Abstract
The synthesis of virulence factors and other extracellular proteins responsible for pathogenicity in Staphylococcus aureus is under the control of the agr locus. A secreted agr-encoded peptide, AgrD, processed from the AgrD gene product, is known to be an effector of self-strain activation and cross-strain inhibition of the agr response. Biochemical analysis of AgrD peptides isolated from culture supernatants has suggested that they contain an unusual thiol ester-linked cyclic structure. In the present work, chemical synthesis is used to confirm that the mature AgrD peptides contain a thiolactone structure and that this feature is absolutely necessary for full biological activity. The AgrD synthetic thiolactone peptides exhibited biological activity in vivo in a mouse protection test. Structure-activity studies have allowed key aspects of the peptide structure involved in the differential activation and inhibition functions to be identified. Accordingly, we propose a model for activation and inhibition of the agr response in which the former, but not the latter, involves specific acylation of the agr transmembrane receptor, AgrC.
Staphylococcus aureus is among the major nosocomial pathogens that are now under increasing risk of developing resistance to all currently available therapeutics. Consequently, there is a pressing need to identify new types of antibacterial agents, and it has been suggested that interference with the expression of virulence may represent a promising antibacterial modality (1, 2). As it happens, S. aureus is a very good candidate for such an approach as it uses a global regulator, agr, activated by secreted autoinducing peptides, to control the expression of most virulence genes (3, 4). These peptides serve as ligands for a signal receptor, AgrC, that initiates this agr response through a classical two-component signaling pathway. The autoinducer–receptor pair shows considerable interstrain sequence variation, which affects the specificity of the receptor–ligand interaction. On the basis of this autoinducer–receptor specificity, S. aureus strains can be placed in at least four different groups (refs. 4–6; A. Figuereido and R.P.N., unpublished data). Remarkably, each of the secreted autoinducing peptides can activate the agr response within the same group and inhibit the agr response in strains belonging to the other groups. This inhibition constitutes a novel form of bacterial interference (5) because it affects the expression of a specific group of genes rather than inhibiting growth, which is the usual manifestation of bacterial interference.
The agr locus contains two divergent promoters, P2 and P3. There are four genes, (agrA–D) in the P2 operon that code for the cytosolic, transmembrane, and extracellular components of a density-sensing/autoinduction circuit (4). The agrD gene product is a propeptide that is processed and secreted through AgrB, an integral membrane protein. The resultant mature autoinducing peptide (AIP) is thought to bind to the transmembrane receptor coded by AgrC. Binding of the AIP triggers a standard two-component signal-transduction pathway in which the AgrC receptor becomes autophosphorylated on a histidine residue (6), presumably leading to the subsequent trans-phosphorylation of the AgrA gene product. Phosphorylated AgrA is assumed to activate transcription from the agr P2/P3 promoters (6). The RNA transcript from the P3 promoter is responsible for the up-regulation of secreted virulence factors as well as the down-regulation of surface proteins (3, 7).
The agr AIPs consist of 7–9 residues and although the sequences are highly variable among the groups, all contain a conserved cysteine 5 aa from the C terminus. Mass spectrometric analysis of AgrD peptides isolated from culture supernatants indicated a mass discrepancy of −18 Da compared with the predicted masses based on the peptide sequences (5). This observation, combined with the lack of activity of a linear synthetic group I peptide (2, 5) and the presence of the absolutely conserved cysteine residue in the AgrD peptides, has led to the suggestion that these secreted peptides contain an intramolecular thiol ester linkage between the cysteine sulfhydryl group and the C terminus (5). Consistent with this thiolactone structure, the addition of hydroxylamine to a purified AgrD peptide was observed to abolish its biological activity (5), whereas iodoacetic acid had no effect. In preliminary studies, we chemically synthesized a very small quantity of the thiolactone derivative of the AgrD peptide predicted from the group III agrD DNA sequence and found that this product had agr inhibitory activity for groups I and II, but no activation activity (5). Because, as described below, only cyclic derivatives have activity and most mutants are inhibitory, we consider this result to represent preliminary confirmation of the thiolactone structure and the lack of activation activity to represent uncertainty in the primary sequence of the group III peptide.
Because the AgrD peptides are relatively hydrophobic and are present at extremely low concentrations, it has not been possible to isolate large quantities from culture supernatants. Consequently, very little is known about the biochemistry of the AgrD/AgrC interaction. For example, the potency of the AgrD peptide in either activating (within S. aureus strains of the same group) or inhibiting (in S. aureus strains from other groups) the agr response is unknown. Equally, it is unknown whether the putative thiolactone structure within the AgrD peptides is required for activation of the agr response, inhibition of the agr response, or both.
In this study, we confirm the presence of the thiolactone moiety through total chemical synthesis, produce enough material for testing in an animal model, and perform structure-activity studies on the AgrD–AgrC interaction. We propose a model for the molecular nature of AgrC receptor–AgrD peptide interaction that accounts for the differential mechanisms of activation and inhibition of the agr response.
MATERIALS AND METHODS
Synthesis of the AgrD Cyclic Thioester Peptides and Analogues.
Peptides were synthesized manually on preloaded t-butoxycarbonyl–amino acyl-3-mercapto-propionamide–polyethylene glycol-poly-(N,N-dimethylacrylamide) (Boc–AA–[COS]–PEGA) supports (8) according to the in situ neutralization/O-benzotriazol-1-yl-N,N,N′,N′,-tetramethyluronium hexafluorophosphate (HBTU) activation protocol for Boc solid-phase peptide synthesis (9). After chain assembly, peptides were treated with HF for 1 hr at 0°C to give the corresponding fully unprotected peptide–[COS]–PEGA resins, which were then washed with cold Et2O and then CH3CN/H2O containing 0.1% trifluoroacetic acid. The alkyl–thiol ester linkage between the peptide and the resin is completely stable to anhydrous HF (8). Unprotected peptides then were chemoselectively cyclized and simultaneously cleaved from the support by swelling the beads in a mixture of 0.1 M sodium phosphate buffer (pH 7.0) and acetonitrile (80:20). After a 12-hr reaction, the beads were removed by filtration and washed with 0.1% trifluoroacetic acid in water, and the peptides were purified from the filtrate by reverse-phase HPLC. In each case, the yield of the final cyclization/cleavage reaction was between 60% and 90%, as determined by HPLC analysis of the crude reaction mixtures and integration of the peaks. After purification, the products were characterized as the expected thiolactone-peptides by electrospray MS, chemical reactivity to neutral hydroxylamine, and two-dimensional 1H NMR spectroscopy. All purified peptides were >95% homogeneous as indicated by reverse-phase HPLC, MS, and NMR analysis.
Synthesis of the Lactone and Lactam AgrDII Peptide Analogues.
The protected peptides Z-Gly-Val-Asn-Ala-Ser(tButyl)-Ser(Benzyl)-Ser(Benzyl)-Leu-Phe and Z-Gly-Val-Asn-Ala-DAPA(Boc)-Ser(Benzyl)-Ser(Benzyl)-Leu-Phe corresponding to the AgrDII sequence with a Cys-5→Ser mutation [lactone] and a Cys→diaminopropionic acid (DAPA) mutation [lactam] respectively, were synthesized on a Wang-resin by using a fluorenylmethoxycarbonyl Nα protection strategy with HBTU activation protocols. After chain assembly, the peptides were cleaved from the support and the Ser-5 or DAPA-5 side chain deprotected by treatment with a trifluoroacetic acid/anisole/water mixture (90:5:5) for 4 hr. The partially protected peptide–αcarboxylates then were dissolved in N,N-dimethylformamide (DMF) (0.5 mg/ml) and treated with benzotriazol-1-yl-oxytripyrrolidinophosphonium hexafluorophosphate (PyBop; Nova Biochem) (5 eq) and in the case of the lactone precursor, a catalytic amount of dimethylaminopyridine. The cyclization reactions were monitored with HPLC, which indicated 2 hr to be sufficient for complete reaction. The remaining protecting groups then were removed by treatment with HF and the desired peptides purified by reverse-phase HPLC and characterized by MS and two-dimensional 1H NMR spectroscopy.
Synthesis of the Linear Thiol Ester Peptide.
The peptide GVNAASSLF was assembled on an 3-mercaptopropionamide–PEGA resin (8) by using Boc solid-phase peptide synthesis (this corresponds to the AgrDII sequence with the single cysteine residue mutated to an alanine). After synthesis and global deprotection, the peptide–[COS]–PEGA beads were swollen in a buffer containing 0.1 M sodium phosphate (pH 7.0) and ethanethiol (2% vol/vol), and the cleavage reaction was allowed to proceed overnight. The desired ethylαthiol ester peptide then was purified from the supernatant by reverse-phase HPLC and characterized by electrospray MS and two-dimensional 1H NMR.
In Vitro Biological Activity Assay.
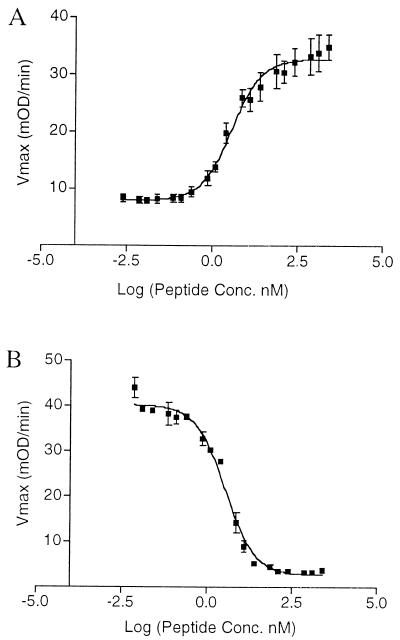
Assays were performed by using groups I (RN6390B), II (SA502A), and III (RN8463) S. aureus strains (5), each containing an agr P3-blaZ (β-lactamase) fusion plasmid (4). Cells were grown in CYGP medium [case amino acid and yeast extracts (1% of each), 100 mM NaCl, 100 mM sodium glycophosphate, and 0.5% glucose, pH 7.4] at 37°C to either early exponential phase (for agr activation studies) or mid-exponential phase (for agr inhibition studies). To the cultured cells then were added either buffer solution (negative control), the appropriate cell supernatant containing the natural AgrD peptide as prepared in ref. 2 (positive control), or the synthetic peptide solution in 20 mM Tris⋅HCl buffer at pH 6.0. Synthetic peptide solutions of known concentration were prepared by diluting an appropriate amount of a stock solution (concentration determined by amino acid analysis) into the assay buffer. The cultures then were incubated at 37°C with shaking for either 55 min (activation) or 80 min (inhibition), and β-lactamase activity then was assayed by using the nitrocefin spectrophotometric method modified as described in ref. 2. ED50 and IC50 values were extracted from the sigmoidal dose-response curves (e.g., Fig. 2) by using the program prism (GraphPad, San Diego). All assays were performed at least in triplicate.
Figure 2.
Synthetic thiolactone peptides are biologically active. Shown are representative data for activation (A) and inhibition (B) of the agr response by a synthetic thiotactone peptide. Degree of activity based on β-lactamase activity shown as a plot of Vmax versus peptide concentration. (A) Activation of the agr response in group II S. aureus cells by synthetic AgrDII. (B) Inhibition of the agr response in group I S. aureus cells by synthetic AgrDII.
In Vivo Mouse Protection Test.
The inhibitory activity of synthetic AgrDII peptide on the ability of a group I strain to cause a s.c. abscess was assayed in a mouse protection test (10). Mid-exponential-phase cultures grown in CY broth of either strain RN6390B (agr+) or RN6911 (agr-null) were centrifuged, and the bacteria were resuspended in physiological saline + 10 mM sodium phosphate (pH 7.4) at approximately 109 cells per ml and held on ice until use. Hairless mice (6- to 8-wk-old, strain SKH1(hrhr)Br from Charles River Labs, average weight 25 g) were injected s.c. in the flank area with approximately 108 colony-forming units. Either 5 or 10 μg of a synthetic preparation of the AgrDII autoinducing peptide (total body concentration of 200 or 400 nM for a 25-g mouse or about 2× and 4× the IC100 shown in Fig. 2B) diluted from 10 mM acetate buffer (pH 5.0) were included with the bacteria for six of the 10 RN6390 animals. Lesions were measured daily for 10 days.
RESULTS
Synthesis and Biological Activity of the AgrD Thiolactone Peptides.
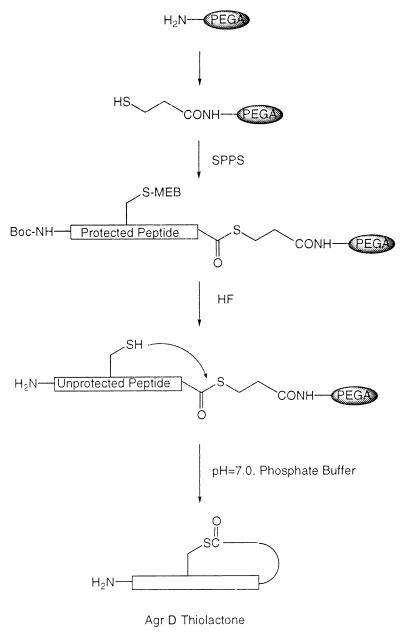
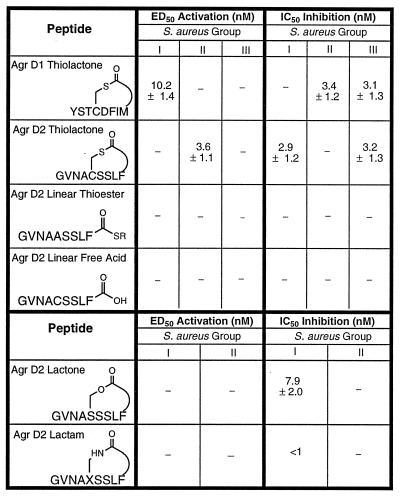
A synthetic strategy was developed that involved solid-phase cyclization as the final synthetic step and allowed the desired thiolactone peptides to be prepared in good yield (Fig. 1). After purification, each thiolactone peptide was fully characterized as described in Materials and Methods. The synthetic AgrDI and AgrDII peptides were found to activate the agr response only within their own S. aureus class and inhibit the agr response only in S. aureus strains from the other two classes (Table 1). [The group III peptide exhibited only inhibitory activities, consistent with our previous observations (5)]. Further studies revealed a dose-dependent relationship between the amount of peptide added to the culture supernatant and the degree of activation/inhibition of the agr response (Fig. 2). Analysis of the resulting sigmoidal response curves indicated that the ED50 and IC50 values (for activation and inhibition the agr response, respectively) were in the low nanomolar range (Table 1). Moreover, these data indicate that there is a critical threshold concentration of the AgrD peptide required for activation of the agr response. This is consistent with the density-sensing/autoinduction mechanism previously proposed (4).
Figure 1.
Chemical synthesis of AgrD thiolactone-containing autoinducing peptides via a solid-phase intramolecular chemical ligation strategy. Key to this process is the ability to prepare a fully unprotected peptide immobilized on a solid support through a reactive thiol ester bond. Simply swelling such an unprotected peptide-[thioester]-resin in aqueous buffer results in a chemoselective ligation reaction and concomitant cleavage of the peptide from the support. This final step is made possible because of the excellent swelling properties of the PEGA support in water (21).
Table 1.
Biological activity of the synthetic AgrD peptides as determined in an in vitro agr P3/β-lactamase fusion assay
Dashes indicate no activity. x = DAPA.
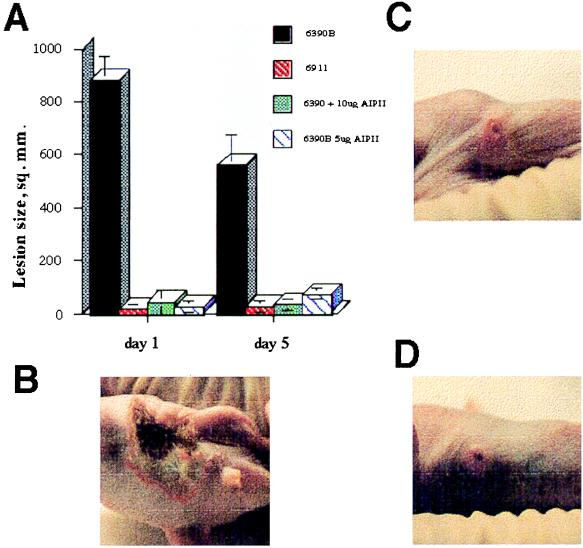
The inhibitory activity of synthetic AgrDII peptide on the ability of a group I strain to cause a s.c. abscess was assayed in a mouse protection test (10). At the dosages administered (5 and 10 μg per mouse or 2× and 4× the IC100 shown in Fig. 2B), the s.c. abscess caused by the injection of approximately 108 colony-forming units of strain RN6390B was dramatically attenuated, nearly to the extent caused by deletion of the entire agr region. The overall results are summarized in Fig. 3A, and typical lesion sizes at day 5 are shown in Fig. 3 B–D.
Figure 3.
Attenuation of the staphylococcal skin abscess by a synthetic thiolactone-containing peptide. (A) Effect of inhibitory peptide on murine skin abscesses. Six- to eight-wk-old hairless mice were injected s.c. in the flank area on day zero with approximately 108 colony-forming units of S. aureus from an exponential-phase culture grown in CY broth. The size of the resulting s.c. abscesses were measured with dividers every 24 hr; sizes were calculated by assuming that lesions were approximately elliptical and of approximately uniform depth. Four mice were injected with strain RN6390B (agr+), four with RN6911 (agr-null), three with RN6390B + 5 μg of synthetic AIPII (group II thiolactone peptide), and three with RN6390B + 10 μg of synthetic AIPII. In each case the peptide was included with the bacteria being injected. The graph shows the data plotted as average lesion size, with error bars representing the SD. The differences among the sizes of the lesions induced by RN6911 and by RN6390B plus the peptide are not significant. (B–D) Typical lesion sizes at day 5 are shown. (B) RN6390B; (C) RN6911; (D) RN6390B + 5 μg of synthetic AIPII.
Importantly, we were unable to detect any activation/inhibition activity in vitro with linear carboxylate synthetic peptides corresponding to the AgrDI and AgrDII sequences (Table 1). The restriction of biological activity to the thiolactone-peptides serves to confirm that this unusual posttranslational modification is present within the secreted AgrD peptides.
Structure-Activity Studies on the AgrDII Peptide.
The convenience of our synthetic approach made it possible to vary the chemical structure of the AgrD peptides systematically, thus enabling detailed structure-activity studies to be performed (summarized in Table 1). By using the AgrDII peptide as the test system, we focused on the following questions: (i) Which amino acids within the sequence are most important for affinity/selectivity? (ii) What is the role of the thiolactone structure in activation and inhibition of the agr response?
To address the first of these issues, we performed alanine scanning substitution on the group II AgrD peptide. Each of the alanine-modified AgrDII peptide analogues was prepared and characterized as before, and in each case the purified peptide was assayed for its ability to activate or inhibit the agr response in group I and II S. aureus strains. Analysis of the results, summarized in Fig. 4, revealed that amino acids within both the ring and the tail regions of the peptide (Asn-3, Leu-8, and Phe-9) are critical for activation of the agr response. In contrast, inhibition of the agr response appears to depend solely on amino acids within the ring structure of the peptide (Leu-8 and Phe-9). Several of the alanine-modified peptides have the interesting property of being potent inhibitors of the agr response (even more so than wild-type peptides), although they are unable to activate the response.
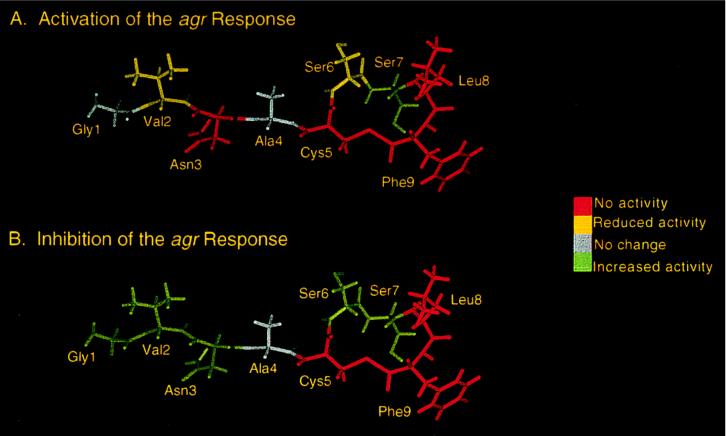
Figure 4.
Effect of replacing each residue within the AgrDII peptide sequence with alanine on activation (A) and inhibition (B) activity. The results are superimposed on a model of the AgrDII peptide created by using the program discover (Biosym Technologies, San Diego). The ED50 values (activation, group II cells) were as follows: Ala-1, 3.2 ± 2.2 nM; Ala-2, 73.7 ± 1.9 nM; Ala-3, no activity; Ala-6, 33.6 ± 1.5 nM; Ala-7, <1 nM; Ala-8, no activity; Ala-9, no activity. The IC50 values (inhibition, group I cells) were as follows: Ala-1, <1 nM; Ala-2, <1 nM; Ala-3, <1 nM; Ala-6, ≪1 nM; Ala-7, <1 nM; Ala-8, no activity; Ala-9, no activity. None of the peptides activated the agr response in group I S.aureus strains or inhibited the agr response in group II strains.
In addressing the role of the thiolactone moiety, we first examined whether a linear thiol ester analogue of the AgrDII peptide had biological activity. As with the linear carboxylate AgrDII peptide analogue described earlier, the linear thiol ester peptide was unable either to activate or inhibit the agr response even when added to cultured cells at micromolar concentrations (Table 1). This result suggests that the cyclic structure present within the AgrD peptides is indispensable for biological activity.
Thiol ester groups are moderately good acylating agents, a property that is used in several biological processes (11–13). It is intriguing to speculate that on receptor binding, the thiolactone present in the AgrD peptides serves as an acyl donor for the covalent modification of a specific residue within AgrC. We thus were interested in the effect of replacing the thiol ester group in the AgrDII peptide with both ester (lactone) and amide (lactam) bonds. The former of these analogues should be significantly less reactive than the wild-type peptide [in particular, thiol esters are significantly more reactive toward nitrogen nucleophiles that oxygen esters (14)], whereas the latter will be completely unreactive in acyl-transfer reactions. Furthermore, the lactone analogue of the AgrDII peptide also should be essentially isosteric to the thiolactone. Synthesis of the desired AgrDII lactone and lactam analogues was achieved via solution cyclization of a partially protected intermediate, followed by global side-chain deprotection. As with the linear peptides described above, both the purified lactone and lactam analogues were unable to activate the agr response in either group I or group II S. aureus strains (Table 1). However, both analogues were potent cross-strain inhibitors of the agr response as demonstrated in group I S. aureus strains. Clearly, the reactive thiol ester bond is necessary for activation of the agr response, but is not necessary for inhibition.
DISCUSSION
Total chemical synthesis has been used to confirm that the AgrD autoinducing peptides from S. aureus contain a functionally important thiolactone structure. A thiolactone structure recently has been observed in an agr-encoded peptide produced by an S. epidermidis strain (15). Synthetic access to these S. aureus AIPs has allowed in vivo and in vitro assays to be performed and has permitted an initial structure-activity analysis of the system. Analysis of the biological properties of the various AgrDII peptide analogues prepared in this study revealed the following: (i) activation of the agr response is extremely sensitive to both the amino acid sequence and the chemical/stereochemical nature of the AgrD peptide; and (ii) inhibition of the agr response is sensitive to the backbone stereochemistry and the amino acid sequence of the AgrD peptide but is not affected by changes in the chemical reactivity of the cyclic linkage. These observations suggest that activation and inhibition of the agr response occur through two different chemical mechanisms.
We propose the following model in which an AgrD peptide binds its own AgrC receptor (i.e., from the same S. aureus class) in a different manner than an AgrC receptor from another class (Fig. 5). This binding may involve two slightly different orientations of the peptide within the receptor-binding pocket, although more than one discrete binding site within the AgrC receptor cannot be excluded. We further hypothesize that in the intraclass association, highly specific side-chain interactions between AgrC and AIP results in positioning of the thiol ester linkage of the peptide adjacent to a nucleophilic group within the receptor. Juxtaposition would lead to a trans-acylation reaction and activation of the agr response through an associated conformational change in the receptor. The thiol ester linkage is sufficiently reactive to participate in this trans-acylation step and would explain its presence within the AgrD peptide rather than one of the more common stereochemical constraints such as disulfide- or amide-bond formation. Although the precise location of the putative nucleophile is currently unknown, it is worth noting that deletion mutagenesis on AgrC seems to indicate that the third extracellular loop of the receptor is sufficient for AgrD peptide binding and receptor activation (6).
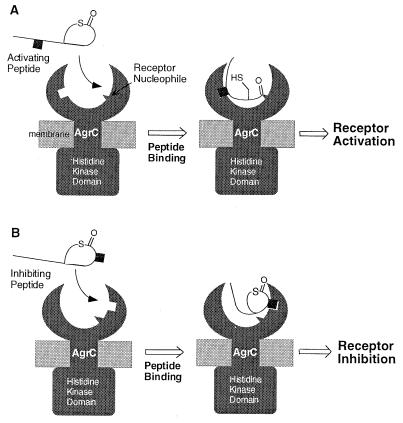
Figure 5.
Proposed model for the activation and the inhibition of the agr response. (A) Activation of the agr response occurs when an AgrD peptide interacts with an AgrC receptor from the same S. aureus class. Specific AgrD–AgrC interactions lead to the thiol ester group within the peptide being positioned close to a nucleophile within the receptor. Subsequent trans-acylation leads to a signal-transducing conformational change in the receptor, which may include homodimerization and trans-phosphorylation of the histidine kinase domains. (B) Inhibition of the agr response occurs via an interclass, noncovalent interaction that serves to exclude the strain’s own activating peptide from the receptor.
The AgrC receptor contains a cytoplasmic histidine kinase domain at its C terminus (5). Given that cytoplasmic histidine kinases are known to undergo trans-phosphorylation via dimerization (16–19), it seems likely that binding/reaction of the activating AgrD peptide results in the formation of an active AgrC homodimer, which in turn leads to histidine trans-phosphorylation (6). It remains to be seen whether AgrD peptide binding induces receptor dimerization in a manner analogous to polypeptide hormones (20) or whether it results in a conformational change in a preformed AgrC dimer. It also is pertinent to note at this time that autophosphorylation in cis cannot be dismissed on the basis of the studies presented here.
In contrast to the activation situation, an interclass receptor–ligand interaction (i.e., inhibition) would not lead to a thiol ester-nucleophile juxtaposition, because of the absence of specific interactions involving the tail region of the AgrD peptide. Thus, inhibition of the agr response would not involve a signal-transducing trans-acylation step, but rather a noncovalent binding interaction that would serve to exclude the strain’s own autoinducing AgrD peptide from the receptor-binding pocket. It is important to note that the lactone and lactam AgrDII analogues, although being able to inhibit in the appropriate interclass fashion, do not inhibit self. Inhibition by default, therefore, does not occur when the unreactive lactone/lactam AgrD analogue encounters its own receptor. This evidence further supports our proposed model in which activation and inhibition occur through discrete mechanisms.
It is also interesting to note that the thiolactone structure is likely to limit the in vivo half-life of the peptide; the thiol ester linkage will undergo slow hydrolysis at physiological pH leading to the generation of an inactive linear peptide. The biological significance of such a short-term in vivo efficacy is as yet unknown.
In summary, ready synthetic access to AgrD peptides has allowed us to demonstrate biological activity with respect to the agr response both in vivo and in vitro. The demonstration that it is possible to prepare synthetic AgrD analogues that are able to inhibit but not activate the agr response is extremely significant in terms of exploiting the agr response as a route to novel therapeutics, a long-term goal of our program. Moreover, the ability to adapt our solid-phase synthetic strategy easily to combinatorial-type synthesis may be advantageous in the rapid identification of active compounds.
Acknowledgments
We thank Graham Cotton, Julio Camarero, and Peter Model for useful discussions and comments on this manuscript. This work was supported by National Institutes of Health Grants GM55843–01 (T.W.M), AI30138–09 (R.P.N.), and A142783 (R.P.N.), the Pew Foundation Scholarship (T.W.M), and a Burroughs Wellcome graduate fellowship (P.M).
ABBREVIATIONS
- agr
accessory gene regulator
- AIP
autoinducing peptide
- Boc
t-butoxycarbonyl
- DAPA
diaminopropionic acid
- HBTU
O-benzotriazol-1-yl-N,N,N′,N′,-tetramethyluronium hexafluorophosphate
- PEGA
polyethylene glycol-poly-(N,N-dimethylacrylamide)
Note Added in Proof:
It recently has been reported that a 38-kDa protein is the agr activator for S. aureus group I strains and that a linear heptapeptide, YSPWTNF, is a potent inhibitor of the agr response (22). We and others (S. Foster, personal communication; J. Larrick, personal communication) have been unable to confirm these results.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Roychoudhury S, Zielinski N A, Ninfa A J, Allen N E, Jungheim L N, Nicas T I, Chakrabarty A M. Proc Natl Acad Sci USA. 1993;90:965–969. doi: 10.1073/pnas.90.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ji G, Beavis R, Novick R P. Proc Natl Acad Sci USA. 1995;92:12055–12059. doi: 10.1073/pnas.92.26.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Novick R P, Ross H F, Projan S, Kornblum J, Krieswirth B, Moghazeh S. EMBO J. 1993;12:3967–3975. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Novick R P, Projan S, Kornblum J, Kreiswirth B, Moghazeh S. Mol Gen Genet. 1995;248:446–458. doi: 10.1007/BF02191645. [DOI] [PubMed] [Google Scholar]
- 5.Ji G, Beavis R, Novick R P. Science. 1997;276:2027–2030. doi: 10.1126/science.276.5321.2027. [DOI] [PubMed] [Google Scholar]
- 6.Lina G, Jarraud S, Ji G, Greenland T, Pedraza A, Etienne J, Novick R P, Vandensch F. Mol Microbiol. 1998;28:655–62. doi: 10.1046/j.1365-2958.1998.00830.x. [DOI] [PubMed] [Google Scholar]
- 7.Morfeldt E, Taylor D, von Gabain A, Arvidson S. EMBO J. 1995;14:4569–4577. doi: 10.1002/j.1460-2075.1995.tb00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Camarero J A, Cotton G C, Adeva A, Muir T W. J Peptide Res. 1998;51:303–316. doi: 10.1111/j.1399-3011.1998.tb00428.x. [DOI] [PubMed] [Google Scholar]
- 9.Schnölzer M, Alewood P, Jones A, Alewood D, Kent S B H. Int J Pept Protein Res. 1997;40:180–193. doi: 10.1111/j.1399-3011.1992.tb00291.x. [DOI] [PubMed] [Google Scholar]
- 10.Barg N, Bunce C, Wheeler L, Reed G, Musser J. Infect Immun. 1992;60:2636–2640. doi: 10.1128/iai.60.7.2636-2640.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Law S K, Dodds A W. Protein Sci. 1997;6:263–274. doi: 10.1002/pro.5560060201. ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu M-Q, Perler F B. EMBO J. 1996;15:5146–5153. ;. [PMC free article] [PubMed] [Google Scholar]
- 13.Porter J A, Young K E, Beachy P A. Science. 1996;274:255–259. doi: 10.1126/science.274.5285.255. [DOI] [PubMed] [Google Scholar]
- 14.Bruice T C, Benkovic S J. In: Bioorganic Mechanisms. Breslow R, Karplus M, editors. New York: Benjamin; 1966. pp. 259–297. [Google Scholar]
- 15.Otto M, Sübmuth R, Jung G, Götz F. FEBS Lett. 1998;424:89–94. doi: 10.1016/s0014-5793(98)00145-8. [DOI] [PubMed] [Google Scholar]
- 16.Yang Y, Inouye M. Proc Natl Acad Sci USA. 1991;88:11057–11061. doi: 10.1073/pnas.88.24.11057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swanson R V, Bourret R B, Simon M I. Mol Microbiol. 1993;8:435–441. doi: 10.1111/j.1365-2958.1993.tb01588.x. [DOI] [PubMed] [Google Scholar]
- 18.Ninfa E G, Atkinson M R, Kamberov E S, Ninfa A J. J Bacteriol. 1993;175:7024–7032. doi: 10.1128/jb.175.21.7024-7032.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Egger L A, Park H, Inouye M. Genes Cells. 1997;2:167–184. doi: 10.1046/j.1365-2443.1997.d01-311.x. [DOI] [PubMed] [Google Scholar]
- 20.Wells J A. Proc Natl Acad Sci USA. 1996;93:1–6. doi: 10.1073/pnas.93.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mendal M. Tetrahedron Lett. 1992;33:3077–3080. [Google Scholar]
- 22.Balaban N, Goldkorn T, Nhan R T, Dang L B, Scott S, Ridgely R M, Rasooly A, Wright S C, Larrick J W, Rasooly R, Carlson J R. Science. 1998;280:438–440. doi: 10.1126/science.280.5362.438. [DOI] [PubMed] [Google Scholar]