Abstract
Bacteria exist in a variety of morphologies, but the relationship between cellular forms and biological functions remains poorly understood. We show that stalks (prosthecae), cylindrical extensions of the Caulobacter crescentus cell envelope, can take up and hydrolyze organic phosphate molecules and contain the high-affinity phosphate-binding protein PstS, but not PstA, a protein that is required for transport of phosphate into the cytoplasm. Therefore, uptake, hydrolysis, and periplasmic binding of a phosphate source can take place in the stalk, but high-affinity import must take place in the cell body. Furthermore, by using analytical modeling, we illustrate the biophysical advantage of the stalk as a morphological adaptation to the diffusion-limited, oligotrophic environments where C. crescentus thrives. This advantage is due to the fact that a stalk is long and thin, a favorable shape for maximizing contact with diffusing nutrients while minimizing increases in both surface area and cell volume.
Keywords: Caulobacter, diffusion, morphology, protein localization, stalk
Bacteria exhibit an amazing diversity of shapes and sizes. Bacterial cells can be round, cylindrical, curved, or coiled. Some are shaped like a flat square, others like a star; some are branched, and some have projections of the cell surface. Exactly how bacterial shapes are generated is not known, and the purposes of a specific cell shape are not always clear. Yet, in most cases, specific shapes are precisely reproduced at every generation. Cell shape changes can also occur during the life cycle of many bacterial species, such as the transformation of Sinorhizobium from rods to branched cells (Y forms) after colonization of legume root nodules. Although these morphological transformations are thought to play important roles in the life cycles of bacteria, the advantages of cell shape changes remain essentially speculative (1).
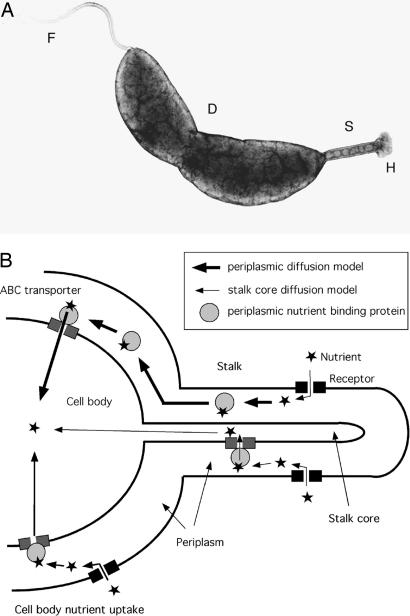
Here, we investigate the function of prosthecae, cell envelope extensions that are present in a morphologically diverse group of Gram-negative bacteria (2). Unlike flagella or pili, prosthecae are true extensions of the cell proper, possessing both peptidoglycan and cell membranes. In the aquatic bacterium Caulobacter crescentus, prosthecae are referred to as stalks. The function of Caulobacter stalks is not known, but one common hypothesis is that they facilitate the acquisition of nutrients from the environment (3, 4). This hypothesis is consistent with the observation that stalks dramatically elongate under phosphate starvation conditions, with the capacity to reach lengths of >30 μm (5, 6), a length that is approximately equivalent to that of 15 cell bodies. We show that the stalk of C. crescentus (Fig. 1A) can transport a nutrient molecule from the extracellular space into its periplasm and that the stalk constitutes a biophysically efficient morphological adaptation to environments where nutrient uptake is limited by diffusion.
Fig. 1.
Micrograph of a C. crescentus cell and models of nutrient uptake. (A) Transmission electron micrograph of C. crescentus. F, flagellum; S, stalk; H, adhesive holdfast; D, division site. (B) Models of nutrient uptake by the stalk and cell body. The model in the top part of the diagram (thick arrows) illustrates uptake of a nutrient molecule into the stalk periplasm, its binding to a periplasmic nutrient-binding protein, the diffusion of the nutrient-binding protein from the stalk periplasm to the cell body periplasm, and the uptake of the nutrient into the cell body cytoplasm by an ABC transporter. This periplasm-to-periplasm model is consistent with our results. The model on the bottom part of the diagram (thin arrows) illustrates nutrient uptake into the stalk core, followed by its diffusion into the cell body cytoplasm. Nutrient uptake in the cell body is also shown (intermediate-thickness arrows).
Results
Stalks Can Take Up and Hydrolyze Fluorescein Diphosphate (FDP).
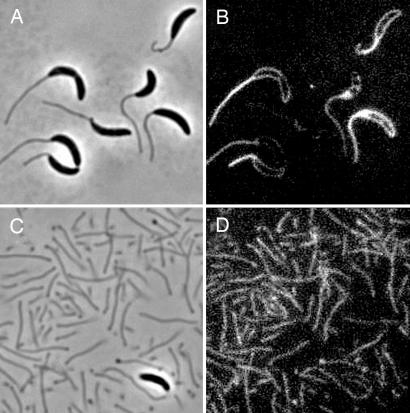
To determine whether stalks are capable of nutrient uptake, we incubated cells with FDP, a fluorogenic substrate for the periplasmic enzyme alkaline phosphatase. Fluorescein is liberated when the phosphate-ester bonds of FDP are cleaved; thus, the uptake and hydrolysis of this substrate can be monitored by using epifluorescent microscopy. Two to 5 min after addition of FDP, the perimeter of the cell body and its attached stalk began emitting fluorescence simultaneously (Fig. 2B), suggesting that FDP uptake occurs at a similar rate in the stalk and the cell body. The peripheral fluorescence emission in cell bodies was consistent with the reaction occurring in the periplasm. Similarly, purified stalks began emitting fluorescence within 2–5 min (Fig. 2D), indicating that the stalk alone is capable of FDP uptake and hydrolysis. Stalk fluorescence increased simultaneously along the length of the stalk, indicating that FDP uptake occurred throughout the stalk. Stalks and cells incubated with purified fluorescein never fluoresced (data not shown), indicating that the emission seen in the stalks upon exposure to FDP was not due to a secreted alkaline phosphatase or the uptake and/or nonspecific association of unconjugated fluorescein with the cellular surface. The ability of the stalk to take up FDP but not fluorescein suggests that the stalk possesses receptors or porins with specificity for organic phosphate.
Fig. 2.
FDP uptake by cells and stalks. (A and C) Phase contrast. (B and D) Fluorescence. Cells (B) and stalks (D) after incubation with FDP are shown.
The Stalk Is Compartmentalized from the Cell Body.
The organophosphate compound FDP is imported into the stalk periplasm and cleaved by alkaline phosphatase to release inorganic phosphate. What is the fate of this periplasmic phosphate molecule? In cells, periplasmic phosphate is transported across the inner membrane (IM) by the high-affinity phosphate ATP-binding cassette (ABC) transporter (PstSCAB) (6), which is composed of a high-affinity periplasmic phosphate-binding protein, PstS; two IM channel proteins, PstA and PstC; and a cytoplasmic traffic ATPase, PstB. In one scenario, the phosphate released in the stalk periplasm is bound by PstS, and the phosphate–PstS complex diffuses through the stalk periplasm to the cell body periplasm to be taken up through the IM by PstCAB (Fig. 1B). Alternatively, phosphate could be transported across the stalk IM by PstSCAB and then diffuse along the core of the stalk to the cytoplasm of the cell body to be metabolized (Fig. 1B). The first scenario requires that only PstS is present in the stalk periplasm, whereas the second scenario requires that the entire PstSCAB transporter and ATP be present in the stalk.
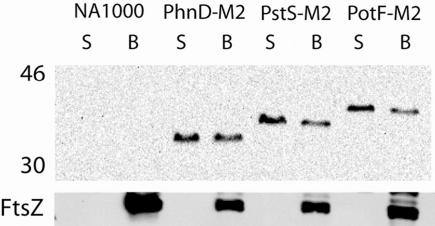
To determine whether PstS is present in the stalk periplasm and to examine the relative abundance of periplasmic nutrient-binding proteins in the stalk relative to the cell body, M2 epitope fusions were made to PstS (CC1515); a putative phosphonate-binding protein, PhnD (CC0362); and a putative putrescine-binding protein, PotF (CC3137). Western blot analysis showed that purified stalks contain significant levels of all three periplasmic binding proteins (Fig. 3). The relative level of the periplasmic nutrient-binding proteins was higher in the stalks than in cell bodies, as would be expected, because stalk fractions contain very little cytoplasmic protein (7). The abundant cytoplasmic protein FtsZ was undetectable in the stalk fractions, indicating that there was no significant contamination of the stalk fractions by cell bodies (Fig. 3, lower blot).
Fig. 3.
Periplasmic nutrient-binding protein localization in the stalk. Western blot analysis of periplasmic binding proteins in stalk (S) and cell body (B) fractions. PstS (CC1515) is the high-affinity phosphate-binding protein, PhnD (CC0362) is a putative phosphonate-binding protein, and PotF (CC3137) is a putative putrescine-binding protein. Lower blot shows the same membrane reprobed with an antibody raised against the abundant cytoplasmic protein FtsZ. Molecular weights are indicated to the left. NA1000 is the isogenic wild-type control.
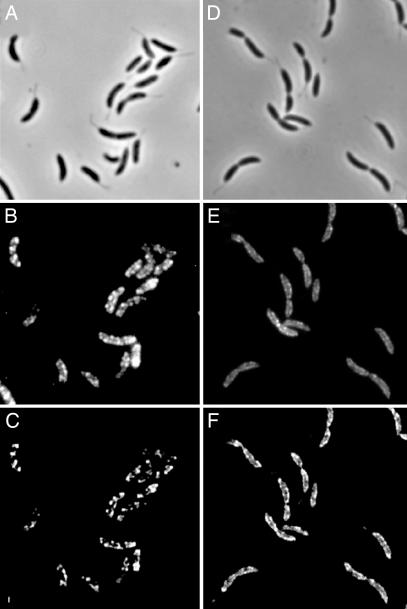
We next asked whether the stalk contained other components of the phosphate ABC transporter by using a PstA–GFP fusion. Fluorescence was easily detected in the cell body (Fig. 4E and F) but not in stalks. Western blot analysis confirmed that PstA–GFP is found in cell membranes and intact cells but is virtually undetectable in stalk fractions (Fig. 6, which is published as supporting information on the PNAS web site). Because PstS is present in the stalk and PstA is absent, phosphate taken up in the stalk periplasm, either as a free molecule or bound to PstS, must diffuse from the stalk periplasm to the cell body periplasm to be transported into the cytoplasm by the phosphate ABC transporter.
Fig. 4.
ExbB and PstA localize to the cell body but not the stalk. (A and D) Phase contrast. (B and E) Fluorescence. (C and F) 2D deconvolved fluorescence. Localization of ExbB–M2 in YB4058 (B and C) and PstA–GFP in YB4062 (E and F) is shown.
The absence of PstA–GFP from the stalk led us to ask whether other IM proteins were present or absent from the stalk. Immunofluorescence localization of an M2 epitope fusion to the IM protein ExbB, a protein that is required for TonB-dependent transport across the outer membrane (OM) of Gram-negative bacteria, showed that ExbB–M2 was associated with the cell body but was not detectable in the stalk (Fig. 4 B and C) even after long exposures. The absence of two IM proteins, PstA and ExbB, from the stalk is consistent with our previous 2D gel analysis of the stalk proteome, which had identified many periplasmic and OM proteins but essentially no IM proteins (7). Because the IM proteins involved in nutrient uptake are hydrophobic and therefore difficult to identify by using 2D gel electrophoresis, we used 2D liquid chromatography tandem MS (2D LC-MS) to obtain a more complete proteomic profile of purified stalks. The proteomic 2D LC-MS profiling suggested that, in the stalk, OM and periplasmic proteins are more abundant than IM and cytoplasmic proteins (Table 1, which is published as supporting information on the PNAS web site). These data indicate that the stalk organelle is at least partially compartmentalized from the cell body with respect to protein composition.
Stalks Confer a Biophysical Advantage to C. crescentus When Nutrient Uptake Is Limited by Diffusion.
C. crescentus stalks may have evolved to enhance nutrient uptake to the cell. Why would growing a stalk be a more advantageous strategy for enhancing nutrient uptake than simply adding more nutrient receptors to the cell body? In the environments where they are found, such as lakes, ponds, or water-saturated soil, C. crescentus cells often attach to surfaces by means of a holdfast found at the distal tip of the stalk (Fig. 1A) and typically are not subject to fluid flow. In such diffusion-limited environments, nutrient molecules depend on diffusion for contact with their receptors. Microscopically, the random-walk trajectory of a diffusing molecule explores a given region in space well before wandering away (8). As a result, adding a second receptor to the cell surface will double the rate of uptake only if it is well separated from the first receptor with respect to its size, because a diffusing molecule in the vicinity of the second receptor might instead be absorbed by the first one if they are placed “too close” to each other. This phenomenon is illustrated in Fig. 7, which is published as supporting information on the PNAS web site.
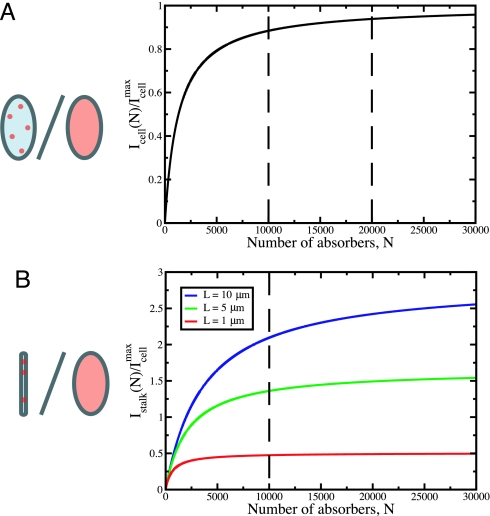
A mathematical analysis (see Supporting Text, which is published as supporting information on the PNAS web site) of diffusive uptake of nutrients to the C. crescentus cell body and stalk following Berg and Purcell (9) demonstrates the saturation of uptake capability with increasing numbers of receptors. Considering a range consistent with published abundances of some bacterial OM nutrient receptors (10, 11), we find that doubling the number of receptors on the cell body from 10,000 to 20,000 leads to an increase of only 5% (from 83% to 88%) in the rate of nutrient absorption relative to the maximum rate that is obtained when the entire cell surface is absorptive (Fig. 5A). However, if the additional 10,000 receptors are placed on a stalk instead of the cell body, for stalks of length 1, 5, and 10 μm, the overall rate of uptake is increased by 50%, 140%, and 210%, respectively (Fig. 5B). Thus, incorporating additional receptors on a stalk is a dramatically more efficient strategy for increasing nutrient uptake capacity than incorporating new receptors on a cell body surface among those receptors already synthesized.
Fig. 5.
Rate of diffusive uptake by discrete absorbers. (A) The number of particles per unit time absorbed by the cell body, Icell(N), is plotted for bcell = 0.25 μm and Lcell = 1 μm, in units of the maximum rate of uptake by the cell body, Icellmax. The radius of an individual absorber is s = 1 nm, a typical size for transport proteins (26). By increasing the number of absorbers on the cell body from N = 10,000 to N = 20,000, the rate of uptake is increased by only 5% of the maximum rate. (B) The number of particles per unit time absorbed by the stalk, Istalk(N), is plotted for bstalk = 50 nm and Lstalk = 1 μm (red), 5 μm (green), and 10 μm (blue). Addition of N = 10,000 absorbers to stalks of length 1, 5, and 10 μm increases the rate of uptake by ≈50%, 140%, and 210% of Icellmax. The absorbing surface is represented in red in the figures (Left).
In mitigating the saturation to the maximum rate of uptake, is elaborating a stalk preferable to elongating the cell itself? Underlying the results given above and as is easily shown (see ref. 9 and Supporting Text), the rate of nutrient uptake in diffusion-limited environments is proportional to the effective linear dimension of a structure rather than to its surface area. This dependence is in contrast to systems with fluid flow or mixing, such as in the highly evaginated surface of the intestinal epithelium, where uptake is proportional to surface area. For example, a 5.45-μm-long stalk with a diameter of 0.1 μm has the same surface area as a cell body of typical dimensions used in our analysis (1 μm in length and 0.5 μm in diameter). However, because it is longer, the maximum rate of uptake by the stalk is ≈1.8 times that of the cell body. In other words, even when a stalk and cell body have the same surface area, the stalk has a higher uptake capacity because it is longer. In Fig. 8, which is published as supporting information on the PNAS web site, we demonstrate that the rate of nutrient uptake is comparable for elongated cells and stalked cells of the same total length (where, for the stalked cells, the cell body is of fixed, typical size, and the stalk has variable length). However, when normalized to either surface area (Fig. 9A, which is published as supporting information on the PNAS web site) or volume (Fig. 9B), the maximum rate of uptake is significantly greater for stalked cells than for elongated cells. Therefore, adding nutrient receptors on a long, thin appendage such as a stalk should be a more advantageous strategy for enhancing nutrient uptake than increasing cell length and placing the additional receptors on the new surface. Indeed, a stalked cell with the same surface area as an elongated cell of length 2 μm has 1.8 times the maximum rate of uptake of the elongated cell because of the fact that the stalked cell, in addition to having a cell body of length 1 μm, is composed of a stalk of length 2.4 μm. We conclude that elongating a stalk is an efficient strategy for a cell to increase its rate of diffusive nutrient uptake and that this advantage is further enhanced by minimizing the energetic cost of increasing both surface area and volume. Finally, we note that once a linear structure such as a stalk has been elaborated under diffusion-limited conditions to increase the rate of nutrient uptake, the presence of the additional absorptive surface area will do likewise under flow conditions as well.
Discussion
Our data indicate that with respect to protein complement, the stalk is at least partially compartmentalized from the cell body, consistent with the role of the stalk as a specialized nutrient uptake organelle. We envision three scenarios that could explain the absence of IM and cytoplasmic proteins from the stalk. The first scenario is that there is a physical barrier transecting the cytoplasm and IM at the stalk–body junction. A second plausible, albeit more complicated, scenario for the compartmentalization is that certain proteins are actively targeted to the stalk, whereas other proteins are excluded. This model requires that there be stalk-specific sorting signals for proteins, and possibly a stalk-specific subset of secretion proteins, and thus seems less probable. Our preferred scenario is that stalks are compartmentalized by a combination of stalk structure and protein localization. Because the stalk IMs (12) are separated by ≈40 nm (L. Comolli and H. McAdams, personal communication), a space that is large enough for cytoplasmic proteins to enter, perhaps some as yet undefined barrier prevents the diffusion of cytoplasmic proteins into the stalk core. We hypothesize that most IM proteins do not enter the stalk because they are localized to specific structures or protein complexes in the cell body and thus are not free to diffuse. This hypothesis would be consistent with the localization of ExbB–M2 and PstA–GFP (see Fig. 4 B–F), which are detected in a pattern that is reminiscent of the helical pattern seen for peptidoglycan incorporation (13), OM protein incorporation (14), lipopolysaccharide incorporation (14), binding of bacteriophage λ to the OM protein maltose porin, LamB (15), and the bacterial cytoskeletal protein MreB (15–17). One common feature among the proteins and processes mentioned above is that they are associated with the transport of molecules across bacterial membranes. The intriguing correlation between molecule transport and the observed localization patterns mentioned above warrants further examination, because it suggests that, at least in some bacteria, many cellular processes, including energy-dependent nutrient uptake, take place at or near the same specific sites in the envelope.
The compartmentalization between the stalk and the cell body suggests that nutrient uptake by the stalk is passive, because this type of transport does not require IM or cytoplasmic proteins to provide the energy required. Given that C. crescentus lives in oligotrophic environments where the concentration of extracellular nutrients is very low, our results beg the following question: Is passive transport sufficient for efficient nutrient uptake by the stalk in oligotrophic environments? For passive diffusion to be sufficient, the level of periplasmic nutrients must be maintained at lower levels than those found in the environment, and at least two mechanisms would ensure this scenario. First, the periplasmic binding proteins will bind free nutrients with high affinity in the periplasm and thus remove them from the effective concentration. Second, the cell body IM, equipped with active ABC transporters, will act as a nutrient sink, generating a concentration gradient along the length of the stalk. Recent FLIP (fluorescence loss in photobleaching) experiments on the later stages of cell division in C. crescentus suggest that the periplasmic spaces of the stalk and cell body are contiguous (see ref. 18 and Fig. 5 A and C). Therefore, it is likely that nutrient molecules absorbed by the stalk OM and bound to periplasmic nutrient-binding proteins diffuse from the stalk periplasmic space into the cell body periplasmic space. Once steady state is achieved in the periplasmic space, the presence of the stalk effectively increases the concentration of nutrient molecules in the cell body periplasm, leading to a greater absorption rate across the cell body IM that is equal to the absorption rate by the stalk OM. The time scale for reaching steady state in the periplasm is estimated to be ≈17 and 42 min for stalks with lengths of 1 and 5 μm, respectively (see Supporting Text). This time scale is short compared with the duration of the C. crescentus cell cycle, which is on the order of hours under nutrient-deprived conditions. Hence, the stalk would quickly enhance the rate of nutrient uptake into the cytoplasm by an amount that is equal to the stalk’s rate of uptake from the environment.
One question that is particularly intriguing is how nutrient molecules might be transported across the C. crescentus OM, particularly in stalks. Interestingly, the C. crescentus genome lacks sequence homologs to any of the major general porins (such as OmpF and OmpC) that are found in many other bacteria. There are 65 putative TonB-dependent receptors in C. crescentus, and TonB-dependent receptors make up a predominance of the proteins identified in stalk fractions, as shown in this study and in ref. 7. However, because ExbB, an IM protein that is required for TonB-dependent transport, is absent from the stalks, they are unlikely to be capable of TonB-dependent transport, at least as it is currently understood. In the future, it will be of interest to determine what proteins are required for the uptake of nutrients by the stalk as well as whether the mechanism is passive or requires energy.
In summary, the increase in stalk length upon phosphate starvation (6), the ability of stalks to take up and hydrolyze a phosphate ester compound, the presence of high-affinity periplasmic nutrient-binding proteins in the stalk, the protein compartmentalization between the stalk and cell body, and the advantage of placing additional nutrient receptors on a linear appendage while minimizing increases in surface area and volume all support the hypothesis that stalk formation is a morphological adaptation that enhances nutrient uptake in oligotrophic environments.
Materials and Methods
Bacterial Strains, Plasmids, Oligonucleotides, and Growth Conditions.
The strains and plasmids used in this study are provided in Table 2, which is published as supporting information on the PNAS web site. All cultures were grown at 30°C in either PYE (19) or HIGG (20) medium containing 120 μM phosphate. When appropriate, liquid cultures were supplemented with 5 μg/ml kanamycin. The cultures from which stalks were harvested were grown to stationary phase. Oligonucleotides used in this study are listed in Table 3, which is published as supporting information on the PNAS web site.
Sample Preparation.
All steps were carried out on ice or at 4°C. To collect stalks by shearing, cultures were grown in HIGG medium containing 120 μM phosphate, chilled on ice for 10–20 min, and blended for 3 min in an ice-cold Waring blender as described in refs. 21 and 22. This stalk removal method does not lead to significant leakage of periplasmic or cytoplasmic proteins from samples, and cell bodies have no detectable loss in viability (22). For stalk purification, the cells were pelleted by centrifugation at 10,410 × g for 15 min. The supernatants, excluding the loose pellet, were again centrifuged at 10,410 × g for 15 min to remove more cell bodies. The supernatants, excluding the loose pellets, were divided into Oakridge tubes and centrifuged at 38,720 × g for 30 min to pellet the stalks. To further enrich for stalks free of cell bodies, only the fluffy outer portion of the pellet in each tube was resuspended. The first pellet was resuspended in 0.5 ml of doubly distilled H2O (ddH2O), and the suspension from this tube was transferred to the next until all of the pellets were resuspended. The suspension was examined microscopically to determine the extent of cell body contamination, which was generally very rare and ranged from one to five cell bodies per 2,000 stalks (data not shown). To prepare the cell bodies relatively free of unattached stalks, one of the pellets from the first centrifugation was resuspended in ≈250 ml of high basal salts without phosphate buffer (20), and this suspension was centrifuged at 10,410 × g for 10 min. The supernatant was poured off, and the pellet was washed once with ddH2O and resuspended in 10 ml of ddH2O. Microscopic examination confirmed that the bodies had only one to two free stalks per 200 cell bodies (data not shown).
Microscopy.
Phase-contrast and epifluorescence microscopy were performed on a Nikon Eclipse E800 light microscope equipped with a ×100 Plan Apo oil objective and a cooled charge-coupled device camera (model 1317, Princeton Instruments, Trenton, NJ). Images were captured and analyzed by using metamorph 4.6 imaging software (Universal Imaging, Downingtown, PA). 2D deconvolution was performed by using the metamorph software “nearest neighbors” function with the following settings: filter size, 9; scaling factor, 0.97; result scale, 5; suppress noise, on; autoscale result, on.
In Vivo Experiments with FDP.
To examine the ability of cells and stalks to hydrolyze the chromogenic substrate FDP (Molecular Probes), FDP was added to a final concentration of 100 μM, and samples were mounted on a slide and observed under ×1,000 magnification by using phase and fluorescence with an FITC filter. Two to 5 min passed from the addition of substrate to the time of image capture. The experiment with fluorescein was performed by incubating samples with 100 μM fluorescein (Molecular Probes) for 2 min to 1 h and mounting samples as before.
Western Blot Analysis.
The M2 epitope-tagged proteins were detected by using a 1:750 or 1:1,000 dilution of anti-M2 polyclonal antibody conjugated to horseradish peroxidase (Sigma). The GFP-tagged proteins were detected by using a 1:5,000 dilution of monoclonal anti-GFP antibody (Clontech) and a 1:20,000 dilution of horseradish peroxidase-conjugated goat anti-mouse secondary antibody (Bio-Rad) preincubated for 15 min with NA1000 acetone powders (23). Western blot images were captured with a Kodak Image Station 440 CF.
Immunofluorescence Microscopy.
Immunofluorescence microscopy was performed by fixing cells in 2.5% (vol/vol) formaldehyde and 30 mM PBS (pH 7.5). All centrifugations were carried out at 7,000 rpm in a Sorvall Biofuge Pico table-top microcentrifuge for 7 min at room temperature. All washes were in a 1.0-ml volume unless indicated otherwise. The cells were washed twice with PBS and incubated with 100 μl of 0.1% (vol/vol) Triton X-100 in PBS for 45 min at room temperature. The cells were washed three times with PBS and resuspended in 100 μl of 5.0 mM EDTA in PBS for 20 min at room temperature. Lysozyme (Sigma) was added to a final concentration of 1.0–2.0 ng/μl, and the cells were monitored by microscopy until they became slightly transparent. The cells were washed three times with PBS to remove the lysozyme and incubated with 100 μl of blocking reagent [0.5% (wt/vol) Roche Molecular Biochemicals Blocking Reagent in PBS] at 37°C for 30 min. Anti-M2 polyclonal antibody (Sigma) was added to a final concentration of 1:250, and the cells were returned to 37°C for 1 h. The cells were washed three times with PBS containing 0.05% (vol/vol) Tween 20 and then resuspended in 100 μl of blocking reagent containing a 1:100 dilution of FITC-conjugated, affinity-purified goat anti-rabbit secondary antibody (Jackson ImmunoResearch) and incubated at 37°C for 30 min. Cells were then washed three times with PBS containing 0.05% (vol/vol) Tween 20 and resuspended in 5.0 μl of Slowfade Reagent A (Molecular Probes). Samples were kept on ice in the dark before microscopy.
2D Liquid Chromatography Tandem MS Analysis.
Stalks were purified as described above, except that they were resuspended in 0.25–0.5 ml of TE (10 mM Tris/1 mM EDTA, pH 8.0) or doubly distilled H2O. Also, stalk samples were incubated for 30 min to 1 h on ice with 16 μg/ml lysozyme (Sigma). The concentration of protein in the purified stalks was determined by the method of Bradford (24). One hundred fifty micrograms of stalk protein in 50 mM ammonium bicarbonate and 0.5–1.0% (wt/vol) RapiGest (Waters) was boiled for 5 min. The sample was allowed to cool to room temperature, dithiolthreitol was added to a final concentration of 5 mM, and the sample was placed at 60°C for 30 min. The sample was cooled to room temperature, iodacetamide was added to a final concentration of 15 mM, and the sample was placed in the dark for 30 min at room temperature. Trypsin (Sigma) was added at a ratio of 1 μg of trypsin::50 μg of stalk protein, and the sample was incubated at 37°C for a minimum of 16 h. To remove the RapiGest, HCl was added to a final concentration of 40 mM, and the sample was placed at 37°C for 30 min. This step precipitated the detergent. To remove the detergent and any debris present, the samples were centrifuged for 40 min at 30,000 rpm by using a TLA 100 rotor in a Beckman Optima TLX ultracentrifuge at 4°C.
The strong cation exchange column used in all experiments was 10 cm × 254 μm i.d. and was packed with 5-μm silica beads with a benzene sulfonic acid functional group (Phenomenex, Torrance, CA). In all cases, a 1-h linear gradient from 0 to 200 mM sodium perchlorate in 0.1% formic acid and 50% acetonitrile was used. The flow rate was 5 μl/min in each experiment. Approximately 20 μg of tryptic digest, as determined from a Bradford assay, was injected onto the SCX column in each experiment. Column effluent was collected manually in 2-min fractions for the duration of the gradient. Each fraction was subsequently diluted to reduce the acetonitrile concentration by adding 40 μl of 0.1% trifluoroacetic acid before reverse-phase analysis.
The reverse-phase column used in all experiments was packed with 5-μm C18 derivatized silica beads and was 10 cm × 254 μm. The diluted fractions collected in the strong cation exchange dimension were injected onto the reverse-phase column without further purification. The gradient in all reverse-phase experiments increased linearly from 5% to 40% of 0.1% formic acid in acetonitrile. The six ion-exchange fractions with the highest mass spectrometric ion yield were separated with a 6-h gradient. The six fractions with the next highest ion yield were separated with a 2-h gradient. All others were fractionated with a 1-h gradient. The reverse-phase column effluent was directly analyzed by using an LCQ Deca XP ion trap mass spectrometer (Thermo Electron, San Jose, CA) with an electrospray source.
The mass spectrometer was operated in data-dependent mode with three MS/MS spectra recorded for every MS spectrum. In this mode, the three most intense peaks in the MS spectrum are chosen for fragmentation. These parent masses are then placed on an exclusion list for 3 min. Mass spectra were collected for the duration of each reverse-phase experiment by using the xcalibur software package operated in centroid mode.
sequest Analysis.
The results of all experiments were analyzed by using the sequest search algorithm. The data were searched against a database of C. crescentus proteins obtained from SwissProt with the sequence for Gallus gallus egg white lysozyme manually inserted, which yielded a total of 3,750 proteins in the database. Because the sample was reduced and alkylated, all cysteine residues were assumed to be acetamidated. All analyses assumed tryptic enzyme activity with a maximum of two missed cleavages. To be considered acceptable peptide identification, the crosscorrelation values had to be at least 2.0, 2.5, or 3.5 for charge states of +1, +2, and +3, respectively. These criteria have been previously shown to be quite stringent (25). All mass spectra meeting these minimum criteria were also manually validated. Peptides that contained overlapping sequence coverage due to missed cleavages were counted as separate peptide identifications. At least two peptides from a protein had to be detected in order for that protein to be considered identified.
Supplementary Material
Acknowledgments
Y.V.B. dedicates this article to the loving memory of his father, Omer Brun (1926–2006), who playfully taught a young boy to seek scientific explanations to natural phenomena. We thank Howard Berg for helpful discussions; members of Y.V.B.’s laboratory, Daniel Kearns, Howard Berg, Harley McAdams, Kevin Young, Ned Wingreen, Volkmar Braun, Lucy Shapiro, and Clay Fuqua for critical reading of early versions of the manuscript; and L. Comolli and H. McAdams for communicating results before publication. This work was supported by National Institutes of Health Grants GM51986 (to Y.V.B.) and GM61336 (to J.P.R. and Y.V.B.) and by National Institutes of Health Predoctoral Fellowship GM07757 (to J.K.W.).
Abbreviations
- FDP
fluorescein diphosphate
- IM
inner membrane
- OM
outer membrane
- ABC
ATP-binding cassette.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
See Commentary on page 11435.
References
- 1.Cabeen M. T., Jacobs-Wagner C. Nat. Rev. Microbiol. 2005;3:601–610. doi: 10.1038/nrmicro1205. [DOI] [PubMed] [Google Scholar]
- 2.Poindexter J. S. In: The Prokaryotes. Balows A., Trüper H. G., Dworkin M., Harder W., Schleifer K.-H., editors. Vol. 3. New York: Springer; 1992. pp. 2176–2196. [Google Scholar]
- 3.Poindexter J. S. In: Current Perspectives in Microbial Ecology. Klug M. J., Reddy C. A., editors. Washington, DC: Am. Soc. Microbiol.; 1984. pp. 33–40. [Google Scholar]
- 4.Poindexter J. S. Microbiol. Rev. 1981;45:123–179. doi: 10.1128/mr.45.1.123-179.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmidt J. M., Stanier R. Y. J. Cell Biol. 1966;28:423–436. doi: 10.1083/jcb.28.3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gonin M., Quardokus E. M., O’Donnol D., Maddock J., Brun Y. V. J. Bacteriol. 2000;182:337–347. doi: 10.1128/jb.182.2.337-347.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ireland M. M., Karty J. A., Quardokus E. M., Reilly J. P., Brun Y. V. Mol. Microbiol. 2002;45:1029–1041. doi: 10.1046/j.1365-2958.2002.03071.x. [DOI] [PubMed] [Google Scholar]
- 8.Berg H. C. Random Walks in Biology. Princeton: Princeton Univ. Press; 1993. [Google Scholar]
- 9.Berg H. C., Purcell E. M. Biophys. J. 1977;20:193–219. doi: 10.1016/S0006-3495(77)85544-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benz R. Annu. Rev. Microbiol. 1988;42:359–393. doi: 10.1146/annurev.mi.42.100188.002043. [DOI] [PubMed] [Google Scholar]
- 11.Newton S. M., Allen J. S., Cao Z., Qi Z., Jiang X., Sprencel C., Igo J. D., Foster S. B., Payne M. A., Klebba P. E. Proc. Natl. Acad. Sci. USA. 1997;94:4560–4565. doi: 10.1073/pnas.94.9.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poindexter J. L. S., Bazire G. C. J. Cell Biol. 1964;23:587–607. [Google Scholar]
- 13.Daniel R. A., Errington J. Cell. 2003;113:767–776. doi: 10.1016/s0092-8674(03)00421-5. [DOI] [PubMed] [Google Scholar]
- 14.Ghosh A. S., Young K. D. J. Bacteriol. 2005;187:1913–1922. doi: 10.1128/JB.187.6.1913-1922.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibbs K. A., Isaac D. D., Xu J., Hendrix R. W., Silhavy T. J., Theriot J. A. Mol. Microbiol. 2004;53:1771–1783. doi: 10.1111/j.1365-2958.2004.04242.x. [DOI] [PubMed] [Google Scholar]
- 16.Figge R. M., Divakaruni A. V., Gober J. W. Mol. Microbiol. 2004;51:1321–1332. doi: 10.1111/j.1365-2958.2003.03936.x. [DOI] [PubMed] [Google Scholar]
- 17.Kruse T., Moller-Jensen J., Lobner-Olesen A., Gerdes K. EMBO J. 2003;22:5283–5292. doi: 10.1093/emboj/cdg504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Judd E. M., Comolli L. R., Chen J. C., Downing K. H., Moerner W. E., McAdams H. H. J. Bacteriol. 2005;187:6874–6882. doi: 10.1128/JB.187.20.6874-6882.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poindexter J. S. Bacteriol. Rev. 1964;28:231–295. doi: 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poindexter J. S., Hagenzieker J. G. J. Bacteriol. 1982;150:332–347. doi: 10.1128/jb.150.1.332-347.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones H. C., Schmidt J. M. J. Bacteriol. 1973;116:466–470. doi: 10.1128/jb.116.1.466-470.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jordan T. L., Porter J. S., Pate J. L. Arch. Mikrobiol. 1974;96:1–16. doi: 10.1007/BF00590158. [DOI] [PubMed] [Google Scholar]
- 23.Maddock J. R., Shapiro L. Science. 1993;259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- 24.Bradford M. M. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 25.Cargile B. J., Bundy J. L., Stephenson J. L., Jr. J. Proteome Res. 2004;3:1082–1085. doi: 10.1021/pr049946o. [DOI] [PubMed] [Google Scholar]
- 26.Klebba P. E., Newton S. M. Curr. Opin. Microbiol. 1998;1:238–247. doi: 10.1016/s1369-5274(98)80017-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.