Abstract
Cyclic nucleotide phosphodiesterases (PDEs) constitute a large superfamily of enzymes regulating concentrations of intracellular second messengers cAMP and cGMP through PDE-catalyzed hydrolysis. Although three-dimensional x-ray crystal structures of PDE4 and PDE5 have been reported, it is uncertain whether a critical, second bridging ligand (BL2) in the active site is H2O or HO− because hydrogen atoms cannot be determined by x-ray diffraction. The identity of BL2 is theoretically determined by performing molecular dynamics simulations and hybrid quantum mechanical/molecular mechanical (QM/MM) calculations, for the first time, on the protein structures resolved by x-ray diffraction. The computational results confirm our previous suggestion, which was based on QM calculations on a simplified active site model, that BL2 in PDE4 should be HO−, rather than H2O, serving as the nucleophile to initialize the catalytic hydrolysis of cAMP. The molecular dynamics simulations and QM/MM calculations on PDE5 demonstrate for the first time that the BL2 in PDE5 should also be HO− rather than H2O as proposed in recently published reports on the x-ray crystal structures, which serves as the nucleophile to initialize the PDE5-catalyzed hydrolysis of cGMP. These fundamental structural insights provide a rational basis for future structure-based drug design targeting PDEs.
INTRODUCTION
Cyclic nucleotide phosphodiesterases (PDEs) constitute a large superfamily (with at least 11 different gene families, i.e., PDE1 to PDE11) of structurally related, functionally distinct, and highly regulated enzymes (1). Most PDE families comprise more than one gene (∼20 PDE genes), which generate multiple protein products (>50 PDE proteins) via alternative mRNA splicing or use of different promoters/transcription initiation sites (2). PDEs regulate physiological processes by degrading intracellular second messengers, cyclic adenosine 3′,5′-monophosphate (cAMP) and cyclic guanosine 3′,5′-monophosphate (cGMP), through PDE-catalyzed hydrolysis (3–12). PDE4, PDE7, and PDE8 are highly specific for cAMP, whereas PDE5, PDE6, and PDE9 are highly specific for cGMP. PDE1, PDE2, PDE3, PDE10, and PDE11 exhibit dual specificity with greater or lesser preference for cAMP or cGMP (3). Thus, PDEs are clinical targets for such biological disorders as retinal degeneration, congestive heart failure, depression, asthma, erectile dysfunction, and inflammation (5,13–18).
Selective inhibitors of PDEs have already been shown or are expected to exert beneficial effects in a number of therapeutic areas, including stimulation of myocardial contractility, inhibition of mediator release, inhibition of platelet aggregation, cancer chemotherapy, analgesia, and treatment of depression, Parkinson's disease, and learning and memory disorders (14,16,19–41). For example, selective inhibitors of PDE4 may be used as new antidepressants, memory-enhancing drugs, and novel antiasthmatic and antiinflammatory agents for the treatment of chronic obstructive pulmonary disease (COPD), asthma, and other respiratory diseases (42). Selective inhibitors of PDE5, such as the well-known sildenafil (Viagra), vardenafil (Levitra), and tadalafil (Cialis), have been used to treat male erectile dysfunction (ED) (43–49). Understanding the protein structures, particularly the active site structures, and catalytic mechanism will provide a solid basis for rational design of novel, more potent inhibitors of PDEs for therapeutic treatment of a number of human diseases.
PDE families share a similar active site structure. In particular, a conserved carboxyl-terminal catalytic domain contains a histidine-rich motif [HD(X2)H(X)4N] and two divalent metal ion-binding sites (3,50,51). A divergent amino-terminal domain confers isoform-specific regulatory properties. Xu et al. (52) first reported a three-dimensional (3D) x-ray crystal structure of the catalytic domain of human phosphodiesterase 4B2B (PDE4). In the reported x-ray crystal structure, the active site contains a cluster of two divalent metal ions, denoted by Me1 and Me2. Me1 should be a Zn2+ ion based on the observed geometry of the metal-coordinating ligands, the anomalous x-ray diffraction behavior, the existing biochemical evidence, and the known high affinity of PDE4 for zinc. Me2 is most likely Mg2+ (53,54), but the possibility of Me2 = Mn2+ or Zn2+ cannot be ruled out (52). According to the 3D x-ray crystal structure reported by Xu et al. (52), in the PDE4 active site Asp-392 residue coordinates Me1 through an Oδ atom, His-238 and His-274 residues coordinate Me1 through the nitrogen atoms (denoted by Nɛ) on the side chain, and four solvent water molecules coordinate Me2 through the O atoms. In addition, there are two bridging ligands. One bridging ligand is clearly Asp-275, whose two oxygen atoms (denoted by Oδ) on the side chain respectively coordinate Me1 and Me2. However, it was uncertain whether the second bridging ligand (BL2) is a water molecule or a hydroxide ion because hydrogen atoms cannot be determined by an x-ray diffraction technique regardless of the resolution of the x-ray crystal structure. Such a structural problem is also difficult to solve by using other existing experimental approaches. For example, biochemical experiments would not be able to directly determine whether BL2 should be a water molecule or a hydroxide ion without using any hypothesis. Nuclear magnetic resonance (NMR), in principle, might be a potentially useful approach to solve such a structural problem, but no NMR study on such a bridging ligand (water molecule versus hydroxide ion) has ever been reported as far as we know.
In addition, we note that some interesting model compounds have been synthesized and reported in literature for mimicking bimetallic active sites of PDE or other similar metalloenzymes (55). Of particular interest, the dizinc model compounds synthesized have either a tightly bridged Zn-O(H)-Zn or a more loosely bridged Zn-(H)O…HO(H)-Zn unit. The more loosely bridged Zn-(H)O…HO(H)-Zn unit might be interesting for understanding the detailed catalytic process, although the study presented here concerns only the resting state of the enzymes. As far as the resting state is concerned, the x-ray crystal structures of PDE reported so far all demonstrated that BL2 in the PDE active sites should be either a water molecule or a hydroxide ion.
Xu et al. (52) further considered the possible position of substrate cAMP in the PDE4 active site to discuss the catalytic mechanism based on the PDE4 structure in which Me1 = Zn2+ and Me2 = Mg2+. They concluded from a model of cAMP docked in the PDE4 active site that a water molecule coordinating one or both metal ions could act as the nucleophile in the catalytic hydrolysis because they considered the second bridging ligand (BL2) to be a water molecule (52). Obviously, compared to a water molecule coordinating to one metal ion, the postulated bridging water molecule should be a worse nucleophile, whereas a possible bridging hydroxide ion should be a better nucleophile. For this reason, if BL2 is a water molecule, the nucleophile is likely a water molecule coordinating one metal ion (Me2). If BL2 is a hydroxide ion, the nucleophile is likely the bridging hydroxide ion. Thus, it is a key step for determining the nucleophile in the catalytic hydrolysis and understanding the catalytic mechanism to identify the structural form of BL2. The structural form of this critical bridging ligand should also affect the enzyme binding with substrates or inhibitors. Our previous first-principles quantum mechanical (QM) calculations (56) on simplified active site models of PDE4 indicated that a hydroxide ion can bridge the two positively charged metal ions, whereas a water molecule can coordinate to only one of the two metal ions. The QM calculations (56) were performed by using gradient-corrected density functional theory (DFT) with Becke's three-parameter hybrid exchange functional and the Lee-Yang-Parr correlation functional (B3LYP) (57–59) in combination with the 6-31G* basis set (60,61). The computational results suggest that BL2 in the active site of the reported x-ray crystal structure of PDE4 might be a hydroxide ion rather than a water molecule, to serve as the nucleophile to initialize the catalytic degradation of the intracellular second messenger. However, the effects of the protein environment were not accounted for in the QM calculations. It is unclear whether the protein environment affects the structural identity of BL2 or not.
Sung et al. (62) reported 3D x-ray crystal structures of the catalytic domain (residues 537 to 860) of a cGMP-specific human PDE5 complexed with three drug molecules, i.e., sildenafil (Viagra), tadalafil (Cialis), and vardenafil (Levitra). Most recently reported x-ray crystal structures of PDE4 and PDE5 have also demonstrated similar active site structures (63,64). All of these crystal structures consistently demonstrate that the active site of PDE5 is located at the center of the C-terminal helical bundle domain. Summarized in Table 1 are the geometric parameters from both the first (52,62) and newer (64,65) x-ray crystal structures. The substrate pocket is ∼10 Å deep, with a narrow opening and a wide inner space, giving a total volume of ∼330 Å3. It is composed of four subsites: a metal-binding site (M site), core pocket (Q pocket), hydrophobic pocket (H pocket), and lid region (L region). Overall, the M site and Q pocket are similar to those of PDE4, but the H pocket and the L region show significant structural differences compared to PDE4. The M site contains both a zinc ion (Me1) and a second metal ion (Me2, likely Mg2+) and is surrounded by helices α6, α8, α9, α10, and α12 (3). The first bridging ligand was clearly Asp-654. However, the second bridging ligand (BL2) was described as a water molecule (62–66).
TABLE 1.
Some key internuclear distances (Å) involving the metal ions in the QM/MM-optimized geometries* of PDE4 and PDE5 structures in comparison with the corresponding distances in the x-ray crystal structures
| PDE4
|
PDE5
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| BL2 = OH−
|
BL2 = H2O
|
BL2 = OH−
|
BL2 = H2O
|
|||||||
| Internuclear distances in PDE4/5 active site§ | Me2 = Mg2+ | Me2 = Zn2+ | Me2 = Mn2+ | Me2 = Mg2+ | Expt.† x-ray | Me2 = Mg2+ | Me2 = Zn2+ | Me2 = Mn2+ | Me2 = Mg2+ | Expt.‡ x-ray |
| Me1-O (HO− or H2O) | 1.94 (2.05) | 1.93 | 1.93 | 3.70 | 1.90 (2.14) | 1.98 | 1.98 | 1.98 | 3.58 | 2.54 (2.05) |
| Me1-O (Asp-275/654) | 2.20 (2.09) | 2.63 | 3.76 | 2.00 | 2.20 (2.16) | 2.23 | 2.45 | 2.27 | 1.98 | 2.06 (2.11) |
| Me1-O (Asp-392/764) | 2.08 (2.17) | 1.96 | 1.92 | 1.91 | 2.20 (2.08) | 2.02 | 1.98 | 2.02 | 1.97 | 2.07 (2.08) |
| Me1-N (His-238/617) | 2.02 (2.01) | 2.01 | 2.02 | 1.98 | 2.00 (2.15) | 2.03 | 2.01 | 2.02 | 1.97 | 2.09 (2.14) |
| Me1-N (His-274/653) | 2.08 (2.05) | 2.05 | 2.04 | 2.01 | 2.10 (2.15) | 2.07 | 2.05 | 2.05 | 1.99 | 2.09 (2.14) |
| Me2-O (HO− or H2O) | 1.94 (2.05) | 1.86 | 1.95 | 2.01 | 2.40 (2.11) | 1.96 | 1.89 | 2.00 | 2.00 | 2.37 (2.07) |
| Me2-O (Asp-275/654) | 2.05 (2.05) | 2.07 | 3.27 | 2.05 | 2.40 (2.18) | 2.08 | 1.93 | 2.13 | 2.04 | 2.10 (2.11) |
| Me2-O (W1) | 2.28 (2.27) | 3.49 | 3.61 | 2.18 | 2.49 (2.19) | 2.21 | 2.06 | 2.33 | 2.29 | 2.29 (2.11) |
| Me2-O (W2) | 2.18 (2.16) | 2.31 | 2.24 | 2.14 | 2.34 (2.15) | 2.26 | 3.70 | 2.38 | 2.25 | 2.39 (2.13) |
| Me2-O (W3) | 2.19 (2.15) | 2.21 | 2.44 | 2.17 | 2.57 (2.21) | 2.22 | 4.44 | 2.46 | 2.14 | 2.29 (2.10) |
| Me2-O (W4) | 2.11 (2.11) | 2.08 | 2.16 | 2.12 | 2.18 (2.17) | 2.06 | 2.00 | 2.15 | 2.09 | 2.28 (2.12) |
The geometries were fully optimized by performing the QM/MM calculations at the B3LYP/6-31G*:Amber level. The internuclear distances given in parentheses for PDE4 (BL2 = HO−) refer to another PDE4 (BL2 = HO−) structure (with Asp-392 residue protonated) obtained from the QM/MM geometry optimization using the initial structure of PDE4 (BL2 = H2O); during the geometry optimization, a proton was transferred from BL2 (i.e., H2O) to an Oδ atom of Asp-392 side chain so that the Asp-392 side chain is protonated (see text for the detailed description).
Experimental values from the first x-ray crystal structure of PDE4 (pdb code 1F0J (52)). The corresponding values in the parentheses are experimental values from the latest x-ray crystal structure of PDE4 (pdb code 1XOM (64)).
Experimental values from the first x-ray crystal structure of PDE5 (pdb code 1RKP (62)). The corresponding values in the parentheses are experimental values from the latest x-ray crystal structure of PDE5 (pdb code 1TBF (66)).
Numbering of the active site residues used here is based on PDE4B2B/PDE5A. W1, W2, W3, and W4 represent the water molecules coordinating the second metal ion (Me2).
The background summarized above reveals that it is still unclear whether BL2, a critical bridging ligand, in the active site of PDE5 and PDE4 should be H2O or HO−. Further, it is also unknown whether the structural form (H2O or HO−) of BL2 in the PDE5 active site is the same as that in the PDE4 active site or not and whether the specific protein environment can alter the structural form of BL2 or not. Reasonable answers to these fundamental structural questions are crucial for understanding the catalytic mechanisms of PDEs, for studying enzyme–ligand (substrate or inhibitor) binding, and for future rational design of novel drugs targeting PDEs. For example, different answers to this question may point to different possible nucleophiles attacking the phosphorus center of the substrate (cAMP or cGMP) to initialize the catalytic reaction. In addition, the structural identity of BL2 should also affect the PDE binding with substrates and inhibitors, as different structural forms (neutral H2O or negatively charged HO−) of BL2 would provide different electrostatic potentials affecting PDE binding with a substrate or inhibitor. Thus, a drug design and discovery effort based on an incorrect structural identity of BL2 could lead only to meaningless predictions.
To answer the above crucial structural questions, we have carried out extensive molecular dynamics (MD) simulations and hybrid quantum mechanical/molecular mechanical (QM/MM) calculations, to our knowledge for the first time, on the entire catalytic domains of PDE4 and PDE5. These MD simulations and QM/MM calculations accounting for the specific protein environmental effects provide consistent answers to these structural questions.
COMPUTATIONAL STRATEGY AND METHODS
Although it is unknown whether the second bridging ligand (BL2) in the active site of PDE4 and PDE5 should be H2O or HO−, the available x-ray crystal structures (52,62–66) have clearly provided the relative positions of the heavy atoms. In particular, all of the x-ray crystal structures clearly show that BL2 bridges the two metal ions in the active site for both PDE4 and PDE5. Based on these x-ray crystal structures, we wanted to further examine which structural form (H2O or HO−) of BL2 can bridge the two metal ions in the active site for each PDE family (PDE4 and PDE5) and, therefore, can lead to a 3D structure that is consistent with the corresponding x-ray crystal structures. To solve this structural problem, we performed two sets of MD simulations and QM/MM calculations for each PDE. The two sets of modeling studies started from the same x-ray crystal structure for each PDE. However, BL2 was set to H2O in one set of modeling studies, whereas BL2 was set to HO− in the other set of modeling studies. These two sets of modeling studies allow us to determine which structural form (HO− or H2O) of BL2 leads to a protein structure being consistent with the corresponding x-ray crystal structure for each PDE. Such a computational strategy can complement the x-ray diffraction techniques to answer such kinds of complex questions of protein structure that cannot be answered by the x-ray diffraction experiment alone.
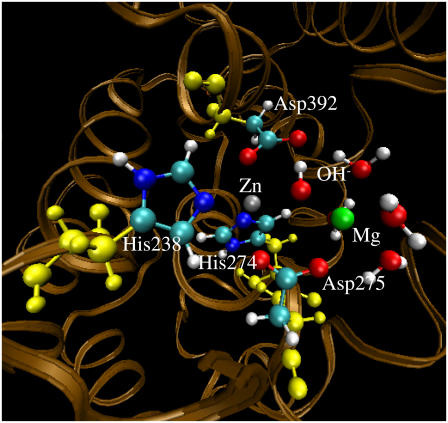
The standard protonation states at physiological condition (pH ∼7.4) were set to all ionizable residues, and the proton was set on the Nɛ atom for all His residues. As seen in Figs. 1–4, His-238 and His-274 of PDE4 and His-617 and His-653 of PDE5 all coordinate the Zn2+ ion through the other nitrogen atom (denoted by Nη) on the side chain. The Amber7 program suite (67) was used to perform all the MD simulations in this study. The partial atomic charges for the nonresidue atoms of hydroxide ion were calculated by using the restricted electrostatic potential (RESP) fitting protocol implemented in the Antechamber module of the Amber7 program after electrostatic potential (ESP) calculations at ab initio HF/6-31G* level. Each aforementioned initial structure was neutralized by adding counterions and was solvated in a rectangular box of TIP3P water molecules with a minimum solute–wall distance of 10 Å. Specifically, the net charge is −19 for the PDE4 (BL2 = HO−) structure, −18 for the PDE4 (BL2 = H2O) structure, 0 for the PDE5 (BL2 = HO−) structure, and +1 for the PDE5 (BL2 = H2O) structure. Hence, 19 Na+ ions were added to neutralize the solvated PDE4 (BL2 = HO−) system, 18 Na+ ions were added to neutralize the solvated PDE4 (BL2 = H2O) system, and one Cl− was added to neutralize the solvated PDE5 (BL2 = H2O) system. The total number of atoms in each solvated protein structure for the MD simulations is more than 40,000, although the total number of atoms of each PDE with the substrate is only ∼5000.
FIGURE 1.
Geometry of the PDE4 structure (with Me1 = Zn2+, Me2 = Mg2+, and BL2 = HO−) optimized by the QM/MM calculation at the B3LYP/6-31G*:Amber level. The high-layer atoms are in the PDE4 active site represented by the balls in colors other than yellow. The yellow balls represent the low-layer atoms of the residues coordinating the metal ions.
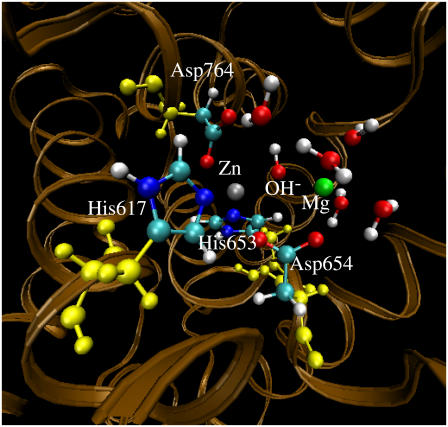
FIGURE 4.
Geometry of the PDE4 structure (with Me1 = Zn2+, Me2 = Mg2+, and BL2 = H2O) optimized by the QM/MM calculation at the B3LYP/6-31G*:Amber level. The high-layer atoms are represented by the balls in colors other than yellow. The yellow balls represent the low-layer atoms of the residues coordinating the metal ions.
FIGURE 2.
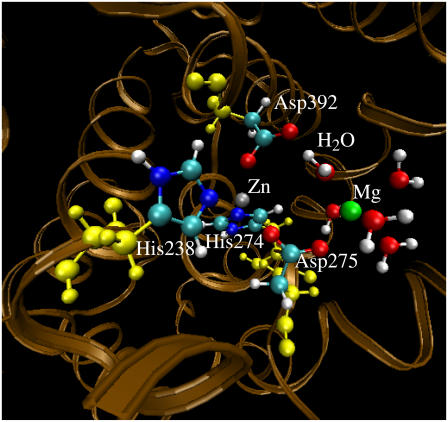
Geometry of the PDE4 structure (with Me1 = Zn2+, Me2 = Mg2+, and BL2 = H2O) optimized by the QM/MM calculation at the B3LYP/6-31G*:Amber level. The high-layer atoms are represented by the balls in colors other than yellow. The yellow balls represent the low-layer atoms of the residues coordinating the metal ions.
FIGURE 3.
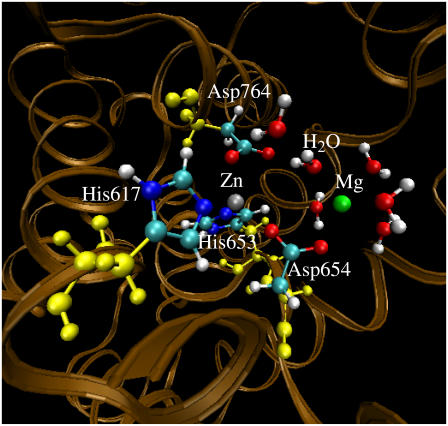
Geometry of the PDE5 structure (with Me1 = Zn2+, Me2 = Mg2+, and BL2 = HO−) optimized by the QM/MM calculation at the B3LYP/6-31G*:Amber level. The high-layer atoms are represented by the balls in colors other than yellow. The yellow balls represent the low-layer atoms of the residues coordinating the metal ions.
The general procedure for carrying out the MD simulations in water is essentially the same as that used in our previous MD simulations on other similar protein systems containing a bimetallic active site (68–70). The nonbonded models were used for the metal ions. The MD simulations in this study were performed by using the Sander module of the Amber7 program. The protein–solvent system was optimized before the simulation as follows. First, the protein was frozen, and the solvent molecules with counterions were allowed to move during a 3000-step minimization. Second, all the atoms were allowed to relax by a 3000-step full minimization. After full relaxation, the protein was frozen, and the solvent molecules with the counterions were allowed to move during a 2500-step MD simulation. Then, with all the atoms in relaxation, the system was slowly heated to 250 K in 20 ps and then to 298.15 K in 80 ps. The production MD simulation at T = 298.15 K was kept running until we believed that a stable MD trajectory had been obtained for each of the simulated structures. The time step used for the MD simulations was 2 fs. Periodic boundary conditions in the constant pressure and temperature (NPT) ensemble (i.e., isothermal-isobaric ensemble) at T = 298.15 K with Berendsen temperature coupling (71) and P = 1 atm with isotropic molecule-based scaling (71) were applied. The SHAKE algorithm (72) was used to fix all covalent bonds containing a hydrogen atom. The nonbonded pair list was updated every 10 steps. The particle mesh Ewald (PME) method (73) was used to treat long-range electrostatic interactions. A residue-based cutoff of 10 Å was utilized for the noncovalent interactions. The coordinates of the simulated systems were collected every 1 ps during the production MD stages.
The ONIOM approach (74–76) implemented in the Gaussian03 program (77) was used to fully optimize geometries of the PDE4 and PDE5 structures. Two layers were defined in each of our ONIOM calculations: the high layer (including the metal ions and atoms from all ligands coordinating the metal ions; see below for the specific high-layer atoms depicted as the balls in Figs. 1–4) was treated quantum mechanically at the B3LYP/6-31G* level, and the low layer was treated molecular mechanically by using the Amber force field as used in our MD simulations with the Amber7 program. The low layer included all of the amino acid residues, the two metal ions, BL2, and all water molecules coordinating the metal ions; the other solvent water molecules were neglected in the QM/MM calculations. Some missing force field parameters (including the RESP charges for HO− and van der Waals parameters for Zn2+ and Mg2+) were added before running the QM/MM calculations with ONIOM approach. The RESP charges used for HO− were calculated at the HF/6-31G* level to be −1.206 and 0.206 for the O and H atoms, respectively. The van der Waals parameters for Zn2+ and Mg2+ came from the “parm99.dat” data file of the Amber force field (67); these parameters were missing in the “parm96.dat” data file adopted by the Gaussian03 (77).
The initial PDE4 and PDE5 structures used in the MD simulations and QM/MM calculations were built from the corresponding x-ray crystal structures (52,63) deposited in the Protein Data Bank (78) (Protein Data Bank (pdb) code: 1FOJ for PDE4 and 1RKP for PDE5). All of the MD simulations and QM/MM calculations started from the same x-ray crystal structures in terms of the coordinates of the nonhydrogen atoms. The MD simulations and QM/MM calculations were performed on an HP Superdome supercomputer (a shared-memory system with a total of 256 processors and parallel computing) at University of Kentucky Center for Computational Sciences and on a 34-processor IBM x335 Linux cluster (with parallel computing) in our own lab.
RESULTS AND DISCUSSION
As mentioned above, in both proteins PDE4 and PDE5, Me1 is known to be Zn2+ and Me2 is most likely Mg2+. Unless explicitly stated otherwise, all MD simulations and QM/MM calculations discussed below were carried out for the protein structures in which Me1 = Zn2+ and Me2 = Mg2+.
MD-simulated structures
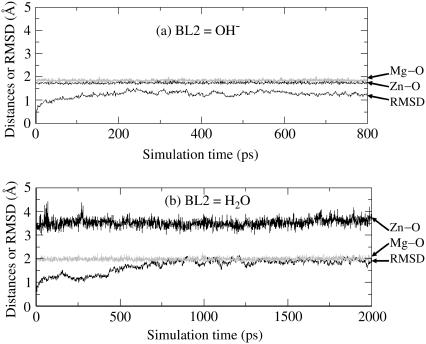
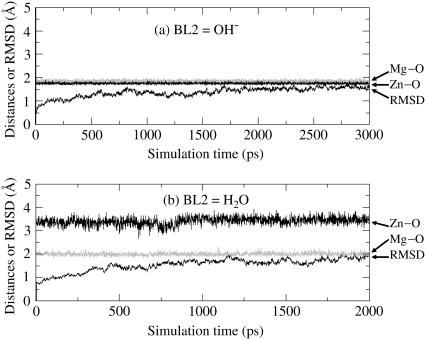
The trajectory of the MD simulation on the solvated PDE4(BL2=HO−) system was quickly stabilized after ∼200 ps. Thus, the MD simulation was stopped at 800 ps. It needed a little longer time to obtain a stable MD trajectory for each of other solvated protein systems. So, the other MD simulations were stopped at either 2000 ps or 3000 ps. Depicted in Figs. 5 and 6 are plots of the simulated key internuclear distances between the metal ions and the oxygen atom (O) in the examined second bridging ligand (BL2 = HO− or H2O) versus the simulation time, along with root mean-square deviation (RMSD) of the positions of backbone atoms in the simulated structure from those in the initial structure. As seen in Figs. 5 and 6, the RMSD values are all smaller than 2.0 Å for all of the MD trajectories, demonstrating that the backbones of the proteins PDE4 and PDE5 did not dramatically change in going from their crystal structures to the protein structures in water. Specifically, the average RMSD value is 1.30 Å for PDE4 (BL2 = HO−) over 200–800 ps, 1.88 Å for PDE4 (BL2 = H2O) over 1000–2000 ps, 1.55 Å for PDE5 (BL2 = HO−) over 2000–3000 ps, and 1.73 Å for PDE5 (BL2 = H2O) over 1000–2000 ps. The RMSD values associated with BL2 = H2O are systematically larger than the RMSD values associated with BL2 = HO−, implying that the protein structures with BL2 = HO− might be slightly more stable than the corresponding structures with BL2 = H2O.
FIGURE 5.
Plots of the key internuclear distances (Å) versus the simulation time in the MD-simulated PDE4 structures in water. Zn-O refers to the distance between the Zn2+ ion and the BL2 (the second bridging ligand) oxygen. Mg-O represents the distance between the Mg2+ ion and the BL2 oxygen. RMSD represents the root mean-square deviation (Å) of the simulated positions of PDE4 backbone atoms from those in the initial x-ray crystal structure. BL2 = HO− (a) or H2O (b).
FIGURE 6.
Plots of the key internuclear distances (Å) versus the simulation time in the MD-simulated PDE5 structures. Zn-O refers to the distance between the Zn2+ ion and the BL2 (the second bridging ligand) oxygen. Mg-O represents the distance between the Mg2+ ion and the BL2 oxygen. RMSD represents the root mean-square deviation (Å) of the simulated positions of PDE5 backbone atoms from those in the initial structure. BL2 = HO− (a) or H2O (b).
A survey of the simulated distances depicted in Figs. 5 and 6 reveals a remarkable difference between the structures simulated with BL2 = HO− and with BL2 = H2O. When BL2 was considered to be HO−, the simulated average distances between the BL2 oxygen and two metal ions (Zn2+ and Mg2+) in both PDE4 and PDE5 are close to ∼2.0 Å, showing that a hydroxide ion as BL2 coordinated to the two metal ions simultaneously in the simulated PDE4 and PDE5 structures. When BL2 was regarded as H2O, in the simulated structures of PDE4 and PDE5, the average distance between the BL2 oxygen and Mg2+ (Me2) is 1.99 Å in PDE4 and 2.01 Å in PDE5, whereas the average distance between the BL2 oxygen and Zn2+ (Me1) is 3.54 Å in PDE4 and 3.45 Å in PDE5. The BL2 oxygen coordinated to only the second metal ion (Me2) and left the first metal ion (Me1) during the MD simulations of the PDE4 and PDE5 structures, when BL2 was regarded as H2O. So, only the structures simulated with BL2 = HO− are qualitatively consistent with the corresponding x-ray crystal structures for both PDE4 and PDE5. The MD simulations suggest that BL2 should be a hydroxide ion rather than a water molecule.
The remaining question is whether the MD simulations based on classical force field parameters are or are not reliable to answer such a complex structural question. Nevertheless, as discussed below, the results obtained from the MD simulations are supported by the results obtained from the QM/MM calculations in which the QM-treated atoms (the high layer) include the metal ions and all of the atoms coordinating the metal ions.
Geometries optimized by the QM/MM calculations on the systems in which Me1 = Zn2+ and Me2 = Mg2+
Some important internuclear distances in the QM/MM-optimized geometries of PDE4 and PDE5 structures are summarized in Table 1, in comparison with the corresponding experimental data in the x-ray crystal structures. The optimized protein structures are depicted in Figs. 1–4, where the high-layer atoms are highlighted by the balls.
As seen in Table 1, the QM/MM-optimized geometries are qualitatively consistent with the corresponding MD-simulated structures discussed above. For both PDE4 and PDE5, only a hydroxide ion (considered as BL2) can bridge the two positively charged metal ions, whereas a water molecule (considered as BL2) can coordinate to only one metal ion and left the other metal ion during the geometry optimizations. Specifically, when BL2 = H2O, we carefully tested using different initial geometries in the geometry optimizations, including starting from the optimized geometries of the corresponding proteins with BL2 = HO−, but an additional proton was added to the hydroxide oxygen. In turned out that all of the geometry optimizations using different initial geometries eventually led to qualitatively the same geometries concerning whether BL2 coordinates one or two metal ions in the active site. For a special test of the QM/MM geometry optimization on the proposed PDE4 (BL2 = H2O) structure, we first performed a partial geometry optimization with two critical internuclear distances fixed at the experimental values in the x-ray crystal structure: one is the internuclear distance between Zn2+ (Me1) and the BL2 oxygen, and the other is that between Mg2+ (Me2) and the BL2 oxygen. However, during such a partial geometry optimization, a proton in BL2 left the BL2 oxygen and went to form a covalent bond with the Asp-392 Oδ atom, which does not coordinate the metal ions. The partial geometry optimization was followed by a full geometry optimization. The finally optimized O(BL2)–H(BL2) and Oδ(Asp-392)–H(BL2) distances are 1.74 and 1.00 Å, respectively. So, the partial optimization of the proposed PDE4 (BL2 = H2O) structure followed by the full optimization actually led to another PDE4 (BL2 = HO−) structure in which the Asp-392 side chain is protonated. The key geometric parameters in this optimized PDE4 (BL2 = HO−) geometry are also summarized in Table 1 for comparison.
When BL2 = HO−, the optimized distances between the BL2 oxygen and Zn2+ (Me1) and between the BL2 oxygen and Mg2+ (Me2) in PDE4 are all ∼1.94 Å, and the corresponding distances optimized in PDE5 are 1.98 and 1.96 Å, respectively. These QM/MM-optimized distances are reasonably close to the corresponding experimental values in the x-ray crystal structures, as seen in Table 1.
When BL2 = H2O, the optimized distance between the BL2 oxygen and Mg2+ (Me2) is 2.01 Å in PDE4 and 2.00 Å in PDE5, whereas the optimized distance between the BL2 oxygen and Zn2+ (Me1) is 3.70 Å in PDE4 and 3.58 Å in PDE5. The optimized distances between the BL2 oxygen and Zn2+ (Me1) are considerably longer than the experimental values (1.90 to 2.54 Å) in the x-ray crystal structures.
The protein structures optimized by the QM/MM calculations are consistent with the MD simulations and confirm that BL2 in the PDE active site should be HO−, rather than H2O, for both PDE4 and PDE5.
It should be pointed out that the QM/MM-optimized geometric parameters summarized in Table 1 for PDE4 are all qualitatively consistent with the corresponding parameters reported in the earlier QM study (56) of the simplified PDE4 model system in terms of the coordination of the two metal ions. However, the specific internuclear distances optimized by performing the QM/MM calculations at the B3LYP:Amber level, accounting for effects of the protein environment, are significantly different from the corresponding distances optimized at the B3LYP/6-31G* level for the simplified PDE4 active site model neglecting effects of the protein environment. For example, the Me1-O(HO−) and Me2-O(HO−) distances optimized at the B3LYP/6-31G* level for the simplified PDE4 model system are ∼1.95 and ∼1.99 Å, respectively, whereas the corresponding Me1-O(HO−) and Me2-O(HO−) distances optimized at the QM/MM(B3LYP:Amber) level accounting for effects of the protein environment are all ∼1.94 Å. Thus, for more accurate determination of the protein structures, effects of the protein environment should be accounted for. Thus, we performed the QM/MM geometry optimizations for PDE5 structures, in addition to the MD simulations, in the computational study presented here.
Geometries of PDE4 and PDE5 with different metal ions
All of the above-mentioned MD simulations and QM/MM calculations were performed for the PDE4 and PDE5 structures in which Me1 = Zn2+ and Me2 = Mg2+. As discussed above, Me1 is clearly Zn2+, and, therefore, there is no question concerning the identity of Me1. Me2 is most likely Mg2+, but the possibilities of Me2 = Zn2+ or Mn2+ cannot be ruled out. We also wanted to know whether the second bridging ligand (BL2) in the active site of PDE4 and PDE5 could be H2O or not when Me2 was replaced by Zn2+ or Mn2+. So, we also optimize the geometries of the possible PDE4 and PDE5 structures in which Me1 = Zn2+ and Me2 = Zn2+ or Mn2+ by carrying out the QM/MM calculations at the B3LYP/6-31G*:Amber level. The optimized geometries of the PDE4 and PDE5 structures (with Me1 = Zn2+, Me2 = Mg2+, and BL2 = HO−) were used as the starting structures (with the necessary replacement of Me2 and BL2) to optimize the geometries of the corresponding PDE4 and PDE5 structures (with Me1 = Zn2+, Me2 = Zn2+ or Mn2+, and BL2 = HO− or H2O).
The QM/MM geometry optimizations with Me1 = Zn2+ and Me2 = Zn2+ or Mn2+ led to protein structures similar to the corresponding structures optimized with Me2 = Mg2+ concerning the binding of BL2 with the metal ions in both PDE4 and PDE5. Only HO− as BL2 can bridge the two positively charged metal ions. When BL2 = H2O, during the geometry optimization process, the BL2 oxygen gradually left one of the two metal ions and coordinated only to the other metal ion in the active site no matter whether Me2 = Zn2+ or Mn2+ (the geometries are not shown). Thus, our conclusion concerning the identity of BL2 does not change when Mg2+ (as Me2) in the active site is replaced by Zn2+ or Mn2+ for both PDE4 and PDE5.
Some key internuclear distances in the optimized geometries of the PDE4 and PDE5 structures (with Me1 = Zn2+, Me2 = Zn2+ or Mn2+, and BL2 = HO−) are also summarized in Table 1 for comparison. As seen in Table 1, all of the distances in the optimized geometries of the PDE4 structures (with Me1 = Zn2+, Me2 = Zn2+ or Mn2+, and BL2 = HO−) are very close to the corresponding distances in the optimized geometry (with Me1 = Zn2+, Me2 = Mg2+, and BL2 = HO−), except the distance between Me2 and the oxygen atom of a water molecule (W1). The optimized Me2-W1 distance is 2.28 Å when Me2 = Mg2+, 3.49 Å when Me2 = Zn2+, and 3.61 Å when Me2 = Mn2+. These results indicate that W1 left Me2 when it was changed from Mg2+ to Zn2+ or Mn2+ in the PDE4 active site. Apparently, the optimized PDE4 structure with Me2 = Mg2+ is in the best agreement with the x-ray crystal structures of PDE4, which further supports the assumption of Me2 = Mg2+ in the PDE4 active site.
Concerning the optimized geometries of the PDE5 structures with different identities of Me2, as seen in Table 1, the internuclear distances in the PDE5 structure with Me2 = Zn2+ and Mn2+ are all very close to the corresponding distances in the PDE5 structure with Me2 = Mg2+, except the distances between Me2 and the oxygen atoms of water molecules W2 and W3 in the active site of PDE5 with Me2 = Zn2+. The Me2–O(W2) distance optimized at the B3LYP/6-31G*:Amber level is 2.26 Å when Me2 = Mg2+, 3.70 Å when Me2 = Zn2+, and 2.38 Å when Me2 = Mn2+. The Me2–O(W3) distance optimized at the B3LYP/6-31G*:Amber level is 2.22 Å when Me2 = Mg2+, 4.44 Å when Me2 = Zn2+, and 2.46 Å when Me2 = Mn2+. The optimized geometries of PDE5 with Me2 = Mg2+ and with Me2 = Mn2+ are all consistent with the x-ray crystal structures of PDE5, and, therefore, both Me2 = Mg2+ and Me2 = Mn2+ are possible in the PDE5 active site.
So, all of the MD and QM/MM results consistently reveal that for both PDE4 and PDE5, the second bridging ligand in the active site of the reported x-ray crystal structures should be a hydroxide ion, rather than a water molecule, no matter whether the uncertain second metal ion is Mg2+, Zn2+, or Mn2+. This conclusion provides a valuable structural basis for future rational design of drugs targeting PDE4 and PDE5.
CONCLUSION
Molecular dynamics (MD) simulations and hybrid quantum mechanical/molecular mechanical (QM/MM) calculations on phosphodiesterase (PDE) family 4 (PDE4) structure including the protein environment resolved by the x-ray diffraction confirm our previous suggestion, which was based on QM calculations on a simplified active site model neglecting the protein environment, that the second bridging ligand in the PDE4 active site should be a hydroxide ion rather than a water molecule, which serves as the nucleophile to initialize the PDE4-catalyzed hydrolysis of substrate cAMP. The MD simulations and QM/MM calculations on PDE5 including the protein environment resolved by the x-ray diffraction demonstrate, for the first time to our knowledge, that the second bridging ligand in the PDE5 active site should also be a hydroxide ion rather than the water molecule proposed in recent publications reporting the x-ray crystal structures of PDE5, which serves as the nucleophile to initialize the PDE5-catalyzed hydrolysis of substrate cGMP. These conclusions stand no matter whether the uncertain second metal ion is Mg2+, Zn2+, or Mn2+ in both PDE4 and PDE5 in light of the QM/MM calculations on the PDE4 and PDE5 structures with different metal ions. All of the computational results consistently indicate that the effects of the protein environment do not qualitatively change the identity of the second bridging ligand, implying that the second bridging ligand in the active site of other proteins in the PDE superfamily could also be a hydroxide ion rather than a water molecule, which serves as the nucleophile to initialize the PDE-catalyzed hydrolysis of the intracellular second messenger cAMP or cGMP. These fundamental structural insights provide a rational basis for future structure-based design of drugs targeting proteins in the PDE superfamily.
Acknowledgments
The research was supported in part by the National Institutes of Health (grant R01DA013930 to C.-G. Zhan), by National Natural Science Foundation of China, and by the Center for Computational Sciences at University of Kentucky.
Ying Xiong and Hai-Ting Lu contributed equally to this work.
References
- 1.Francis, S. H., I. V. Turko, and J. D. Corbin. 2001. Cyclic nucleotide phosphodiesterases: relating structure and function. Prog. Nucleic Acid Res. Mol. Biol. 65:1–52. [DOI] [PubMed] [Google Scholar]
- 2.Manganiello, V. 2003. Cyclic nucleotide phosphodiesterase 5 and sildenafil: Promises realized. Mol. Pharmacol. 63:1209–1211. [DOI] [PubMed] [Google Scholar]
- 3.Karnam, S. M., Z. Huiping, and M. M. Gabriel. 2002. PKA-dependent activation of PDE3A and PDE4 and inhibition of adenylyl cyclase V/VI in smooth muscle. Am. J. Physiol. Cell Physiol. 282:C508–C517. [DOI] [PubMed] [Google Scholar]
- 4.Callahan, S. M., N. W. Cornell, and P. V. Dunlap. 1995. Purification and properties of periplasmic 3′/5′-cyclic nucleotide phosphodiesterase—a novel zinc-containing enzyme from the marine symbiotic bacterium vibrio-fischeri. J. Biol. Chem. 270:17627–17632. [DOI] [PubMed] [Google Scholar]
- 5.Conti, M., S. L. C. Jin, L. Monaco, D. R. Repaske, and J. V. Swinnen. 1991. Hormonal-regulation of cyclic-nucleotide phosphodiesterases. Endocr. Rev. 12:218–234. [DOI] [PubMed] [Google Scholar]
- 6.Houslay, M. D. 1998. Adaptation in cyclic AMP signalling processes: A central role for cyclic AMP phosphodiesterases. Semin. Cell Dev. Biol. 9:161–167. [DOI] [PubMed] [Google Scholar]
- 7.Conti, M., and S. L. C. Jin. 2000. The molecular biology of cyclic nucleotide phosphodiesterases. Prog. Nucleic Acid Res. Mol. Biol. 63:1–38. [DOI] [PubMed] [Google Scholar]
- 8.Mehats, C., C. B. Andersen, M. Filopanti, S. L. C. Jin, and M. Conti. 2002. Cyclic nucleotide phosphodiesterases and their role in endocrine cell signaling. Trends Endocrinol. Metab. 13:29–35.11750860 [Google Scholar]
- 9.Park, J. Y., F. Richard, S. Y. Chun, J. H. Park, E. Law, K. Horner, S. L. C. Jin, and M. Conti. 2003. Phosphodiesterase regulation is critical for the differentiation and pattern of gene expression in granulosa cells of the ovarian follicle. Mol. Endocrinol. 17:1117–1130. [DOI] [PubMed] [Google Scholar]
- 10.Zhang, H. T., J. D. Steketee, S. L. C. Jin, M. Conti, and J. M. O'Donnell. 2003. Deficiency of the PDE4B phosphodiesterase enzyme alters amphetamine-induced hyperactivity and stereotypic behavior. FASEB J. 17:A206–A207. [Google Scholar]
- 11.Houslay, M. D., and D. R. Adams. 2003. PDE4 cAMP phosphodiesterases: modular enzymes that orchestrate signalling cross-talk, desensitization and compartmentalization. Biochem. J. 370:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamato, S., R. Shoda, J. Akiyama, N. Uemura, M. Tokuhara, T. Shimizu, and K. Matsueda. 2003. Phosphodiesterase inhibitor enhances nerve-mediated relaxation of the human stomach. Gastroenterology. 124:A579–A579. (Abstr.) [Google Scholar]
- 13.Teixeira, M. M., R. W. Gristwood, N. Cooper, and P. G. Hellewell. 1997. Phosphodiesterase (PDE)4 inhibitors: Anti-inflammatory drugs of the future? Trends Pharmacol. Sci. 18:164–170. [DOI] [PubMed] [Google Scholar]
- 14.Torphy, T. J., and C. Page. 2002. Phosphodiesterases: the journey towards therapeutics. Trends Pharmacol. Sci. 21:157–159. [DOI] [PubMed] [Google Scholar]
- 15.Rotella, D. P. 2002. Phosphodiesterase 5 inhibitors: Current status and potential applications. Nat. Rev. Drug Discov. 1:674–682. [DOI] [PubMed] [Google Scholar]
- 16.Reffelmann, T., and R. A. Kloner. 2003. Therapeutic potential of phosphodiesterase 5 inhibition for cardiovascular disease. Circulation. 108:239–244. [DOI] [PubMed] [Google Scholar]
- 17.Zhang, K. Y., P. N. Ibrahim, S. Gillette, and G. Bollag. 2005. Phosphodiesterase-4 as a potential drug target. Expert Opin. Ther. Targets. 9:1283–1305. [DOI] [PubMed] [Google Scholar]
- 18.Houslay, M. D., M. Sullivan, and G. B. Bolger. 1998. The multienzyme PDE4 cyclic adenosine monophosphate-specific phosphodiesterase family: intracellular targeting, regulation, and selective inhibition by compounds exerting anti-inflammatory and antidepressant actions. Adv. Pharmacol. 44:225–342. [DOI] [PubMed] [Google Scholar]
- 19.Adachi, H., H. Ishihara, and T. Nagakura. 1998. Effects of a novel phosphodiesterase 5 inhibitor, E4010, on cardiohemodynamics and plasma hormones in a porcine model of heart failure. Naunyn Schmiedebergs Arch. Pharmacol. 358:36100. (Abstr.) [Google Scholar]
- 20.Chevalier, E., F. Petoux, A. Langlois, M. Chovet, and A. Doherty. 1999. Protective effects of CI-1018, a novel selective type IV phosphodiesterase inhibitor on a rate sepsis model. Gastroenterology. 116:G3776. (Abstr.) [Google Scholar]
- 21.Mukai, E., H. Ishida, S. Fujimoto, M. Kajikawa, J. Fujita, Y. Tsuura, Y. Yamada, and Y. Seino. 1999. The novel hypoglycemic agent JTT-608 is a phosphodiesterase inhibitor. Diabetologia 42(Suppl):462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Montana, J., N. Cooper, H. Dyke, J. Oxford, R. Gristwood, C. Lowe, H. Kendall, G. Buckley, R. Maxey, J. Warneck, J. Gregory, L. Gowers, F. Galleway, R. Wills, R. Naylor, B. Tudhalar, K. Broadley, and H. Danahay. 1999. Activity of D4418, a novel phosphodiesterase 4 (PDE4) inhibitor, effects in cellular and animal models of asthma and early clinical studies. Am. J. Resp. Crit. Care Med. 159(Suppl):A624–A624. [Google Scholar]
- 23.Hirose, H., S. Mashiko, T. Kimura, F. Ishida, N. Mochizuki, T. Nishibe, and M. Nishikibe. 2000. Antithrombotic activity of NSP-513, a novel selective phosphodiesterase 3 inhibitor, on femoral arterial thrombosis induced by physical stenosis and electrical current: Comparison of antithrombotic and hemodynamic effects. J. Cardiovasc. Pharmacol. 35:586–594. [DOI] [PubMed] [Google Scholar]
- 24.Aoki, M., M. Kobayashi, J. Ishikawa, Y. Saita, Y. Terai, K. Takayama, K. Miyata, and T. Yamada. 2000. A novel phosphodiesterase type 4 inhibitor, YM976 (4-(3-chlorophenyl)-1,7-diethylpyrido[2,3-d]pyrimidin-2(1H)-one), with little emetogenic activity. J. Pharmacol. Exp. Ther. 295:255–260. [PubMed] [Google Scholar]
- 25.Inoue, H., K. Yano, T. Ikeo, T. Noto, and K. Kikkawa. 2001. T-1032, a novel specific phosphodiesterase type 5 inhibitor, increases venous compliance in anesthetized rats. Eur. J. Pharmacol. 422:109–114. [DOI] [PubMed] [Google Scholar]
- 26.Billah, M. M., N. Cooper, M. Minnicozzi, J. Warneck, P. Wang, J. A. Hey, W. Kreutner, C. A. Rizzo, S. R. Smith, S. Young, R. W. Chapman, H. Dyke, N. Y. Shih, J. J. Piwinski, F. M. Cuss, J. Montana, A. K. Ganguly, and R. W. Egan. 2002. Pharmacology of N-(3,5-dichloro-1-oxido-4-pyridinyl)-8-methoxy-2-(trifluoromethyl)-5-quinoline carboxamide (SCH 351591), a novel, orally active phosphodiesterase 4 inhibitor. J. Pharmacol. Exp. Ther. 302:127–137. [DOI] [PubMed] [Google Scholar]
- 27.Ukita, T., M. Sugahara, Y. Terakawa, T. Kuroda, K. Wada, A. Nakata, Y. Ohmachi, H. Kikkawa, K. Ikezawa, and K. Naito. 1999. Novel, potent, and selective phosphodiesterase-4 inhibitors as antiasthmatic agents: Synthesis and biological activities of a series of 1-pyridylnaphthalene derivatives. J. Med. Chem. 42:1088–1099. [DOI] [PubMed] [Google Scholar]
- 28.Ukita, T., Y. Nakamura, A. Kubo, Y. Yamamoto, M. Takahashi, J. Kotera, and T. Ikeo. 1999. 1-Arylnaphthalene lignan: A novel scaffold for type 5 phosphodiesterase inhibitor. J. Med. Chem. 42:1293–1305. [DOI] [PubMed] [Google Scholar]
- 29.Hersperger, R., K. Bray-French, L. Mazzoni, and T. Muller. 2000. Palladium-catalyzed cross-coupling reactions for the synthesis of 6,8-disubstituted 1,7-naphthyridines: A novel class of potent and selective phosphodiesterase type 4D inhibitors. J. Med. Chem. 43:675–682. [DOI] [PubMed] [Google Scholar]
- 30.Ukita, T., Y. Nakamura, A. Kubo, Y. Yamamoto, Y. Moritani, K. Saruta, T. Higashijima, J. Kotera, M. Takagi, K. Kikkawa, and K. Omori. 2001. Novel, potent, and selective phosphodiesterase 5 inhibitors: Synthesis and biological activities of a series of 4-aryl-1-isoquinolinone derivatives. J. Med. Chem. 44:2204–2218. [DOI] [PubMed] [Google Scholar]
- 31.van der Mey, M., H. Boss, D. Couwenberg, A. Hatzelmann, G. J. Sterk, K. Goubitz, H. Schenk, and H. Timmerman. 2002. Novel selective phosphodiesterase (PDE4) inhibitors. 4. Resolution, absolute configuration, and PDE4 inhibitory activity of cis-tetra- and cis-hexahydrophthalazinones. J. Med. Chem. 45:2526–2533. [DOI] [PubMed] [Google Scholar]
- 32.Manley, P. W., M. Acemoglu, W. Marterer, and W. Pachinger. 2003. Large-scale Negishi coupling as applied to the synthesis of PDE472, an inhibitor of phosphodiesterase type 4D. Org. Process Res. Dev. 7:436–445. [Google Scholar]
- 33.Friesen, R. W., Y. Ducharme, R. G. Ball, M. Blouin, L. Boulet, B. Cote, R. Frenette, M. Girard, D. Guay, Z. Huang, T. R. Jones, F. Laliberte, J. J. Lynch, J. Mancini, E. Martins, P. Masson, E. Muise, D. J. Pon, P. K. S. Siegl, A. Styhler, N. N. Tsou, M. J. Turner, R. N. Young, and Y. Girard. 2003. Optimization of a tertiary alcohol series of phosphodiesterase-4 (PDE4) inhibitors: Structure-activity relationship related to PDE4 inhibition and human ether-a-go-go related gene potassium channel binding affinity. J. Med. Chem. 46:2413–2426. [DOI] [PubMed] [Google Scholar]
- 34.Murata, K., T. Sudo, M. Kameyama, H. Fukuoka, M. Mukai, Y. Doki, Y. Sasaki, O. Ishikawa, Y. Kimura, and S. Imaoka. 2001. Cyclic AMP specific phosphodiesterase activity and colon cancer cell motility. Clin. Exp. Metastasis 18:599–604. [DOI] [PubMed] [Google Scholar]
- 35.Weishaar, R. E., M. H. Cain, and J. A. Bristol. 1985. A new generation of phosphodiesterase inhibitors - multiple molecular-forms of phosphodiesterase and the potential for drug selectivity. J. Med. Chem. 28:537–545. [DOI] [PubMed] [Google Scholar]
- 36.Feldman, A. M., and D. M. McNamara. 2002. Reevaluating the role of phosphodiesterase inhibitors in the treatment of cardiovascular disease. Clin. Cardiol. 25:256–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perez-Torres, S., R. Cortes, M. Tolnay, A. Probst, J. M. Palacios, and G. Mengod. 2003. Alterations on phosphodiesterase type 7 and 8 isozyme mRNA expression in Alzheimer's disease brains examined by in situ hybridization. Exp. Neurol. 182:322–334. [DOI] [PubMed] [Google Scholar]
- 38.Nagakura, A., M. Niimura, and S. Takeo. 2002. Effects of a phosphodiesterase IV inhibitor rolipram on microsphere embolism-induced defects in memory function and cerebral cyclic AMP signal transduction system in rats. Br. J. Pharmacol. 135:1783–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prickaerts, J., W. C. G. van Staveren, A. Sik, M. Markerink-van Ittersum, U. Niewohner, F. J. van der Staay, A. Blokland, and J. de Vente. 2002. Effects of two selective phosphodiesterase type 5 inhibitors, sildenafil and vardenafil, on object recognition memory and hippocampal cyclic GMP levels in the rat. Neuroscience. 113:351–361. [DOI] [PubMed] [Google Scholar]
- 40.Bourtchouladze, R., R. Lidge, R. Catapano, J. Stanley, S. Gossweiler, D. Romashko, R. Scott, and T. Tully. 2003. A mouse model of Rubinstein-Taybi syndrome: Defective long-term memory is ameliorated by inhibitors of phosphodiesterase 4. Proc. Natl. Acad. Sci. USA. 100:10518–10522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weishaar, R. E., M. H. Cain, and J. A. Bristol. 1985. A new generation of phosphodiesterase inhibitors - multiple molecular-forms of phosphodiesterase and the potential for drug selectivity. J. Med. Chem. 28:537–545. [DOI] [PubMed] [Google Scholar]
- 42.Soto, F. J., and N. A. Hanania. 2005. Selective phosphodiesterase-4 inhibitors in chronic obstructive lung disease. Curr. Opin. Pulm. Med. 11:129–134. [DOI] [PubMed] [Google Scholar]
- 43.Lincoln, T. M. 2004. Cyclic GMP and phosphodiesterase 5 inhibitor therapies: What's on the horizon? Mol. Pharm. 66:11–13. [DOI] [PubMed] [Google Scholar]
- 44.Burnett, A. L. 2004. The impact of sildenafil on molecular science and sexual health. Eur. Urol. 46:9–14. [DOI] [PubMed] [Google Scholar]
- 45.Kukreja, R. C., R. Ockaili, F. Salloum, C. Yin, J. Hawkins, A. Das, and L. Xi. 2004. Cardioprotection with phosphodiesterase-5 inhibition—a novel preconditioning strategy. J. Mol. Cell. Cardiol. 36:165–173. [DOI] [PubMed] [Google Scholar]
- 46.Abdel-Hamid, I. A. 2004. Phosphodiesterase 5 inhibitors in rapid ejaculation—Potential use and possible mechanisms of action. Drugs. 64:13–26. [DOI] [PubMed] [Google Scholar]
- 47.Bi, Y., P. Stoy, L. Adam, B. He, J. Krupinski, D. Normandin, R. Pongrac, L. Seliger, A. Watsonc, and J. E. Macora. 2004. Quinolines as extremely potent and selective PDE5 inhibitors as potential agents for treatment of erectile dysfunction. Bioorg. Med. Chem. Lett. 14:1577–1580. [DOI] [PubMed] [Google Scholar]
- 48.Rotella, D. P., Z. Sun, Y. Zhu, J. Krupinski, R. Pongrac, L. Seliger, D. Normandin, and J. E. Macor. 2000. N-3-Substituted imidazoquinazolinones: Potent and selective PDE5 inhibitors as potential agents for treatment of erectile dysfunction. J. Med. Chem. 43:1257–1263. [DOI] [PubMed] [Google Scholar]
- 49.Haning, H., U. Niewohner, T. Schenke, M. Es-Sayed, G. Schmidt, T. Lampea, and E. Bischoffb. 2002. Imidazo[5,1-f][1,2,4]triazin-4(3H)-ones, a new class of potent PDE 5 inhibitors. Bioorg. Med. Chem. Lett. 12:865–868. [DOI] [PubMed] [Google Scholar]
- 50.Degerman, E., P. Belfrage, and V. C. Manganiello. 1997. Structure, localization, and regulation of cGMP-inhibited phosphodiesterase (PDE3). J. Biol. Chem. 272:6823–6826. [DOI] [PubMed] [Google Scholar]
- 51.Soderling, S. H., and J. A. Beavo. 2000. Regulation of cAMP and cGMP signaling: new phosphodiesterases and new functions. Curr. Opin. Cell Biol. 12:174–179. [DOI] [PubMed] [Google Scholar]
- 52.Xu, R. X., A. M. Hassell, D. Vanderwall, M. H. Lambert, W. D. Holmes, M. A. Luther, W. J. Rocque, M. V. Milburn, Y. Zhao, H. Ke, and R. T. Nolte. 2000. Atomic structure of PDE4: Insights into phosphodiesterase mechanism and specificity. Science. 288:1822–1825. [DOI] [PubMed] [Google Scholar]
- 53.Liu, S., F. Laliberte, B. Bobechko, A. Bartlett, P. Lario, E. Gorseth, J. Van Hamme, M. J. Gresser, and Z. Huang. 2001. Dissecting the cofactor-dependent and independent bindings of PDE4 inhibitors. Biochemistry. 40:10179–10186. [DOI] [PubMed] [Google Scholar]
- 54.Wang, P., P. Wu, J. G. Myers, A. Stamford, R. W. Egan, and M. M. Billah. 2001. Characterization of human, dog and rabbit corpus cavernosum type 5 phosphodiesterases. Life Sci. 68:1977–1987. [DOI] [PubMed] [Google Scholar]
- 55.Bauer-Siebenlist, B., F. Meyer, E. Farkas, D. Vidovic, and S. Dechert. 2005. Effect of ZnZn separation on the hydrolytic activity of model dizinc phosphodiesterases. Chemistry 11:4349–4360. [DOI] [PubMed] [Google Scholar]
- 56.Zhan, C.-G., and F. Zheng. 2001. First computational evidence for a catalytic bridging hydroxide ion in a phosphodiesterase active site. J. Am. Chem. Soc. 123:2835–2838. [DOI] [PubMed] [Google Scholar]
- 57.Becke, A. D. 1993. Density-functional thermochemistry 3: the role of exact exchange. J. Chem. Phys. 98:5648–5652. [Google Scholar]
- 58.Lee, C., W. Yang, and R. G. Parr. 1988. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron-density. Phys. Rev. B. 37:785–789. [DOI] [PubMed] [Google Scholar]
- 59.Stephens, P. J., F. J. Devlin, C. F. Chabalowski, and M. J. Frisch. 1994. 1994. Ab initio calculation of vibrational absorption and circular-dichroism spectra using density-functional force-fields. J. Phys. Chem. 98:11623–11627. [Google Scholar]
- 60.Hehre, W. J., and L. Radom. P. v. R. Schleyer, and J. A. Pople. 1986. Ab Initio Molecular Orbital Theory. John Wiley & Sons, New York.
- 61.Rassolov, V. A., J. A. Pople, M. A. Ratner, and T. L. Windus. 1998. 6–31G* basis set for atoms K through Zn. J. Chem. Phys. 109:1223–1229. [Google Scholar]
- 62.Sung, B. J., K. Y. Hwang, Y. H. Jeon, J. I. Lee, Y. S. Heo, J. H. Kim, J. Moon, J. M. Yoon, Y. L. Hyun, E. Kim, S. J. Eum, S. Y. Park, J. O. Lee, T. G. Lee, S. Ro, and J. M. Cho. 2003. Structure of the catalytic domain of human phosphodiesterase 5 with bound drug molecules. Nature. 425:98–102. [DOI] [PubMed] [Google Scholar]
- 63.Huai, Q., Y. Liu, S. H. Francis, J. D. Corbin, and H. Ke. 2004. Crystal structures of phosphodiesterases 4 and 5 in complex with inhibitor 3-isobutyl-1-methylxanthine suggest a conformation determinant of inhibitor selectivity. J. Biol. Chem. 279:13095–13101. [DOI] [PubMed] [Google Scholar]
- 64.Card, G. L., B. P. England, Y. Suzuki, D. Fong, B. Powell, B. Lee, C. Lu, M. Tabrizizad, S. Gillette, P. N. Ibrahim, D. R. Artis, G. Bollag, M. V. Milburn, N. S.-H. Kim, J. Schlessinger, and K. Y. J. Zhang. 2004. Structural basis for the activity of drugs that inhibit phosphodiesterases. Structure. 12:2233–2247. [DOI] [PubMed] [Google Scholar]
- 65.Zhang, K. Y. J., G. L. Card, Y. Suzuki, D. R. Artis, D. Fong, S. Gillette, D. Hsieh, J. Neiman, B. L. West, C. Zhang, M. V. Milburn, S.-H. Kim, J. Schlessinger, and G. Bollag. 2004. A glutamine switch mechanism for nucleotide selectivity by phosphodiesterases. Mol. Cell. 15:279–286. [DOI] [PubMed] [Google Scholar]
- 66.Lee, M. E., J. Markowitz, J. O. Lee, and H. Lee. 2002. Crystal structure of phosphodiesterase 4D and inhibitor complex. FEBS Lett. 530:53–58. [DOI] [PubMed] [Google Scholar]
- 67.Case, D. A., D. A. Pearlman, J. W. Caldwell, T. E. Cheatham III, J. Wang, W. S. Ross, C. L. Simmerling, T. A. Darden, K. M. Merz, R. V. Stanton, A. L. Cheng, J. J. Vincent, M. Crowley, V. Tsui, H. Gohlke, R. J. Radmer, Y. Duan, J. Pitera, I. Massova, G. L. Seibel, U. C. Singh, P. K. Weiner, and P. A. Kollman. 2002. AMBER 7, University of California, San Francisco.
- 68.Zhan, C.-G., O. Norberto de Souza, R. Rittenhouse, and R. L. Ornstein. 1999. Determination of two structural forms of catalytic bridging ligand in zinc-phosphotriesterase by molecular dynamics simulation and quantum chemical calculation. J. Am. Chem. Soc. 121:7279–7282. [DOI] [PubMed] [Google Scholar]
- 69.Koca, J., C.-G. Zhan, R. Rittenhouse, and R. L. Ornstein. 2001. Mobility of the active site bound paraoxon and sarin in zinc-phosphotriesterase by molecular dynamics simulation and quantum chemical calculation. J. Am. Chem. Soc. 123:817–826. [DOI] [PubMed] [Google Scholar]
- 70.Koca, J., C.-G. Zhan, R. C. Rittenhouse, and R. L. Ornstein. 2003. Coordination number of zinc ions in the phosphotriesterase active site by molecular dynamics and quantum mechanics. J. Comput. Chem. 24:368–378. [DOI] [PubMed] [Google Scholar]
- 71.Berendsen, H. C., J. P. M. Postma, W. F. van Gunsteren, A. DiNola, and J. R. Haak. 1984. Molecular-dynamics with coupling to an external bath. J. Chem. Phys. 81:3684–3690. [Google Scholar]
- 72.Ryckaert, J. P., G. Ciccotti, and H. C. Berendsen. 1977. Numerical-integration of cartesian equations of motion of a system with constraints—molecular-dynamics of n-alkanes. J. Comput. Phys. 23:327–341. [Google Scholar]
- 73.Darden, T., D. York, and L. Pedersen. 1993. Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 98:10089–10092. [Google Scholar]
- 74.Dapprich, S., I. Komaromi, K. S. Byun, K. Morokuma, and M. J. Frisch. 1999. A new ONIOM implementation in Gaussian98. Part I. The calculation of energies, gradients, vibrational frequencies and electric field derivatives. J. Mol. Struct. (Theochem). 461:1–21. [Google Scholar]
- 75.Vreven, T. and Morokuma, K. 2000. The ONIOM (our own N-layered integrated molecular orbital plus molecular mechanics) method for the first singlet excited (S-1) state photoisomerization path of a retinal protonated Schiff base. J. Chem. Phys. 113:2969–2975. [Google Scholar]
- 76.Vreven, T., K. Morokuma, H. B. Farkas, O. Schlegel, and M. Frisch. 2003. Geometry optimization with QM/MM, ONIOM, and other combined methods. I. Microiterations and constraints. J. Comput. Chem. 24:760–769. [DOI] [PubMed] [Google Scholar]
- 77.Frisch, M. J., G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, J. A. Montgomery Jr., T. Vreven, K. N. Kudin, J. C. Burant, J. M. Millam, S. S. Iyengar, J. Tomasi, V. Barone, B. Mennucci, M. Cossi, G. Scalmani, N. Rega, G. A. Petersson, H. Nakatsuji, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, M. Klene, X. Li, J. E. Knox, H. P. Hratchian, J. B. Cross, C. Adamo, J. Jaramillo, R. Gomperts, R. E. Stratmann, O. Yazyev, A. J. Austin, R. Cammi, C. Pomelli, J. W. Ochterski, P. Y. Ayala, K. Morokuma, G. A. Voth, P. Salvador, J. J. Dannenberg, V. G. Zakrzewski, S. Dapprich, A. D. Daniels, M. C. Strain, O. Farkas, D. K. Malick, A. D. Rabuck, K. Raghavachari, J. B. Foresman, J. V. Ortiz, Q. Cui, A. G. Baboul, S. Clifford, J. Cioslowski, B. B. Stefanov, G. Liu, A. Liashenko, P. Piskorz, I. Komaromi, R. L. Martin, D. J. Fox, T. Keith, M. A. AlLaham, C. Y. Peng, A. Nanayakkara, M. Challacombe, P. M. W. Gill, B. Johnson, W. Chen, M. W. Wong, C. Gonzalez, and J. A. Pople. 2003. Gaussian 03, Revision A.1. Gaussian, Inc., Pittsburgh, PA.
- 78.Bernstein, F. C., T. F. Koetzle, G. J. Williams, E. E. Meyer Jr., M. D. Brice, J. R. Rodgers, O. Kennard, T. Shimanouchi, and M. Tasumi. 1977. Protein data bank—computer-based archival file for macromolecular structures. J. Mol. Biol. 112:535–542. [DOI] [PubMed] [Google Scholar]