Abstract
S100A13 is a member of the S100 protein family that is involved in the copper-dependent nonclassical secretion of signal peptideless proteins fibroblast growth factor 1 and interleukin 1α. In this study, we investigate the effects of interplay of Cu2+ and Ca2+ on the structure of S100A13 using a variety of biophysical techniques, including multi-dimensional NMR spectroscopy. Results of the isothermal titration calorimetry experiments show that S100A13 can bind independently to both Ca2+ and Cu2+ with almost equal affinity (Kd in the micromolar range). Terbium binding and isothermal titration calorimetry data reveal that two atoms of Cu2+/Ca2+ bind per subunit of S100A13. Results of the thermal denaturation experiments monitored by far-ultraviolet circular dichroism, limited trypsin digestion, and hydrogen-deuterium exchange (using 1H-15N heteronuclear single quantum coherence spectra) reveal that Ca2+ and Cu2+ have opposite effects on the stability of S100A13. Binding of Ca2+ stabilizes the protein, but the stability of the protein is observed to decrease upon binding to Cu2+. 1H-15N chemical shift perturbation experiments indicate that S100A13 can bind simultaneously to both Ca2+ and Cu2+ and the binding of the metal ions is not mutually exclusive. The results of this study suggest that the Cu2+-binding affinity of S100A13 is important for the formation of the FGF-1 homodimer and the subsequent secretion of the signal peptideless growth factor through the nonclassical release pathway.
INTRODUCTION
S100 proteins are the largest family within the EF-hand protein super-family (1,2). S100 proteins are acidic proteins of small molecular mass (∼10–12 kDa), devoid of disulfide bonds. They contain two EF-hands, an S100-specific EF-hand located at the N-terminal end followed by a classical Ca2+-binding EF-hand (3–6). S100 proteins have been implicated in Ca2+ homeostasis and a wide array of Ca2+-dependent signaling pathways that regulate important cellular processes such as differentiation, development, and tumor growth (7–9). S100 proteins exhibit distinct cell- and tissue-specific expression and are believed to be involved in a multitude of human diseases such as cystic fibrosis, cardiomyopathy, rheumatoid arthritis, and several types of cancer (10–14).
To date, S100A13 is the only member of the S100 family that is known to exhibit ubiquitous expression in a broad range of tissues (10,14). Unlike other S100 proteins, which exhibit positive cooperativity for the four calcium binding sites in their dimeric structure, S100A13 contains two Ca2+ binding sites with distinctly different affinities (14,15). In addition, in typical S100 proteins, the binding of Ca2+ leads to opening of EF-hand domains, which in turn renders large hydrophobic surface(s) accessible to the solvent (5,16). The solvent-exposed hydrophobic domain(s) provides a binding surface for the protein-protein interaction (2,7,16). In marked contrast, the Ca2+-free apo-form of S100A13 shows solvent-exposed hydrophobic pockets (14,15). Conversely, in the presence of calcium, a drastic decrease in the solvent-accessible nonpolar surface(s) is observed (14,15,17). Therefore, it is believed that the molecular mechanism of activation of S100A13 is distinctly different from that of other S100 proteins. S100A13 is known to be involved in the nonclassical export of signal peptideless proteins such as fibroblast growth factor (FGF-1) and interleukin 1α (15,18,19). Formation of the multi-protein release complex, consisting of FGF-1, S100A13, and the p40 form of synaptotagmin 1 (p40 Syt1) is crucial for the nonclassical secretion of FGF-1 (18).
Several studies have demonstrated that copper ions (Cu2+) play a vital role in the organization of the FGF-1 multi-protein release complex (18,20,21). Depletion of Cu2+ using a specific chelator, tetrathiomolybdate, is shown to inhibit the stress-induced release of FGF-1 (20,21). Cu2+ is believed to be crucial for the formation of the covalent FGF-1 homodimer that is present in the multi-protein complex (18). It has been demonstrated that the homodimerization of FGF-1 mediated by cysteine 30 is critical for FGF-1 release (20,21). Additionally, Cu2+ ions induce noncovalent binding between FGF-1, S100A13, and p40 Syt1 (20). However, the primary source of Cu2+ ions required for the formation of the FGF-1 homodimer is still not clear. Interestingly, in addition to Ca2+, many members of the S100 protein family bind to other metals, such as zinc and copper (22). These transition metal ions are believed to play a regulatory role by modulating the affinity of various S100 proteins for Ca2+ and/or protein targets (22). In this context, in the study described here, we investigate the affinity of S100A13 to Cu2+ ions and describe the structural interplay between Ca2+ and Cu2+ binding to the protein. The results obtained show that S100A13 binds to Cu2+ ions with high affinity. Cu2+ binds to dimeric S100A13 structures at a novel location connecting the N-terminus of one subunit with the hinge region of the other subunit. It appears that S100A13 can act as a source for Cu2+ ions required for the organization of the FGF-1 multi-protein release complex.
EXPERIMENTAL METHODS
Protein expression and purification
The amplified cDNA of S100A13 was inserted into a pGEX expression vector. The expression of S100A13 was carried out in Escherichia coli. 15N-labeled protein was prepared by growing the cells in M9-minimal medium containing 15N-labeled NH4Cl as the sole source of nitrogen. The expressed glutathione S-transferase (GST)-fused S100A13 was purified on a GST column(s). Apo-S100A13 preparations were made by repeated dialysis of the protein against EDTA. The absence of the metal was verified by the isocompetition point mass. The GST tag was eliminated by thrombin (Sigma, St. Louis, MO) cleavage and the protein was repurified by affinity chromatography on GST-sepharose.
Isothermal titration calorimetry (ITC)
Metal binding to S100A13 was characterized by measuring the heat changes during the titration of the relevant metal chloride into the protein solution using Microcal VP titration calorimeter (Northampton, MA). S100A13 and metal chloride solutions were centrifuged and degassed under vacuum condition before titration. The sample cell contained 0.1 mM S100A13 dissolved in 25 mM Tris buffer (pH 7.2) containing 25 mM KCl. Solutions of 2 mM CaCl2/CuCl2 were prepared in the same buffer as used in the cell sample and the pH of the solutions was corrected to 7.2. Titrations were performed by injecting 5- to 10-μL aliquots of 2 mM ligand (CaCl2/CuCl2) into a 0.1-mM solution of S100A13. The titration of S100A13 with the C2A domain was performed at 25°C by dissolving both the proteins in 25 mM Tris buffer (pH 7.2) containing 25 mM KCl. Results of the titration curves were corrected using protein-free buffer control and analyzed using Origin software supplied by Microcal (Northampton, MA).
Fluorescence spectroscopy
Fluorescence experiments were carried out on a Hitachi F2500 spectrofluorimeter (Tokyo, Japan). The Tb3+ titration was carried out using a stock solution of 50 mM TbCl3 containing 25 mM Tris (pH 7.2) and 25 mM KCl. All experiments were performed using a quartz cell with a light path of 10 mm and an excitation wavelength of 280 nm. Tb3+ emission spectra were acquired between 400 nm to 600 nm. The slit widths for emission and excitation were set at 2.5 nm and 10 nm, respectively. All experiments were carried out at a protein concentration of 50 μM.
8-anilino-1-naphthalenesulfonate binding experiments
The 8-anilino-1-naphthalenesulfonate (ANS) binding experiments were performed using a Hitachi F-2500 spectrofluorometer at 25°C. Fluorescence spectra were recorded using an excitation wavelength of 390 nm and an emission wavelength range of 400–600 nm. Ca2+ and Cu2+ were added from a 1-M stock solution of CaCl2/CuCl2 prepared in the storage buffer (pH 7.2) containing 25 mM KCl and 25 mM Tris (pH 7.2). Control experiments with ANS were carried out under the same buffer conditions, but in the absence of the protein. The final concentrations of the protein and ANS used were 50 μM and 250 μM, respectively.
Circular dichroism
The circular dichroism (CD) experiments were performed using a Jasco J-720 spectropolarimeter (Tokyo, Japan). All CD experiments were carried out at 100 μg/mL protein concentration. A 0.2-cm quartz cell was used for all the experiments and the signal was monitored in the wavelength range of 200–250 nm. Each spectrum represents an average of 10 scans.
Proteolytic digestion
Proteolytic digestion experiments were carried out at 25°C using trypsin (Sigma) as the proteolytic enzyme, in the absence or presence of the metal ions (at a metal ion(Cu2+/Ca2+) concentration of 2 mM). Proteolytic digestions were carried out at an enzyme/protein molar ratio of 1:10. The reaction was arrested at regular time intervals by the addition of the gel loading dye (Bio-Rad, Hercules, CA). The degree of proteolytic cleavage was estimated by densitometry of the intensity of the ∼12-kDa band corresponding to the uncleaved S100A13. The intensity of the S100A13 band not subjected to the trypsin treatment was used as a control for 100% protection against trypsin cleavage.
NMR spectroscopy
NMR experiments were performed on a Bruker Avance DMX-700 MHz spectrometer (Billerica, MA) equipped with a 5-mm inverse cryoprobe. The protein samples were prepared in 25 mM d6-Tris HCl containing 25 mM KCl, 2 mM CaCl2/CuCl2, and 0.03% NaN3. The pH of all the samples was corrected to 7.2. All NMR data were acquired at 25°C and the data were analyzed using XWINNMR 3.5 software supplied by Bruker.
Hydrogen-deuterium exchange experiment
The protein samples (Ca2+-bound or Cu2+-bound S100A13 titrated with 2 mM CaCl2) were lyophilized along with 25 mM Tris HCl and 25 mM KCl. The lyophilized powder was dissolved immediately in 0.55 mL of D2O. The pH (7.2) was not corrected for deuterium effects. Data collection was initiated after 10 min of incubation of the protein in D2O. The concentration of S100A13 used was 0.5 mM. The 600 1H-15N heteronuclear single quantum coherence (HSQC) spectra were acquired continuously for a time period of 200 h. All data were analyzed using the Kaleidagraph software (Synergy Software, Philadelphia, PA).
RESULTS
Apo-state of S100A13 binds to both Ca2+ and Cu2+
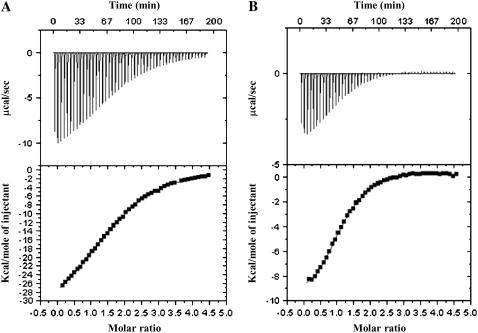
Isothermal titration calorimetry is an important tool for the direct measurement of thermodynamic parameters in various biological interactions (23). As ITC is dependent only on the exchange of heat during a reaction, it is an invaluable tool for monitoring a variety of reactions independent of spectroscopic changes. ITC has been successfully used not only to quantitatively measure binding affinities between two interacting molecules but also to calculate various thermodynamic parameters, such as change in Gibbs free energy (ΔG), enthalpy change (ΔH), entropy change (ΔS), and specific heat capacity (ΔCp), that provide insights into the nature and magnitude of interactions that govern protein-protein or protein-metal interactions (24). It is in this context that we used ITC to characterize the binding affinity of S100A13 to metal ions such as Ca2+ and Cu2+. The binding curve representing the interaction between apo-S100A13 and Ca2+ is almost hyperbolic (Fig. 1 A). The binding between apo-S100A13 and Ca2+ is exothermic and the reaction proceeds with a negative change in enthalpy. Best-fit of the binding isotherm yields apparent binding constant values of ∼8 μM and ∼66 μM (Table 1). The binding curve saturates at a Ca2+/protein dimer ratio of 4:1. Recently, Arnesano et al., studying the affinity of human S100A13 to Cu2+, reported that prior binding of Ca2+ is a prerequisite for the binding of Cu2+ to the protein (17). They suggested that binding of Ca2+ to S100A13 triggers a drastic conformational change creating a novel site for Cu2+. The results of their study imply that Cu2+ cannot bind to apo-S100A13 independently of Ca2+. To verify this observation, we investigated the binding affinity of apo-S100A13 to Cu2+. Interestingly, the binding isotherm of apo-S100A13 with Cu2+ clearly shows that Cu2+ binds to apo-S100A13 (Fig. 1 B) and the binding constant values characterizing the apo-S100A13-Cu2+ interaction are ∼12 μM and ∼55 μM, respectively (Table 1). These results suggest that S100A13 binds to Ca2+ and Cu2+ with almost similar affinities. The binding curve saturates at Cu2+ to the S100A13 monomer ratio of 2:1, suggesting that four Cu2+ ions bind per molecule of the S100A13 dimer (Table 1). Thus, ITC data clearly show that apo-S100A13 binds to both Ca2+ and Cu2+, and the binding of Cu2+ does not require the prior binding of Ca2+ to the protein.
FIGURE 1.
Isothermograms representing the binding of S100A13 to (A) Ca2+ and (B) Cu2+. The upper panels represent the raw data and the bottom panels are the best fits of the raw data. The binding isotherms have been fit to the multiple binding sites model. The concentration of protein used was 100 μM in 25 mM Tris (pH 7.2) containing 25 mM KCl. Appropriate background corrections were made to account for the heats of dilution and ionization. All experiments were performed at 25°C.
TABLE 1.
Binding parameters characterizing the S100A13-metal interation
| Binding constant(μM)
|
||||
|---|---|---|---|---|
| Interaction type | Kd1 | Kd2 | ΔGb(kcal mol−1) | N (number of binding sites per monomer) |
| apo-S100A13 + Ca2+ | 8 ± 1 | 66 ± 1 | −8.065 ± 0.5 | 2 |
| apo-S100A13 + Cu2+ | 12 ± 1 | 55 ± 1 | −7.086 ± 0.5 | 2 |
| Ca2+-bound S100A13 + Cu2+ | 62 ± 1 | 120 ± 1 | −6.126 ± 0.5 | 2 |
| Cu2+-bound S100A13 + Ca2+ | 7.6 ± 1 | 140 ± 1 | −7.808 ± 0.5 | 2 |
Conformational changes monitored by ANS binding
The three-dimensional structures of S100 proteins are characterized by the presence of two EF-hands (2,3). Each EF-hand presents a helix-loop-helix structure (2). Ca2+ binding sites are located in the loop portions of the helix-loop-helix structures. Ca2+ binding decreases the flexibility of residues in the loop portion of the helix-loop-helix structure, and consequently a small β-sheet segment is induced in that region in most S100 proteins, including S100A13 (2,5; Sivaraja et al., unpublished results). In addition, binding of Ca2+ to S100 proteins is known to cause a significant change of the interhelical angle between helix-3 and helix-4 at the C-termini (5). These subtle secondary structural changes in S100A13 could not be clearly detected by conventional spectroscopic probes such as infrared spectroscopy and circular dichroism spectroscopy (15). Therefore, we used ANS binding experiments to monitor the conformational changes induced by metal ions (Ca2+/Cu2+) in S100A13.
ANS is a hydrophobic dye that has been successfully used to detect solvent-accessible hydrophobic surface(s) in proteins. Using ANS binding experiments we had previously shown that there are large areas of solvent-accessible nonpolar surfaces in apo-S100A13 (15). Presence of solvent-exposed nonpolar clefts in apo-S100A13 is manifested by an intense ANS emission signal that is blue-shifted by ∼30 nm (from 520 nm in the absence of the protein to 490 nm in the presence of the protein). Addition of Ca2+ results in a progressive decrease in the emission intensity of the dye, suggesting a subtle conformational change resulting in the increase in the fraction of free ANS in the bulk polar solvent. (Fig. 2 A). This situation is in sharp contrast to that observed in other S100 proteins, wherein Ca2+ binding results in an increase in the solvent accessibility of nonpolar residues located in the cleft between helix-3 and helix-4 (comprising the C-terminal EF-hand (Ridinger et al. (14)). Typically, Ca2+-bound states of S100 proteins recognize and bind to their target proteins through the solvent-exposed hydrophobic surfaces (3). The nonavailability of such solvent-accessible nonpolar surfaces in the Ca2+-bound state of S100A13 suggests that its mechanism of activation and recognition of target protein partners is significantly different from other members of the S100 protein family.
FIGURE 2.
(A) Emission spectra of ANS in the presence of the S100A13 at various concentrations of Ca2+. The inset figure shows the decrease in fluorescence intensity of ANS (at 520 nm) upon increasing the concentration of Ca2+. (B) Emission spectra of ANS in the presence of S100A13 at various concentration of Cu2+. The inset figure depicts the decrease in the ANS emission intensity (at 520 nm) as a function of the concentration of Cu2+. (C) Changes in ANS emission intensity (at 520 nm) upon addition of Ca2+ alone (red) and Cu2+ alone (blue) to S100A13. The portion of the curve indicated in green shows the decrease in the ANS emission intensity when increasing amounts of Ca2+ (0–0.5 mM) are added to the Cu2+-saturated (2 mM) S100A13. The concentrations of S100A13 and ANS used are 50 μM and 250 μM, respectively. Background corrections were made in all the spectra. The dashed line indicates the beginning of the titration of the Cu2+-saturated (2 mM) S100A13 with increasing concentration of Ca2+.
It was interesting to examine whether Cu2+, like Ca2+, could also bring about subtle conformational changes that influence the solvent accessibility of hydrophobic residues in S100A13. Fig. 2 B shows that the emission intensity of ANS at 520 nm gradually decreases with an increase in the concentration of Cu2+. The decrease in emission intensity is accompanied by a progressive red shift in the emission maxima of ANS from 490 nm to 508 nm, suggesting the burial of the nonpolar surface to the interior of the protein molecule. Increasing the Cu2+ concentration beyond 0.2 mM does not produce further appreciable decrease in the ANS emission intensity, indicating the saturation of the Cu2+-mediated structural changes beyond this metal ion concentration (0.2 mM) (Fig. 2, B and C). Although both Ca2+ and Cu2+ promote the burial of the solvent-accessible nonpolar residues to the interior of the protein (S100A13), the degrees of exposure of the nonpolar residues to the solvent are possibly different in the Ca2+- and Cu2+-bound states of S100A13. The exposure of solvent-accessible nonpolar surface(s) is marginally higher in the Cu2+-bound state than that observed in the Ca2+-bound state of S100A13 (Fig. 2 C). Titration of the Cu2+-bound S100A13 with increasing concentration of Ca2+ not only results in a gradual red shift in the emission maxima (from 480 nm to 500 nm) but also in a decrease in the emission intensity of the hydrophobic dye (at 520 nm, Fig. 2 C). Upon addition of 2 mM of Ca2+ to the Cu2+-bound S100A13, the relative emission intensity of ANS (at 520 nm) matches quite well with that obtained upon binding to the protein in the presence of only Ca2+. These results support our conclusion that the degree of solvent accessibility of the nonpolar surfaces in S100A13 is higher in the presence of Cu2+ than that observed in the presence of Ca2+.
Detection of conformational changes using limited trypsin digestion
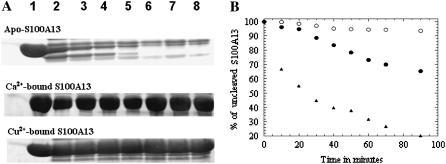
Limited protease digestion analysis is a valuable technique for probing conformational changes induced in a protein upon ligand binding (25). The factors that primarily influence protease-induced cleavage are solvent accessibility of the susceptible sites, their protrusion from the surface of the protein, hydrogen bonds between cleavage sites and the rest of the molecule, flexibility of the site, and its propensity to local unfolding/packing (26). In this background, we used limited trypsin-digestion analysis as a tool to probe the possible structural changes induced in S100A13 by metal ions (Ca2+ and Cu2+). Trypsin generally cleaves proteins at the C-terminal ends of basic amino acids such as lysine and arginine. As S100A13 is rich in lysine and arginine residues, trypsin is an apt choice to monitor the conformational changes induced by the metal ions (Ca2+ and Cu2+). The extent of proteolytic cleavage was monitored from the decrease in intensity of the 12-kDa Coomassie blue-stained band (on sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE)) corresponding to uncleaved S100A13 (Fig. 3 A). Cleavage of apo-S100A13 occurs instantaneously upon addition of trypsin. More than 80% of the protein is cleaved within 70 min of initiation of the cleavage reaction (Fig. 3, A and B). In marked contrast, the Ca2+-bound state of S100A13 is resistant to trypsin cleavage (Fig. 3, A and B). More than 90% of the S100A13 protein remains uncleaved after 70 min of initiation of the trypsin cleavage (Fig. 3). These results show that the protein assumes a more tightly packed conformation upon binding to Ca2+. The degree of susceptibility of Cu2+-bound S100A13 to trypsin cleavage is higher than observed in the Ca2+-bound state of S100A13 (Fig. 3, A and B). In the presence of Cu2+, >30% of the 12-kDa band (on SDS-PAGE) is cleaved after 90 min of the cleavage reaction (Fig. 3 B). The rate of trypsin-induced cleavage of Cu2+-bound S100A13 is significantly higher than that observed in the Ca2+-bound state of S100A13, but lower than in the apo-state of the protein (Fig. 3 B). Results of the trypsin digestion experiment clearly show that structures of the Ca2+ and Cu2+-bound states of S100A13 are subtly different. The Ca2+-bound state of S100A13 appears to be more tightly packed than the Cu2+-bound form of the protein.
FIGURE 3.
(A) SDS-PAGE analysis of the trypsin cleavage products of free S100A13 and S100A13 in the presence of metal ions (2 mM Ca2+/Cu2+). Lanes 1 to 8 indicate the progress of the trypsin-induced cleavage of S100A13 after 0 min, 10 min, 20 min, 30 min, 40 min, 50 min, 60minutes, and 70 min of incubation of the protein with the enzyme. (B) Percentage of uncleaved S100A13 in the absence of metal ions (solid triangles) in the presence of 2 mM Ca2+ (open circles) and in the presence of 2 mM Cu2+ (solid circles). The percentage of uncleaved S100A13 was estimated from the intensity of the 12.5-kDa Coomassie blue-stained band on polyacrylamide gel. The concentrations of S100A13 and trypsin used were 50 μM and 500 μM, respectively.
Conformational changes monitored by 2D NMR spectroscopy
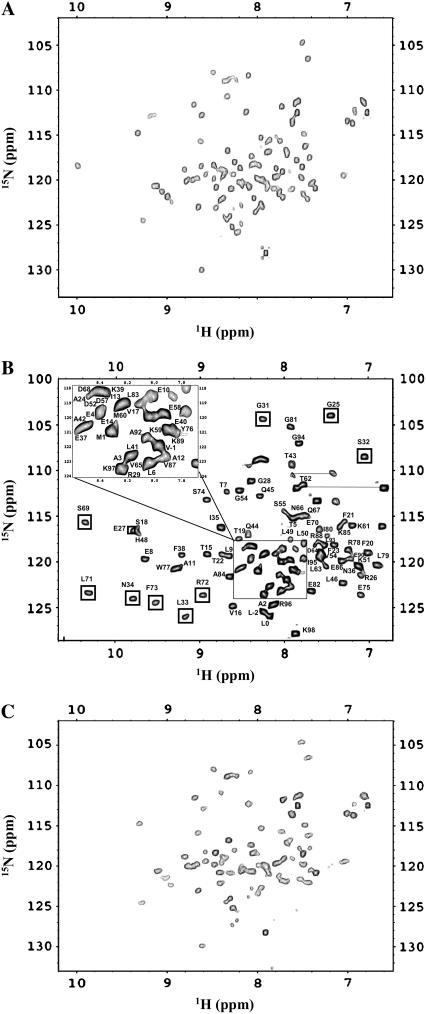
1H-15N HSQC spectrum is a finger-print of the backbone conformation of a protein (27). Each 1H-15N crosspeak in the spectrum represents the microenvironment of an amino acid residue in a given conformation of the protein (27). Therefore, the conformational transitions induced by the metal ions (Ca2+/Cu2+) can be easily traced from the perturbation of the crosspeaks in the 1H-15N HSQC spectrum. The 1H-15N HSQC spectrum of apo-S100A13 is not well-dispersed and many crosspeaks in the spectrum are broadened (Fig. 4 A). We have been able to accomplish only partial assignment of the resonances in apo-S100A13. However, addition of Ca2+ drastically increases the dispersion of crosspeaks in the 1H-15N HSQC spectrum (Fig. 4 B). All the crosspeaks in the 1H-15N HSQC spectrum of the Ca2+-bound state of S100A13 have been unambiguously assigned (28). The three-dimensional solution structure of the Ca2+-bound S100A13 has been recently solved at high resolution (15). As mentioned earlier, β-sheet conformation is induced in the Ca2+-binding site located in the loop portion of the helix-loop-helix structure (in the EF-hands). Residues involved in the calcium binding site in S100A13 include 24–29 and 34–37 in the N-terminal EF-hand, and 64–68 and 71–74 in the C-terminal EF-hand. Most of these residues constitute the new β-sheet segment induced upon Ca2+-binding to S100A13. The crosspeaks of residues involved in the beta-sheet show good chemical shift dispersion in the 1H-15N HSQC spectrum. The transformation of the structureless loop to the β-sheet conformation is reflected in the emergence of new crosspeaks from the severely overlapped region(s) of the 1H-15N HSQC spectrum of S100A13 (Fig. 4 B). Increase in the interhelical angle between helices (H3 and H4 of the EF-hand) does not appear to cause significant chemical perturbation of crosspeaks in the 1H-15N HSQC spectrum. In summary, binding of Ca2+ to apo-S100A13 causes two major structural changes: 1), induction of β-sheet conformation in the loop region of the helix-loop-helix structure in both EF-hands; and 2), change in the interhelical angle between helices constituting the C-terminal EF-hand.
FIGURE 4.
1H-15N HSQC spectra of (A) apo-S100A13 and (B) S100A13 in the presence of 2 mM CaCl2. (C) S100A13 in the presence of 2 mM CuCl2. 1H-15N HSQC spectra were obtained at a protein concentration of 0.1 mM in 25 mM Tris buffer (pH 7.2) containing 25 mM KCl. The buffer solution was prepared in 90% H2O and 10% D2O. HSQC spectra were acquired at 25°C. The boxed area represents the crosspeaks of residues that are locating in close proximity to the Ca2+-binding site.
Copper (Cu2+) is a paramagnetic and, therefore, if Cu2+ should bind to a protein, then the NMR signals at the metal (Cu2+) binding site would be expected to broaden with the reciprocal of the sixth power of the metal-nucleus distance (29). There have been numerous studies wherein crosspeaks of residues (in the 1H-15N HSQC spectrum) that are within the vicinity of the Cu2+-binding site(s) have been shown to decrease in intensity and subsequently disappear (30,31). Selected crosspeaks in the 1H-15N HSQC spectrum of apo-S100A13, acquired in the presence of 1 mM Cu2+, show a progressive decrease in intensity upon addition of Cu2+. Most of these crosspeaks are located in the crowded region(s) of the 1H-15N HSQC spectrum and hence could not be unambiguously assigned (Fig. 4 C). The dramatic increase in the chemical-shift dispersion of the crosspeaks in the 1H-15N HSQC spectrum observed when Ca2+ binds to the protein (S100A13) is not seen in the 1H-15N HSQC spectra of the Cu2+-bound S100A13. The increased chemical-shift dispersion observed in the presence of Ca2+ is primarily attributed to the induction of β-sheet conformation at the Ca2+ binding site. Therefore, it appears that, unlike Ca2+, the binding of Cu2+ to S100A13 does not induce new β-sheet conformation in the protein. Based on the analysis of 1H-15N chemical-shift perturbation, ITC, and ANS binding data, the following general conclusions could be drawn on the effects of binding of Cu2+ to apo-S100A13. 1), Two Cu2+ ions bind per subunit of apo-S100A13. However, it is not clear if the Cu2+ ions bind at the Ca2+-binding sites or at sites remote from the Ca2+-binding sites. 2), Unlike Ca2+, binding of Cu2+ does not appear to lead to the induction of β-sheet conformation in the helix-loop-helix structure of the two EF-hands. 3), ANS binding data show that Cu2+ binding results in partial burial of the solvent-accessible nonpolar surfaces in the protein. However, the degree of accessibility of the nonpolar surface(s) in the Cu2+-bound S100A13 structure is higher than that observed in the structure of the Ca2+-bound S100A13.
Binding of Ca2+ and Cu2+ to S100A13 is not mutually exclusive
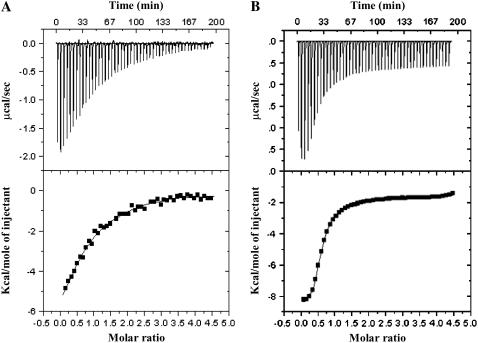
The results discussed thus far clearly show that S100A13 can bind to both Ca2+ and Cu2+. In this context, it would be interesting to understand if these two metal ions compete for the same site(s) in the protein. ITC experiments, monitoring the titration of calcium-bound (2 mM Ca2+) S100A13 with Cu2+ reveal that the binding of the two metal ions (Ca2+ and Cu2+) is not mutually exclusive (Fig. 5 A). Although the apparent binding-constant values characterizing the binding of Cu2+ to the Ca2+-bound S100A13 is marginally lower than that observed in the absence of Ca2+ (Table 1), the number of Cu2+ ions bound to the protein remains the same (two Cu2+ ions per monomer of the Ca2+-bound S100A13). Similarly, the best-fit of the binding curve representing the titration of Cu2+-bound S100A13 with Ca2+ shows that there are ∼2 Ca2+-binding sites per subunit of the Cu2+-bound S100A13 (Fig. 5 B). The binding constant values for the high-affinity and low-affinity Ca2+-binding sites (Kd1) do not change significantly in the presence of saturating amounts of Cu2+ (Table 1). These results clearly suggest that both Ca2+ and Cu2+ can bind simultaneously to S100A13.
FIGURE 5.
Binding curves representing the titration of (A) Ca2+-bound S100A13 with Cu2+, (B) Cu2+-bound S100A13 with Ca2+. S100A13 was prepared in 25 mM Tris buffer (pH 7.2), containing 25 mM KCl. ITC experiments were performed at 25°C and the data were corrected for heats of dilution and ionization. The upper and lower panels represent the raw data and the best fits of the raw data, respectively. The concentration of S100A13 used was 100 μM.
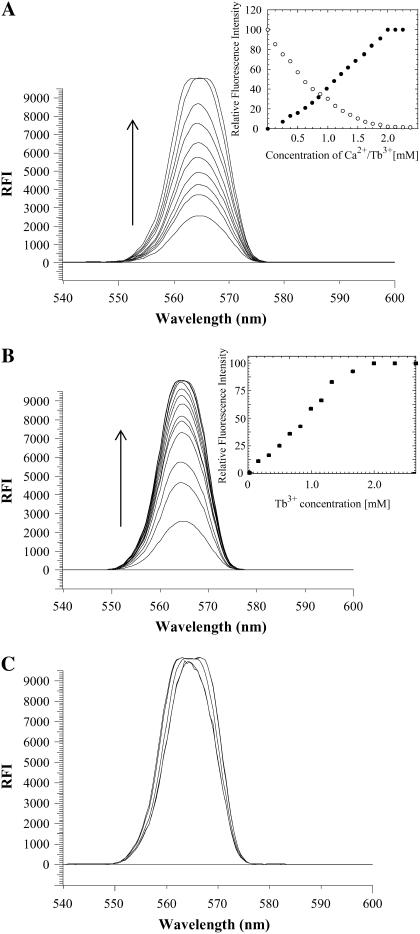
It is interesting to explore whether the two metal ions (Ca2+ and Cu2+) compete for the same binding sites on the protein. Terbium (Tb3+) is a valuable fluorescent probe that is used to investigate Ca2+-binding properties of proteins (32). Tb3+ is known to bind to the Ca2+-binding sites and induce luminescence in the visible region via energy transfer from the aromatic residues in proteins (32). The competition between Ca2+ and Cu2+ ions to bind to S100A13 was probed by fluorescence energy transfer (FRET) experiments using Tb3+. Fluorescence intensity of Tb3+ at 565 nm is observed to progressively increase when S100A13 is titrated with increasing concentrations of TbCl3 (Fig. 6 A). The increase in Tb3+ fluorescence is compounded by a concomitant decrease in the tryptophan fluorescence intensity at 340 nm (data not shown). The fluorescence energy transfer from the aromatic residues in S100A13 to Tb3+ suggests that some of the aromatic residues (Trp and Tyr) are located spatially close to the Ca2+ (or Tb3+) binding sites in the protein. The emission intensity at 565 nm is observed to plateau at a S100A13/Tb3+ ratio of 1:2, additionally confirming that two Ca2+ ions bind per subunit of S100A13 (Fig. 6 A, inset). The Ca2+-to-S100A13 binding stoichiometry obtained from the Tb3+ binding data is consistent with that derived from the ITC experiments. Assuming that the Ca2+ and Tb3+ binding sites on S100A13 are the same, we measured the binding affinity of Ca2+ to S100A13 using the Tb3+-Ca2+ competition assay. S100A13, saturated with 2 mM Tb3+, was titrated with increasing concentrations of Ca2+ and the binding affinity (Kd) of Ca2+ to S100A13 was obtained from the decrease in Tb 3+ fluorescence at 545 nM. The Kd value estimated for the Ca2+-S100A13 interaction from the Tb3+/Ca2+ competition assay (∼25 μM) is in the same range as that obtained from the ITC data (Kd ∼ 8 μM).
FIGURE 6.
Terbium binding to S100A13. (A) Titration of apo-S100A13 with increasing concentration of TbCl3. (Inset) Titration of apo-S100A13 with increasing concentrations of Tb3+ (solid circle); S100A13 saturated with Tb3+ and titrated with increasing concentrations of CaCl2 (open circle) represents change in Tb3+ fluorescence (at 545 nm) in the absence (solid circle) and presence (open circle) of various concentrations of Ca2+. (B) Titration of Cu2+-bound S100A13 with increasing concentrations of Tb3+. (C) Terbium-bound S100A13 titrated with increasing concentrations of CuCl2. The inset figure in each panel shows the change(s) in the Tb3+ emission intensity (at 565 nm) under experimental conditions used in the corresponding panel. The concentration of S100A13 used was 50 μM. All solutions were prepared in 25 mM Tris buffer (pH 7.2, containing 25 mM KCl. The appropriate blank corrections were made in all spectra. The direction of the arrows in A and B indicate the increase in the Tb3+ fluorescence.
The affinity of S100A13 to Tb3+ was examined in the presence of saturating concentrations (2 mM) of Cu2+ to assess whether the Ca2+ (or Tb3+) competes for the Cu2+ sites in the protein. Even at a saturating concentration of Cu2+ (2 mM), the fluorescence at 565 nm steadily increases with an increase in the concentration of Tb3+, suggesting that the binding of Ca2+ (or Tb3+) and Cu2+ to S100A13 are mutually independent (Fig. 6 B). The Ca2+ (or Tb3+)-to-S100A13 binding stoichiometry remains unchanged even in the presence of an excess of Cu2+. On the other hand, S100A13 saturated with Tb3+ (2 mM) and titrated with increasing concentrations of Cu2+ showed no change in the Tb3+ fluorescence intensity at 565 nm (Fig. 6 C). These results clearly indicate that Ca2+ and Cu2+ do not compete with each other for the same binding sites in S100A13. In the event of competition between the metal ions to bind to the same sites on S100A13, the Tb3+ fluorescence (at 565 nm) is expected to show a decrease with an increase in concentration of Cu2+.
Interplay between Ca2+ and Cu2+ binding to S100A13
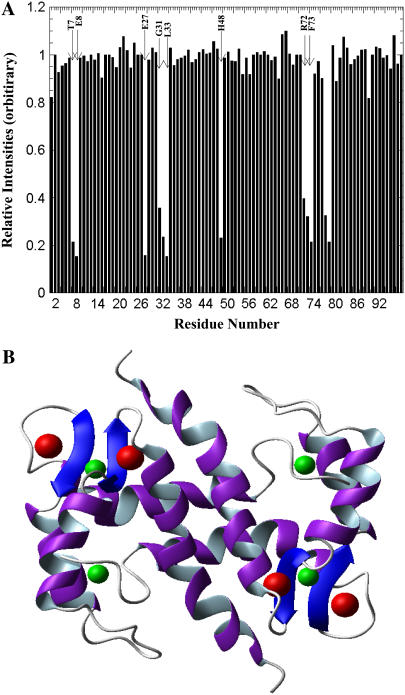
All the resonances (1H, 13C, and 15N) in the Ca2+-bound state of S100A13 have been assigned (28). The three-dimensional solution structure of S100A13 in the Ca2+-bound state has been reported (15). As for all other S100 proteins, S100A13 also has two distinct Ca2+ binding sites. As stated earlier, the Ca2+-binding sites are located in the loops of the two helix-loop-helix motifs. ITC and Tb3+-based fluorescence energy transfer experiments clearly suggest that Cu2+ can bind to S100A13 even in the presence of Ca2+. In this context, it would be interesting to identify the Cu2+ binding sites on the Ca2+-bound S100A13. Increasing addition of paramagnetic Cu2+ results in a progressive decrease in intensity of a few selected crosspeaks in the 1H-15N HSQC spectrum of Ca2+-bound S100A13 (Fig. 7). The residues whose crosspeaks show a significant decrease include Thr-7, Glu-8, Leu-33, Asn-34, His-48, Leu-71, Arg-72, Phe-73, Ser-74, Trp-77, and Arg-78. There is no indication of a major conformational change when Cu2+ binds to Ca2+-bound S100A13. The residues that bind to Cu2+, in the Ca2+-bound S100A13 state, appear to be distributed in two locations. One site comprises of Leu-33, Asn-34, Leu-71, and Arg-72, located on the β-sheet between helix-3 and helix-4 (Fig. 8 A). The residues in the second Cu2+-binding site include Thr-7, Glu-8, and His-48. These residues are situated in the hinge region between helix-2 and helix-3 and constitute an exclusive Cu2+-binding site (Fig. 8 B). Although the first Cu2+ binding site partially overlaps with the Ca2+ binding site, we believe that the binding of the two kinds of ions to the protein (S100A13) is not mutually exclusive. This notion is supported by the results of the ITC and Tb3+ binding experiments, which show that two Cu2+ ions bind per monomer of the Ca2+-bound S100A13. In addition, the isocompetition point mass of the Ca2+-bound S100A13 treated with 2 mM Cu2+ (after exhaustive dialysis) showed that two Cu2+ and two Ca2+ ions are simultaneously bound to the protein (data not shown). These results unambiguously demonstrate that S100A13 has distinct Ca2+- and Cu2+-binding sites and, therefore, the binding of one metal ion does not hamper the protein's binding to the other metal ion.
FIGURE 7.
Two portions of the overlapped 1H-15N HSQC spectra of Ca2+-bound S100A13 (black) and Ca2+-bound S100A13 in the presence of 2 mM Cu2+ (red). S100A13 (0.1 mM) was prepared in 25 mM Tris buffer (pH 7.2, dissolved in 90% H2O + 10% D2O) containing 25 mM KCl.
FIGURE 8.
(A) Chemical shift perturbation plot of Ca2+-bound S100A13 titrated with 2 mM CuCl2. There are two prominent sites (site 1.T7, E8, and H48; site 2.E27, G31, L33, R72, and F73) in the protein at which Cu2+ ions appear to bind. The concentration of S100A13 used was 0.1 mM. The protein was prepared in 25 mM Tris buffer (pH 7.2) containing 25 mM KCl and 90% H2O and 10% D2O. The experiment was performed at 25°C. (B) MOLMOL representation of the backbone fold of S100A13 showing the Cu2+ (green) and Ca2+ (red) ions bound to S100A13.
1H-15N HSQC spectra of Cu2+-bound S100A13 at increasing concentrations of Ca2+ show a progressive increase in the dispersion of the crosspeaks. The 1H-15N HSQC spectrum of Cu2+-saturated S100A13 in the presence of 2 mM Ca2+ mostly resembles the one obtained for the protein (S100A13) in the presence of calcium alone. These results suggest that Ca2+ not only binds to Cu2+-bound S100A13 but also induces a conformational change that is similar to the one observed when Ca2+ binds to the apo-state of the proteins. The effects of Ca2+ and Cu2+ on the structure of S100A13 appear to be distinctly different. Binding of Cu2+ does not result in discernible conformational change(s) in S100A13. On the other hand, Ca2+, upon binding to the apo-S100A13 or the Cu2+-bound S100A13, induces a prominent change(s) in the conformation. Many S100 proteins have been previously shown to bind to transition metals like Zn2+ (22). Just as with Ca2+, binding of Zn2+ is also known to cause large conformational changes in S100 proteins such as S100B, S100A7, and S100A12 (22). Binding of Zn2+ invariably affects Ca2+ binding and increases target protein-protein affinities of S100 proteins. In marked contrast, Cu2+ appears to play a more docile role as it neither induces a conformational change in S100A13 nor increases the binding affinity of Ca2+ to the protein.
Metal binding alters the stability of the protein
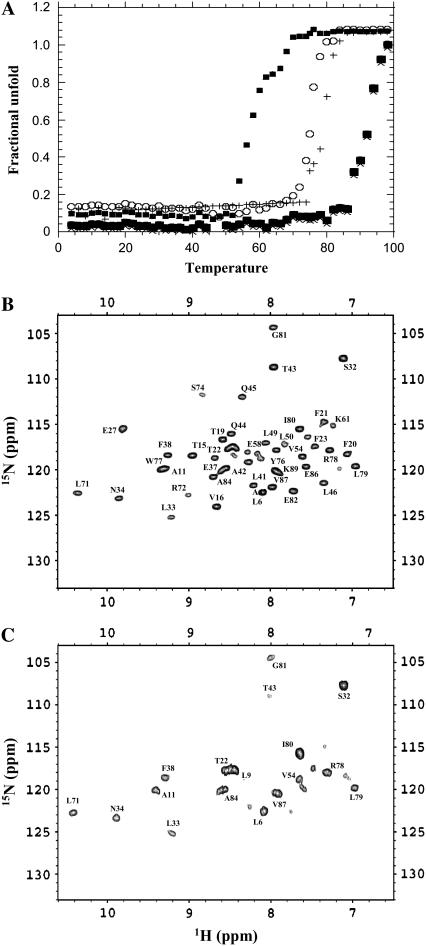
Metals not only induce conformational changes but are also known to alter the stability of proteins (33). Investigation of the effect(s) of metal-ion binding may be important to understanding their physiological role(s) in modulating the functional properties of proteins. In this context, we performed thermal denaturation experiments to examine the influence of Ca2+ and Cu2+ on the thermodynamic stability of apo-S100A13. The far-ultraviolet CD spectrum of S100A13, at 25°C, shows double minima centered at 208 nm and 222 nm (data not shown). These spectral features are consistent with the predominantly helical nature of the protein. Thermal denaturation of apo-S100A13 monitored using ellipticity changes at 222 nm shows that the protein begins to unfold at 68 ± 1°C and complete unfolding occurs beyond 80°C. The apparent Tm (temperature at which 50% of the protein molecules exist in the denatured state(s)) value for the unfolding reaction of apo-S100A13 is estimated to be 76 ± 1°C (Fig. 9 A). Interestingly, in the presence of 2 mM Cu2+, the conformational stability of apo-S100A13 is drastically reduced (Tm = 58 ± 1°C). Addition of Ca2+ (2 mM) to the apo-S100A13 appears to significantly stabilize the protein (Fig. 9 A). The apparent Tm for the unfolding reaction of Ca2+-bound S100A13 is 92 ± 1°C. Treatment of Cu2+-bound S100A13 with 2 mM Ca2+ results in an increase in the thermal stability of the protein. The apparent Tm value of the protein is almost similar (Tm = 90 ± 1°C) to the one observed when apo-S100A13 is treated with 2 mM Ca2+ alone (Fig. 9 A). Surprisingly, the thermodynamic stability of Cu2+-bound S100A13 treated with 2 mM Ca2+ is significantly different from that of Ca2+-bound S100A13 treated with 2 mM Cu2+. The Tm of unfolding of the former is ∼10°C lower than that of the latter (Fig. 9 A). Although two Cu2+ ions and two Ca2+ ions bind to the protein under both of these conditions, the disparities in stabilities observed could be possibly attributed to the subtle differences in the degree of compactness of the structure of the protein under these two conditions. The structure of the Cu2+-bound S100A13 treated with 2 mM Ca2+ appears to be less compact (or less stable) than the structure of the Ca2+-bound S100A13 treated with 2 mM Cu2+. The high conformational flexibility of the Cu2+-bound S100A13 treated with 2 mM Ca2+ is supported by results of the amide proton exchange experiments. Comparison of the 1H-15N HSQC spectra of S100A13 obtained under these two conditions in D2O (after incubation of the protein in D2O for 10 min) clearly shows that most of the residues at the dimer interface and in helix 3 and helix 4 are protected against exchange in S100A13 bound to Ca2+ alone (Fig. 9 B). Interestingly, in Cu2+-bound S100A13 treated with 2 mM Ca2+, most of the amide protons of residues in these regions are exchanged with D2O (Fig. 9 C). The results of the amide proton exchange are in good agreement with the thermal unfolding experiments and suggest that the final conformational stability of S100A13 is dependent not only on the nature of the metal ions but also on the order in which the metal ions bind to the protein.
FIGURE 9.
(A) Thermal unfolding curves of S100A13 monitored by changes in ellipiticity at 222 nm, apo-S100A13 (open circles); S100A13 in 2 mM CaCl2 (solid circles); S100A13 in 2 mM CuCl2 (solid triangles); Cu2+-bound S100A13 treated with 2 mM CaCl2 (plus symbols); and Ca2+-bound S100A13 treated with 2 mM CuCl2 (X). The concentration of S100A13 used was 0.1 mM. All experiments were performed in 25 mM Tris buffer (pH 7.2) containing 25 mM KCl. Blank corrections were made in all data. (B and C) 1H-15N HSQC spectra of S100A13 in 2 mM CaCl2 and Cu2+-bound S100A13 treated with 2 mM CaCl2. The 1H-15N HSQC were acquired after 10 min of initiation of deuterium exchange at 25°C. The concentration of the protein used was 0.5 mM.
S100A13 binds to the C2A domain of synaptotagmin 1 (Syt1)
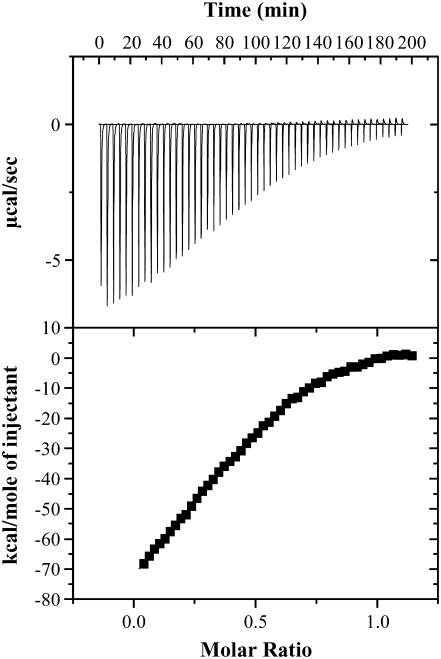
We had previously demonstrated that the C2A domain synaptotagmin 1 has high binding affinity for Cu2+ (Kd(app) is in the micromolar range (38)). Four Cu2+ ions bind per molecule of the C2A domain. No discernible conformational change(s) were observed in the C2A domain upon binding to the metal ion. We performed ITC experiments to examine whether apoS100A13 and the apo-C2A domain interact with each other. The binding isotherm representing the S100A13-C2A titration shows that these proteins interact with each other at a stoichiometric ratio of 1:2 (Fig. 10). Apo-S100A13 binds to the apo-C2A domain with moderate affinity (Kd ∼ 85 μM) and the binding proceeds with the absorption of heat (Fig. 10). The binding affinity does not change significantly with increase in the NaCl concentration (in the range of 50 to 250 mM, data not shown). In addition, titration of S100A13 and the C2A domain in the presence of excess of Cu2+ (2 mM) does appear to affect the binding affinity between these proteins (data not shown). These results suggest that interaction between S100A13 and the C2A domain is not driven by Cu2+.
FIGURE 10.
Binding isotherm representing the titration of apo-S100A13 with the C2A domain. S100A13 and the C2A domain were prepared in 25 mM Tris buffer (pH 7.2), containing 25 mM KCl. The apparent dissociation constant defining the affinity of the apo-S100A13 to the C2A domain is estimated to be in the micromolar range. ITC experiments were performed at 25°C and the data were corrected for heats of dilution and ionization. The upper and lower panels represent the raw data and the best fits of the raw data, respectively.
DISCUSSION
Maciag and coworkers established that Cu2+ is mandatory for the secretion of FGF-1 into the extracellular compartment (18). It is believed that the Cu2+-induced oxidation of the thiol group of Cys-30 leading to the formation of the FGF-1 homodimer, is a crucial step in the nonclassical release of FGF-1. However, the source of Cu2+ that is required for the oxidation of Cys-30, is still not clear. It is well known that free copper is highly reactive and toxic, and accumulation of the free ionic form of copper is expected to have detrimental effects on the cell (34). To overcome this problem, cells have evolved a mechanism involving small copper-binding proteins called the “copper chaperones” (35). These “copper chaperones” receive Cu2+ from cell-surface copper transporters and distribute them to the destination proteins that require Cu2+ for their function(s). Atx1, CCS, and Cox17 are well-studied “copper chaperones” found in the cytoplasm of cells (36). Although there is no experimental evidence of any of the “copper chaperones” interacting with proteins (such as S100A13 or synaptotagmin) that are involved in the multi-protein FGF release complex, it is reasonable to predict that the Cu2+ ions required for the formation of the FGF dimer are transferred from one of the “copper chaperones” to proteins involved in the FGF release complex.
Several members of the S100-family have been reported to bind to transition metals like Cu2+ and Zn2+ (22). Interestingly, S100B and S100A5 play important roles in copper homeostasis as well as in prevention of copper-induced oxidative damage in the brain (3). These S100 proteins, including S100A13, bind to Cu2+ with moderate affinity in the micromolar range (22). Interestingly, unlike the Cu2+-binding proteins (like superoxide dismutase, prion proteins, and the “copper chaperones” such as Atx1 and CCS), the S100 proteins lack the typical copper-binding motif (37). In this context, it may be worth mentioning that RNase A, which also lacks the typical copper-binding motif, binds to Cu2+ (36). Crystal structure of the Cu2+-bound RNaseA revealed that copper binds to the α-amino group of lysine, the carboxyl group of glutamic acid, and the imidazole group of histidine. The Cu2+-binding affinity of S100A13 appears to bear importance in the secretion of FGF-1 through the nonclassical pathway. The Cu2+-binding affinities of the “copper chaperones” and S100A13 are similar (in the micromolar range). Therefore, it is possible that S100A13 is the direct recipient of Cu2+ from the “copper chaperones”. The Cu2+ bound to S100A13 is possibly then transferred to the C2A domain of synaptotagmin 1 (Syt1). This notion of the transfer of Cu2+ from S100A13 to the C2A domain of synaptotagmin is reasonable because 1), ITC experiments show that S100A13 and C2A domain of Syt1 interact with each other with moderate affinity (Fig. 10); 2), the C2A domain binds to Cu2+ with very high affinity in the nanomolar range (38); and 3), ITC and 1H-15N HSQC perturbation experiments suggest that the C2A domain binds to FGF-1 (38). As FGF-1 and the C2A domain of Syt1 are involved in direct interaction, the Cu2+ bound to the C2A domain is possibly utilized to promote the specific oxidation of Cys-30 in FGF-1 to form the homodimer. At this juncture, the mechanism proposed for the Cu2+-induced FGF homodimer is purely speculative. More detailed structural characterization of the binding interfaces between proteins (S100A13 and FGF-1) involved in the multi-protein FGF release complex is required to validate the proposed mechanism. Also, based on the results of this work, the mutation analysis of Cu2+ binding sites of S100A13 will facilitate a better understanding of the role of the metal ion in the nonclassical export of the FGF-1 release complex.
Acknowledgments
This study was supported by research grants from the National Institutes of Health (NIH NCRR COBRE grant P20RR15569), the Department of Energy (DE-FG02-01ER15161), the Arkansas Biosciences Institute, and the National Science Council, Taiwan (NSC 94-2320-B007-005 and NSC 94-2113-M007-036). I.P. and I.G. were supported by National Institutes of Health grants HL35627, HL32348, and RR1555 (Project 1).
References
- 1.Malmendal, A., C. W. Vander Kooi, N. C. Nielsen, and W. J. Chazin. 2005. Calcium-modulated S100 protein-phospholipid interactions. An NMR study of calbindin D9k and DPC. Biochemistry. 44:6502–6512. [DOI] [PubMed] [Google Scholar]
- 2.Bhattacharya, S., C. G. Bunick, and W. J. Chazin. 2004. Target selectivity in EF-hand calcium binding proteins. Biochim. Biophys. Acta. 1742:69–79. [DOI] [PubMed] [Google Scholar]
- 3.Wilder, P. T., K. M. Varney, M. B. Weiss, R. K. Gitti, and D. J. Weber. 2005. Solution structure of zinc- and calcium-bound rat S100B as determined by nuclear magnetic resonance spectroscopy. Biochemistry. 44:5690–5702. [DOI] [PubMed] [Google Scholar]
- 4.Markowitz, J., R. R. Rustandi, K. M. Varney, P. T. Wilder, R. Udan, S. L. Wu, W. D. Horrocks, and D. J. Weber. 2005. Calcium-binding properties of wild-type and EF-hand mutants of S100B in the presence and absence of a peptide derived from the C-terminal negative regulatory domain of p53. Biochemistry. 44:7305–7314. [DOI] [PubMed] [Google Scholar]
- 5.Smith, S. P., and G. S. Shaw. 1998. A change-in-hand mechanism for S100 signalling. Biochem. Cell Biol. 76:324–333. [DOI] [PubMed] [Google Scholar]
- 6.Bhattacharya, S., and W. J. Chazin. 2003. Calcium-driven changes in S100A11 structure revealed. Structure. 11:738–740. [DOI] [PubMed] [Google Scholar]
- 7.Zimmer, D. B., P. Wright Sadosky, and D. J. Weber. 2003. Molecular mechanisms of S100-target protein interactions. Microsc. Res. Tech. 60:552–559. [DOI] [PubMed] [Google Scholar]
- 8.Mueller, A., B. W. Schafer, S. Ferrari, M. Weibel, M. Makek, and C. W. Heizmann. 2005. The calcium-binding protein S100A2 interacts with p53 and modulates its transcriptional activity. J. Biol. Chem. 280:29186–29193. [DOI] [PubMed] [Google Scholar]
- 9.Heizmann, C. W., G. Fritz, and B. W. Schafer. 2002. S100 proteins: structure, functions and pathology. Front. Biosci. 7:1356–1358. [DOI] [PubMed] [Google Scholar]
- 10.Chan, W. Y., C. L. Xia, D. C. Dong, C. W. Heizmann, and D. T. Yew. 2003. Differential expression of S100 proteins in the developing human hippocampus and temporal cortex. Microsc. Res. Tech. 60:600–613. [DOI] [PubMed] [Google Scholar]
- 11.Hsieh, H. L., B. W. Schafer, J. A. Cox, and C. W. Heizmann. 2002. S100A13 and S100A6 exhibit distinct translocation pathways in endothelial cells. J. Cell Sci. 115:3149–3158. [DOI] [PubMed] [Google Scholar]
- 12.Marenholz, I., C. W. Heizmann, and G. Fritz. 2004. S100 proteins in mouse and man: from evolution to function and pathology (including an update of the nomenclature). Biochem. Biophys. Res. Commun. 322:1111–1112. [DOI] [PubMed] [Google Scholar]
- 13.Nelson, M. R., E. Thulin, P. A. Fagon, S. Forsen, and W. J. Chazin. 2002. The EF-hand domain: a globally cooperative structural unit. Protein Sci. 11:198–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ridinger, K., B. W. Schafer, I. Durussel, J. A. Cox, and C. W. Heizmann. 2000. S100A13. Biochemical characterization and subcellular localization in different cell lines. J. Biol. Chem. 275:8686–8694. [DOI] [PubMed] [Google Scholar]
- 15.Sivaraja, V., T. K. S. Kumar, I. Prudovsky, and C. Yu. 2005. Three-dimensional solution structure of a unique S100 protein. Biochem. Biophys. Res. Commun. 335:1140–1148. [DOI] [PubMed] [Google Scholar]
- 16.McClintock, K. A., and G. S. Shaw. 2003. A novel S100 target conformation is revealed by the solution structure of the Ca2+-S100B-TRTK-12 complex. J. Biol. Chem. 278:6251–6257. [DOI] [PubMed] [Google Scholar]
- 17.Arnesano, F., L. Banci, I. Bertini, A. Fantoni, L. Tenori, and M. S. Viezzoli. 2005. Structural interplay between calcium(II) and copper(II) binding to S100A13 protein. Angew. Chem. Int. Ed. Engl. 39:6341–6344. [DOI] [PubMed] [Google Scholar]
- 18.Prudovsky, I., A. Mandinova, R. Soldi, C. Bagala, I. Graziani, M. Landriscina, F. Tarantini, M. Duarte, S. Bellum, H. Doherty, and T. Maciag. 2003. The non-classical export routes: FGF1 and IL-1alpha point the way. J. Cell Sci. 116:4871–4881. [DOI] [PubMed] [Google Scholar]
- 19.Arunkumar, A. I., S. Srisailam, T. K. S. Kumar, K. M. Kathir, Y. H. Chi, H. M. Wang, G. G. Chang, I. Chiu, and C. Yu. 2002. Structure and stability of an acidic fibroblast growth factor from Notophthalmus viridescens. J. Biol. Chem. 277:46424–46432. [DOI] [PubMed] [Google Scholar]
- 20.Landriscina, M., C. Bagala, A. Mandinova, R. Soldi, I. Micucci, S. Bellum, I. Prudovsky, and T. Maciag. 2001. Copper induces the assembly of a multiprotein aggregate implicated in the release of fibroblast growth factor 1 in response to stress. J. Biol. Chem. 276:25549–25557. [DOI] [PubMed] [Google Scholar]
- 21.Landricina, M., R. Soldi, C. Bagala, I. Miccuci, S. Bellum, F. Tarantini, I. Prudovsky, and T. Maciag. 2001. S100A13 participates in the release of fibroblast growth factor 1 in response to heat shock in vitro. J. Biol. Chem. 276:22544–22552. [DOI] [PubMed] [Google Scholar]
- 22.Heizmann, C. W., and J. A. Cox. 1998. New perspectives on S100 proteins: a multi-functional Ca(II)-, Zn(II)- and Cu(II)-binding protein family. Biometals. 11:383–397. [DOI] [PubMed] [Google Scholar]
- 23.Pierce, M. M., C. S. Raman, and B. T. Nall. 1999. Isothermal titration calorimetry of protein-protein interactions. Methods. 2:213–221. [DOI] [PubMed] [Google Scholar]
- 24.Jackson, M., S. Chopra, R. D. Smiley, P. O. A. Maynord, A. Rosowsky, R. E. London, L. Levy, T. I. Kalman, and E. E. Howell. 2005. Calorimetric studies of ligand binding in R67 dihydrofolate reductase. Biochemistry. 37:12420–12433. [DOI] [PubMed] [Google Scholar]
- 25.Gao, X., K. Bain, J. B. Bonanno, M. Buchanan, D. Henderson, D. Lorimer, C. Marsh, J. A. Reynes, J. M. Sauder, K. Schwinn, C. Thai, and S. K. Burley. 2005. High-throughput limited proteolysis/mass spectrometry for protein domain elucidation. J. Struct. Funct. Genomics. 2–3:129–134. [DOI] [PubMed] [Google Scholar]
- 26.Kathir, K. M., T. K. S. Kumar, D. Rajalingam, and C. Yu. 2005. Time-dependent changes in the denatured state(s) influence the folding mechanism of an all β-sheet protein. J. Biol. Chem. 280:29682–29688. [DOI] [PubMed] [Google Scholar]
- 27.Hajuduk, P. J., R. P. Meadows, and S. W. Fesik. 1999. NMR-based screening in drug discovery. Q. Rev. Biophys. 32:211–240. [DOI] [PubMed] [Google Scholar]
- 28.Sivaraja, V., T. K. S. Kumar, and C. Yu. 2005. Resonance assignments for mouse S100A13. J. Biomol. NMR. 3:257. [DOI] [PubMed] [Google Scholar]
- 29.Bertini, I., C. Luchinat, G. Parigi, and R. Pierattelli. 2005. NMR spectroscopy of Paramagnetic metlloproteins. ChemBioChem. 6:1536–1549. [DOI] [PubMed] [Google Scholar]
- 30.Pintacuda, G., and G. Otting. 2002. Identification of protein surfaces by NMR measurements with a paramagnetic Gd(III) chelate. J. Am. Chem. Soc. 3:372–373. [DOI] [PubMed] [Google Scholar]
- 31.Ye, Y., H. W. Lee, W. Yang, S. Shealy, and J. J. Yang. 2005. Probing site-specific calmodulin calcium and lanthanide affinity by grafting. J. Am. Chem. Soc. 11:3743–3750. [DOI] [PubMed] [Google Scholar]
- 32.Jobby, M. K., and Y. Sharma. 2005. Calcium-binding crystallins from Yersinia pestis. Characterization of two single betagamma-crystallin domains of a putative exported protein. J. Biol. Chem. 280:1209–1216. [DOI] [PubMed] [Google Scholar]
- 33.Spinozzi, F., S. Gatto, V. DeFilippis, F. Carsughi, P. DiMuro, and F. Beltramini. 2005. Contribution of the copper ions in the dinuclear active site to the stability of Carcinus aestuarii hemocyanin. Arch. Biochem. Biophys. 439:42–52. [DOI] [PubMed] [Google Scholar]
- 34.Luk, E., L. T. Jensen, and V. C. Culotta. 2003. The many highways for intracellular trafficking of metals. J. Biol. Inorg. Chem. 8:803–809. [DOI] [PubMed] [Google Scholar]
- 35.Pufahl, R. A., C. Singer, K. L. Peariso, S. J. Lin, P. Schimdt, C. Fahrni, V. C. Cullotta, J. E. Penner-Hahn, and T. V. O'Halloran. 1997. Metal ion chaperone function of the soluble Cu(I) receptor Atx1. Science. 278:853–856. [DOI] [PubMed] [Google Scholar]
- 36.Finney, L. A., and T. V. O'Halloran. 2003. Transition metal speciation in the cell: insights from the chemistry of metal ion receptors. Science. 300:931–936. [DOI] [PubMed] [Google Scholar]
- 37.Gaggelli, E., F. Bernardi, E. Molteni, R. Pogni, D. Valensin, G. Valensin, M. Remelli, M. Luczkowski, and H. Kozlowski. 2005. Interaction of the human prion PrP(106–126) sequence with copper(II), manganese(II), and zinc(II): NMR and EPR studies. J. Am. Chem. Soc. 3:996–1006. [DOI] [PubMed] [Google Scholar]
- 38.Rajalingam, D., T. K. S. Kumar, and C. Yu. 2005. C2A domain of synaptotagmin exhibits high binding affinity for copper. Implications in the formation of the multiprotein FGF release complex. Biochemistry. 44:14431–14442. [DOI] [PubMed] [Google Scholar]