to interact with the other mce locus.
Fig. 3.
Genetic interaction map for the mce1 and mce4 loci. Graphic depiction of the identified positive interactions. Predicted suppressor mutations have been excluded, but are noted in Tables 3 and 4. Coloring indicates predicted functional classification (41). The “transport” category is predicated on data presented in this work. Tightly clustered ovals correspond to genes in single predicted transcriptional units.
The first two genes of each mce operon (annotated as yrbEA and yrbEB genes) (11) are similar in both sequence and predicted secondary structure to the transmembrane permease subunits of ABC transporters, suggesting a potential role for these proteins in transmembrane transport. However, all ABC transport systems require an additional nucleotide-binding domain (NBD) subunit to energize transport, and no such gene is encoded in the mce loci. The set of genes that was predicted to positively interact with both mce mutations included rv0655 (Fig. 2C), which encodes a protein homologous to the NBD subunits of ABC transporters (18). rv0655 lacks predicted transmembrane helices and is not encoded near other transporter components. Thus, we hypothesized that the product of this gene might associate with the YrbEAB proteins of both the mce1 and mce4 loci and form transport systems. To indicate a role for this gene in mce function, we will henceforth refer to rv0655 as “mceG.”
Validation of Functional Predictions.
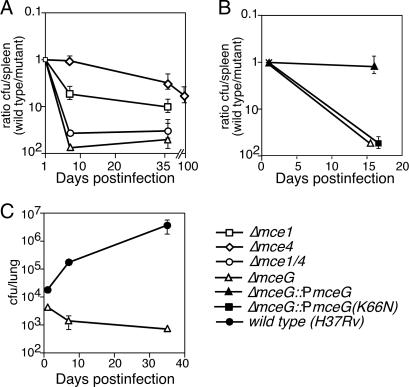
To independently verify these interactions, the in vivo growth rates of mutants lacking either mceG (ΔmceG) or both the mce1 and mce4 loci (Δmce1/4) were compared to single mce mutants by using a competitive model in which mice are infected with a mixture of wild-type and mutant bacteria. Although all mce mutants grew normally in broth culture (data not shown), individual mutations in the mce1 and mce4 loci caused kinetically distinguishable in vivo growth defects (Fig. 4A). The Δmce1 strain showed a pronounced early growth defect that was obvious at 1 week after infection, whereas the Δmce4 strain grew normally for the first week and showed a defect only at later times. Simultaneous mutation of these loci produced an effect that was clearly greater than additive, validating the negative interaction between these loci that was predicted by using TraSH. The deletion of mceG had a similar, but perhaps slightly exaggerated, effect as the mce1/4 mutations. This strain was completely unable to grow in this model and was slowly cleared from the tissue (Fig. 4C). This phenotype was consistent with the loss of both mce1 and mce4 function and was predicted by the observation that mceG interacts with both loci (Fig. 2C).
Fig. 4.
Isolated mce mutants verify the interactions predicted by using TraSH. Each mutant was mixed with a similar number wild type H37Rv, inoculated intravenously into C57BL/6 (A and C) or BALB/c (B) mice, and organs were harvested at the indicated time points. (A and B) The ratio of mutant/wild-type bacteria in the spleens of infected mice plotted over time. (C) Colony-forming units of ΔmceG and wild-type bacteria in the lungs of infected mice plotted individually (same data as A). Log scales are used on each y axis. All mutants grew normally in broth culture in vitro (data not shown).
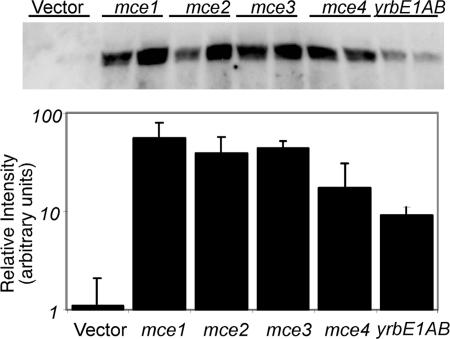
The interaction between MceG and mce-encoded proteins was confirmed biochemically by coexpressing these genes in Mycobacterium smegmatis, a saprophytic relative of M. tuberculosis. When expressed alone from a constitutive promoter, the MceG protein was virtually undetectable in cell lysates (Fig. 5). Coexpression of any mce locus resulted in a 17- to 55-fold increase in MceG protein levels. A similar 13-fold increase was seen when only the YrbEA and YrbEB permease subunits were coexpressed with MceG. The mutual stabilization of members of multiprotein complexes is a well-described phenomenon in bacteria (19), suggesting that physical associations were likely to account for these observations.
Fig. 5.
Stabilization of MceG protein by yrbEAB expression. M. smegmatis constitutively expressing a myc-tagged MceG protein was cotransformed with plasmids expressing the indicated genes. The amount of MceG in exponentially growing cultures of each strain was determined by Western blotting of whole cell extracts. Band intensities are plotted on a log scale. Data from two independent transformants of each strain are shown. “Vector” indicates the vector backbone for the yrbEAB-expressing plasmid.
The importance of mce-meditated transport during infection was investigated by generating a point mutation in the predicted ATP binding site of MceG. Analogous mutations in the “Walker A” motifs of other NBD’s abrogate substrate translocation, but do not affect transporter assembly (20, 21). A epitope-tagged version of this mutant (K66N) mceG allele was expressed at comparable (≈2-fold reduced) levels as the wild-type protein, as determined by Western blotting (data not shown). Although integration of the wild-type allele onto the chromosome completely complemented the in vivo growth defect of the ΔmceG strain, expression of the mutant allele had no effect on the growth of this strain (Fig. 4B).
Discussion
The network of genes found to genetically interact with the mce loci provide several insights into the functions of these genes. Most importantly, the product of the mceG gene was found to be required for the function of multiple mce loci. These data, along with the apparent physical association of MceG with products of each mce locus and the lack of any identifiable paralog of mceG in the genome, lead us to propose that the mce loci encode four distinct ABC-like transport systems that all share this common enzymatic subunit. The requirement for the mce loci during infection appears to be completely attributable to this transport function, as mutation of the ATP-binding site of MceG had the same effect on in vivo growth as the simultaneous inactivation of both the mce1 and mce4 loci.
In addition to the ABC-like transport systems that appear to be formed by MceG and the YrbEA and YrbEB permeases, each mce locus encodes 6–11 genes of unknown function. Mutation of each mce gene produces a phenotype that is similar to the corresponding yrbEAB genes (ref. 1, Tables 1 and 2), suggesting that the encoded proteins function in a coordinated manner, perhaps as subunits of a larger transport apparatus. One product of the mce1 locus, Mce1A, is predicted to be a soluble secreted protein that has been localized on the bacterial surface and reported to promote bacterial uptake into nonphagocytic cells (22). These observations are consistent with a model in which Mce proteins represent components of transport systems that interact with host cells. Structural predictions suggest that some of these proteins may resemble either colicins or β-barrel porins (23, 24), both of which form channels through lipid bilayers. Therefore, it is possible that these proteins may be important for the transit of solutes through hydrophobic barriers such as the mycobacterial envelope or host cell membranes.
In contrast to the growth defect we observe for mce1 mutants in competitive infections, mutations in this locus have also been reported to cause increased bacterial growth in single-strain infection models (14, 16). This “hypervirulent” phenotype was suggested to result from the relatively weak innate immune response that was generated by the mutant bacteria. Although the precise mce1 mutation used in these studies differs from the Δmce1 mutation described here, it is unlikely that this alone could account for the apparent phenotypic differences. We observed similar competitive growth defects for the Δmce1 deletion mutant, for an isolated transposon mutant that disrupts a single gene in this locus (rv0173, data not shown), and for transposon mutations in each gene of the locus using TraSH. Thus, similar competitive growth defects appear to result from any inactivating mutation. The divergent behaviors of mce1 mutants in competitive and single-strain infection models is more likely due to differences in the immune pressures generated by wild type and mce1-deficient bacteria. The results presented here indicate that, in the context of the immune response triggered by wild-type M. tuberculosis, mce1 function is required for bacterial growth.
The mce1 locus appears to be most important immediately after infection, whereas the mce4-encoded transporter is required only at later time points. These kinetically distinct requirements suggest that these transporters serve multiple roles during infection possibly by transporting a number of different substrates. Clues to the identities of these substrates can be found in the interaction data. A large proportion of the genes that were found to positively interact with the mce mutations are predicted to be involved in either transport-related functions or lipid modification, and the latter genes could be involved in substrate metabolism. It is difficult to predict, a priori, whether such genes are involved in biosynthetic or catabolic pathways. Lipid transport is important during in vivo growth both for the export of a variety of complex lipid virulence factors (25–28), and possibly for the import of fatty acids as a source of nutritional carbon (29). The characterization of such interacting genes will aid in elucidating the chemical structures of Mce substrates, the direction of their transport, and their ultimate role during infection.
The interactions reported in this work result from the application of stringent cutoffs to the microarray data, which excluded a large number of potentially significant phenotypic differences. Both computational (30–32) and experimental (33–35) evidence indicates that quantitatively minor genetic interactions are common, and that any gene can interact with hundreds of others. A quantitative method, such as TraSH, could therefore be used in any number of haploid organisms to define robust genetic interaction networks to characterize both individual functional pathways and the higher-order integration of these pathways.
Experimental Procedures
Bacterial Culture and Genetic Manipulations.
M. tuberculosis (H37Rv) was grown on Middlebrook 7H10 agar or in 7H9 broth supplemented with 10% OADC enrichment. Kanamycin or hygromycin were added at 20 or 50 μg/ml, respectively. M. smegmatis, mc2155 (36), and Escherichia coli were grown in LB agar containing 50 μg/ml kanamycin, 50–100 μg/ml hygromycin, 30 μg/ml apramycin, 20 μg/ml chloramphenocol, or 12 μg/ml tetracycline. All genetic deletions were generated in M. tuberculosis by using phage-mediated allelic exchange essentially as described (37). A lox site-flanked hygromycin-chloramphenicol cassette was used to construct the Δmce1 strain (replacement of nucleotides 202507–207411 encompassing mce1D-rv0175), and the ΔmceG strain (nucleotides 751513–752593 are replaced). Δmce4 mutant has the yrbE4A gene replaced by a lox-flanked hygromycin resistance gene (15). The Δmce1/4 double mutant was generated by removing the hygromycin marker from the Δmce4 strain using a counterselectable Cre recombinase-expressing plasmid. This unmarked strain was then transduced with the Δmce1 phage. The MceG complementing plasmid was generated by ligating the mceG ORF into an integrating plasmid downstream of a constitutive “mop” promoter (38). Transformation of M. tuberculosis with this plasmid resulted in single copy integration at the phage L5 att site. The mceG K66N mutation was generated by PCR and verified by sequencing.
TraSH Analysis.
Transposon libraries were made by using the pMycoMarT7 transposon as described (10). Briefly, transposon DNA was introduced into 50-ml broth cultures of M. tuberculosis via transduction. The resulting mixture was plated on 7H10 containing OADC, 20 μg/ml kanamycin, and 0.05% Tween-80. Each library consisted of ≈2 × 105 independent mutants. Groups of four or five C57BL/6 mice (8–10 weeks old) were inoculated intravenously with a mixture of 106 colony-forming units (cfu) of mutant library and an equal number of wild-type H37Rv. At 1 week (for mce1 interactions) or 8 weeks (for mce4 interactions), the mice were killed, and the surviving mutants were collected by plating spleen homogenates on 7H10 containing kanamycin. The ratio of mutant/wild type was determined at this time, and the mutant libraries were ≈10-fold underrepresented relative to wild type. The inoculating libraries were replated in parallel to serve as controls. Each library was collected separately by scraping the plates, chromosomal DNA was isolated from each, and TraSH probe was generated and hybridized to custom microarrays as described (10). Two TraSH probes were generated and hybridized independently for each mutant pool. The data from these microarrays (8–10 per experimental group) were averaged by using GeneSpring software. Spots that did not produce an intensity >300 fluorescence units, were not detected in six of eight replicates, or corresponded to genes that were independently predicted to be required for in vitro growth (39) were discarded. Genetic interactions were identified by comparing the ratios for each gene in the mce-deficient libraries to the corresponding wild-type library using two-way ANOVA testing and a false-discovery rate of 5%. The positive interactions were defined as the genes selected by ANOVA that were also predicted to be required for optimal in vivo growth in a wild-type background (15) and whose ratio decreased by >2-fold in the mutant background. The fraction of these interactions that represent suppressor mutations were defined as those with microarray ratios >2 in the mutant background. The “mce loci” are defined as in ref. 11: mce1, yrbE1A-rv0178; mce2, yrbE2A-mce2F; mce3, yrbE3A-rv1975; mce4, yrbE4A-mce4F.
Animal Infections.
Log phase cultures of bacteria were washed twice in PBS containing 0.05% Tween-80, frozen in 10% glycerol, and titered. Eight- to 10-week-old C57BL/6 mice were inoculated with the 200 μl of PBS containing ≈2 × 105 cfu of wild type and ≈8 × 104 cfu of mutant via the lateral tail vein. At the indicated times, lungs and spleens were removed, homogenized in PBS containing 0.05% Tween-80, and plated on 7H10 agar (for total bacterial counts) or 7H10 agar containing hygromycin (to enumerate cfu of mutant).
M. smegmatis Expression.
To generate the epitope tagged allele of mceG, the following sequence (encoding 6xHis and c-myc tags) was added to the 3′ end of the peptide by PCR: HHHHHHMAEQKLISEEDL. The mceG fusion was cloned downstream of a mycobacterial hsp60 promoter of an episomal shuttle vector, which expresses apramycin resistance to generate pSJ7 (GenBank accession no. DQ823234). To express isolated yrbEAB genes, these ORFs were amplified from the mce1 locus by PCR. This product was cloned into an episomal shuttle vector expressing hygromycin resistance resulting in pCS59 (GenBank accession no. DQ823235). To isolate intact mce loci, a cosmid library of H37Rv chromosomal DNA was generated in a modified supercos-1 vector (39). The ends of random clones were sequenced, and a clone containing each mce locus was isolated. A constitutive hsp60 promoter, a composite hygromycin-chloramphenicol marker, and mycobacterial origin of replication were recombined immediately upstream of each mce locus by using the λ-red recombination system (40). Plasmid sequences are available in GenBank (accession nos. DQ823230–DQ823233). Cultures (100 ml) of M. smegmatis transformed with the indicated plasmids were collected by centrifugation, resuspended in 1 ml of PBS containing protease inhibitor mixture (Sigma, St. Louis, MO), lysed by sonication, heated to 50°C for 10 min in SDS sample buffer containing 1% 2-mercaptoethanol, and separated by 4–20% gradient SDS/PAGE. Proteins were transferred to PVDF membranes, stained with a polyclonal rabbit anti-myc antibody (Abcam, Cambridge, MA) followed by HRP-conjugated anti-rabbit IgG (Promega, Madison, WI), and visualized with ECL reagent (Pierce, Rockford, IL). Light emission was quantified by using a LAS-1000 Plus Gel Documentation System and Intelligent Dark Box II. Images were processed by using the ImageGauge program (Fujifilm, Toyko, Japan)
Supplementary Material
Acknowledgments
We thank Ilona Breiterene for expert technical assistance; John Leong, Doyle Ward, and Jennifer Phillips for critical review of this manuscript; and John Mckinney and James Gomez for insightful conversation. This work was supported by National Institutes of Health Grant AI48704.
Abbreviations
- TraSH
transposon site hybridization
- ABC
ATP-binding cassette
- NBD
nucleotide-binding domain.
Footnotes
References
- 1.Costanzo M. C., Hogan J. D., Cusick M. E., Davis B. P., Fancher A. M., Hodges P. E., Kondu P., Lengieza C., Lew-Smith J. E., Lingner C., et al. Nucleic Acids Res. 2000;28:73–76. doi: 10.1093/nar/28.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ho Y., Gruhler A., Heilbut A., Bader G. D., Moore L., Adams S. L., Millar A., Taylor P., Bennett K., Boutilier K., et al. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 3.Gavin A. C., Bosche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J. M., Michon A. M., Cruciat C. M., et al. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 4.Uetz P., Giot L., Cagney G., Mansfield T. A., Judson R. S., Knight J. R., Lockshon D., Narayan V., Srinivasan M., Pochart P., et al. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 5.Wu L. F., Hughes T. R., Davierwala A. P., Robinson M. D., Stoughton R., Altschuler S. J. Nat. Genet. 2002;31:255–265. doi: 10.1038/ng906. [DOI] [PubMed] [Google Scholar]
- 6.Srinivasan B. S., Caberoy N. B., Suen G., Taylor R. G., Shah R., Tengra F., Goldman B. S., Garza A. G., Welch R. D. Nat. Biotechnol. 2005;23:691–698. doi: 10.1038/nbt1098. [DOI] [PubMed] [Google Scholar]
- 7.Phillips P. C. Genetics. 1998;149:1167–1171. doi: 10.1093/genetics/149.3.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schuldiner M., Collins S. R., Thompson N. J., Denic V., Bhamidipati A., Punna T., Ihmels J., Andrews B., Boone C., Greenblatt J. F., et al. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 9.Pan X., Yuan D. S., Xiang D., Wang X., Sookhai-Mahadeo S., Bader J. S., Hieter P., Spencer F., Boeke J. D. Mol. Cell. 2004;16:487–496. doi: 10.1016/j.molcel.2004.09.035. [DOI] [PubMed] [Google Scholar]
- 10.Sassetti C. M., Boyd D. H., Rubin E. J. Proc. Natl. Acad. Sci. USA. 2001;98:12712–12717. doi: 10.1073/pnas.231275498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cole S. T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S. V., Eiglmeier K., Gas S., Barry C. E., III, et al. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 12.Casali N., Konieczny M., Schmidt M. A., Riley L. W. Infect Immun. 2002;70:6846–6852. doi: 10.1128/IAI.70.12.6846-6852.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arruda S., Bomfim G., Knights R., Huima-Byron T., Riley L. W. Science. 1993;261:1454–1457. doi: 10.1126/science.8367727. [DOI] [PubMed] [Google Scholar]
- 14.Shimono N., Morici L., Casali N., Cantrell S., Sidders B., Ehrt S., Riley L. W. Proc. Natl. Acad. Sci. USA. 2003;100:15918–15923. doi: 10.1073/pnas.2433882100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sassetti C. M., Rubin E. J. Proc. Natl. Acad. Sci. USA. 2003;100:12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gioffre A., Infante E., Aguilar D., De la Paz Santangelo M., Klepp L., Amadio A., Meikle V., Etchechoury I., Romano M. I., Cataldi A., et al. Microbes Infect. 2005;7:325–334. doi: 10.1016/j.micinf.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 17.Tong A. H., Evangelista M., Parsons A. B., Xu H., Bader G. D., Page N., Robinson M., Raghibizadeh S., Hogue C. W., Bussey H., et al. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 18.Marchler-Bauer A., Bryant S. Nucleic Acids Res. 2004;32:W327–W331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berger B. R., Christie P. J. J. Bacteriol. 1994;176:3646–3660. doi: 10.1128/jb.176.12.3646-3660.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henriksen U., Gether U., Litman T. J. Cell Sci. 2005;118:1417–1426. doi: 10.1242/jcs.01729. [DOI] [PubMed] [Google Scholar]
- 21.Sexton J. A., Yeo H. J., Vogel J. P. Mol. Microbiol. 2005;57:70–84. doi: 10.1111/j.1365-2958.2005.04667.x. [DOI] [PubMed] [Google Scholar]
- 22.Chitale S., Ehrt S., Kawamura I., Fujimura T., Shimono N., Anand N., Lu S., Cohen-Gould L., Riley L. W. Cell Microbiol. 2001;3:247–254. doi: 10.1046/j.1462-5822.2001.00110.x. [DOI] [PubMed] [Google Scholar]
- 23.Pajon R., Yero D., Lage A., Llanes A., Borroto C. J. Tuberculosis (Edinburgh) 2006;86:290–302. doi: 10.1016/j.tube.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 24.Das A. K., Mitra D., Harboe M., Nandi B., Harkness R. E., Das D., Wiker H. G. Biochem. Biophys. Res. Commun. 2003;302:442–447. doi: 10.1016/s0006-291x(03)00116-5. [DOI] [PubMed] [Google Scholar]
- 25.Cox J. S., Chen B., McNeil M., Jacobs W. R., Jr. Nature. 1999;402:79–83. doi: 10.1038/47042. [DOI] [PubMed] [Google Scholar]
- 26.Reed M. B., Domenech P., Manca C., Su H., Barczak A. K., Kreiswirth B. N., Kaplan G., Barry C. E., III Nature. 2004;431:84–87. doi: 10.1038/nature02837. [DOI] [PubMed] [Google Scholar]
- 27.Ryll R., Kumazawa Y., Yano I. Microbiol. Immunol. 2001;45:801–811. doi: 10.1111/j.1348-0421.2001.tb01319.x. [DOI] [PubMed] [Google Scholar]
- 28.Converse S. E., Mougous J. D., Leavell M. D., Leary J. A., Bertozzi C. R., Cox J. S. Proc. Natl. Acad. Sci. USA. 2003;100:6121–6126. doi: 10.1073/pnas.1030024100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKinney J. D., Honer zu Bentrup K., Munoz-Elias E. J., Miczak A., Chen B., Chan W. T., Swenson D., Sacchettini J. C., Jacobs W. R., Jr., Russell D. G. Nature. 2000;406:735–738. doi: 10.1038/35021074. [DOI] [PubMed] [Google Scholar]
- 30.Segre D., Deluna A., Church G. M., Kishony R. Nat. Genet. 2005;37:77–83. doi: 10.1038/ng1489. [DOI] [PubMed] [Google Scholar]
- 31.You L., Yin J. Genetics. 2002;160:1273–1281. doi: 10.1093/genetics/160.4.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lenski R. E., Ofria C., Collier T. C., Adami C. Nature. 1999;400:661–664. doi: 10.1038/23245. [DOI] [PubMed] [Google Scholar]
- 33.Elena S. F. J. Mol. Evol. 1999;49:703–707. doi: 10.1007/pl00000082. [DOI] [PubMed] [Google Scholar]
- 34.Elena S. F., Lenski R. E. Nature. 1997;390:395–398. doi: 10.1038/37108. [DOI] [PubMed] [Google Scholar]
- 35.Burch C. L., Turner P. E., Hanley K. A. J. Evol. Biol. 2003;16:1223–1235. doi: 10.1046/j.1420-9101.2003.00632.x. [DOI] [PubMed] [Google Scholar]
- 36.Snapper S. B., Melton R. E., Mustafa S., Kieser T., Jacobs W. R. J. Mol. Microbiol. 1990;4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 37.Bardarov S., Bardarov S., Jr., Pavelka M. S., Jr., Sambandamurthy V., Larsen M., Tufariello J., Chan J., Hatfull G., Jacobs W. R., Jr. Microbiology. 2002;148:3007–3017. doi: 10.1099/00221287-148-10-3007. [DOI] [PubMed] [Google Scholar]
- 38.Guinn K. M., Hickey M. J., Mathur S. K., Zakel K. L., Grotzke J. E., Lewinsohn D. M., Smith S., Sherman D. R. Mol. Microbiol. 2004;51:359–370. doi: 10.1046/j.1365-2958.2003.03844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sassetti C. M., Boyd D. H., Rubin E. J. Mol. Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 40.Datsenko K. A., Wanner B. L. Proc. Natl. Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Camus J. C., Pryor M. J., Medigue C., Cole S. T. Microbiology. 2002;148:2967–2973. doi: 10.1099/00221287-148-10-2967. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.