Abstract
Horse liver alcohol dehydrogenase is a homodimer, the protomer having a coenzyme-binding domain and a catalytic domain. Using all available x-ray structures and 50 ns of molecular dynamics simulations, we investigated the mechanism of NAD+-induced domain closure. When the well-known loop at the domain interface was modeled to its conformation in the closed structure, the NAD+-induced domain closure from the open structure could be simulated with remarkable accuracy. Native interactions in the closed structure between Arg369, Arg47, His51, Ala317, Phe319, and NAD+ were seen to form at different stages during domain closure. Removal of the Arg369 side-chain charge resulted in the loss of the tendency to close, verifying that specific interactions do help drive the domains closed. Further simulations and a careful analysis of x-ray structures suggest that the loop prevents domain closure in the absence of NAD+, and a cooperative mechanism operates between the subunits for domain closure. This cooperative mechanism explains the role of the loop as a block to closure because in the absence of NAD+ it would prevent the occurrence of an unliganded closed subunit when the other subunit closes on NAD+. Simulations that started with one subunit open and one closed supported this.
INTRODUCTION
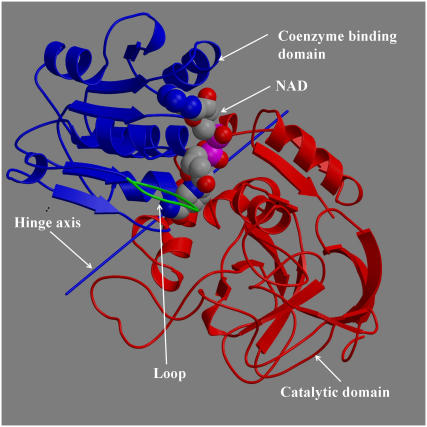
Horse liver alcohol dehydrogenase (LADH) catalyzes the oxidation of alcohol to aldehyde. It is a homodimer with the protomer comprising a C-terminal coenyzme-binding domain and an N-terminal catalytic domain with the active site located at the interdomain cleft (1). The binding of the coenzyme NAD+ in the interdomain cleft induces domain closure whereby the domains rotate ∼10° relative to each other in a classic example of a hinge-bending movement (2,3). This creates the productive binding site for the alcohol substrate (3). The subsequent binding of the alcohol is not thought to cause any appreciable effect on the domain conformation (3). A flexible loop (2,4), situated in the coenzyme-binding domain, contacts the catalytic domain in both open and closed domain conformations. The loop shows a dramatic conformational difference between the open- and closed-domain structures, apparently as a consequence of domain closure. An analysis of LADH x-ray structures, based on a sequential model of binding and domain closure, suggests that to a reasonable approximation, NAD first binds to the coenzyme-binding domain and then induces closure through interactions with specific residues in the catalytic domain (5). This sequential model is supported by kinetics experiments on a human β3β3 LADH, where the association of NADH was observed to occur in two steps, an initial recognition process followed by the NADH-induced isomerization (6). Fig. 1 shows the main features of the LADH protomer.
FIGURE 1.
View of LADH in open conformation.
Most computational studies on the dynamic behavior of LADH have focused on the effect of fluctuations on the reaction itself and have used closed structures of the enzyme (7–9). No molecular dynamics (MD) simulation study has been reported to date on the process of NAD-induced domain closure despite the importance this stage has in the overall reaction process. Here we report the results of free MD simulations of NAD+-induced domain closure. Unlike most sampling-based MD simulations, free MD simulations have the advantage of not presuming the paths for functional transitions. They are not as computationally demanding as sampling simulations (e.g., umbrella sampling) and can therefore be carried out on large systems, such as a fully solvated LADH molecule as we have done here. Unlike sampling simulations, free MD simulations cannot usually give quantitative results for the potential of mean force, but they can sometimes indicate the presence of barriers, and unlike sampling simulations, they can give information on dynamic processes. Although the binding of NAD+ itself to LADH is expected to be beyond the timescales accessible to MD simulation, based on previous simulations of domain proteins (10,11), it is reasonable to expect that the domain closure process itself, induced by NAD+ already bound to the coenzyme-binding domain of the open structure, would be accessible to MD simulation timescales. Based on the sequential model of NAD+ binding and domain closure, most of our simulations started with NAD+ bound to the coenzyme-binding domain with the subunit in the open-domain conformation (as seen in Fig. 1). In some of these starting structures, the loop was modeled to its conformation in the closed structure to assess its effect on the domain closure process.
MATERIAL AND METHODS
X-ray structures
The open structure (protein data bank (PDB) accession code: 1ADG (12)) used has both subunits bound to β-methylene-selenazole-4-carboxyamide adenine dinucleotide (SAD), which was not included in the simulations. This particular open structure was selected because a good proportion of its coenzyme-binding domain interactions with SAD are identical with those that the closed structures have with NAD. This helped in the modeling of NAD into the open structure. The closed structure (PDB accession code: 2OHX (13)) used has both subunits bound to NAD and dimethyl sulfoxide (DMSO), which was not included in the simulations.
Analysis of domain movements
The open and closed structures were analyzed for their domain movements using the program, DynDom (14,15). Two main parameters can be varied in this program: the window length (default value 5 residues) and the minimum domain size (default value 20 residues). With these default values, the domains were defined as residues 1–175 and 318–374 for the catalytic domain and residues 176–290 and 302–317 for the coenzyme-binding domain. The rigid body projection value (11) was used to monitor the extent of closure during the simulations. It measures how far the domain conformation has progressed from the open (a value of 0) to the closed x-ray domain conformation (a value of 1).
Principal component analysis of the trajectories was performed as described previously, where fluctuations are measured not from the average structure but from the starting structure (16,17).
Preparation of starting structures for simulations
The open and closed x-ray structures were used to create the starting conformations. The open starting structures with NAD+ bound to the coenzyme-binding domain were created by superposition as described elsewhere (5). The loop (residues 290–302) in the open-domain structures was modeled to the conformation found in the closed-domain structures by superposition of the coenzyme-binding domains. Likewise, the “mixed state” starting structures (one open subunit, the other closed) were created by superposing the binding domain of one subunit of the closed structure onto the binding domain of one subunit of the open structure to create an open subunit and a closed subunit. All simulations were performed on the dimeric molecule.
MD simulations
All simulations were performed using AMBER 7.0 (18). The protein, prepared as described above, was placed in a rectangular parallelepiped box and was fully solvated with water molecules from a snapshot of TIP3P water (19) equilibrated at room temperature. Whenever possible, crystallographic water molecules were retained. Parameters for NAD+ were taken from previous studies (20,21). A simple point charge model for the Zn2+ ions (22) was used. The Zn2+ ions were liganded by charged cysteines. Histidines were protonated at either the Nδ or Nɛ according to the biochemical evidence whenever available (e.g., His51 and His67, which bond to NAD in the closed structure, had their Nδ protonated, but all remaining histidines were protonated at the Nɛ). Neutrality of the system was maintained by adding chloride counterions.
System preparation involved 200 steps of energy minimization, the first 100 using steepest descent, the last 100 using conjugate gradient. During minimization, nonterminal protein and NAD+ atoms were restrained using a harmonic potential with a force constant of 10 kcal/mol-Å2.
In the MD simulations, periodic boundary conditions were applied, and nonbonded interactions were calculated by the particle mesh Ewald method. The integration time step was 2 fs, and the SHAKE algorithm (23) was used to constrain bonds involving hydrogen atoms. Temperature and pressure were controlled using the weak-coupling method (24).
To prepare for production, after minimization, position restraint MD was performed. Position restraint was applied to nonterminal protein and NAD+ atoms. With a force constant of 1.0 kcal/mol-Å2, 10 ps of simulation was performed at constant volume and at a temperature of 100 K, followed by 10 ps at constant volume at 300 K, followed by 80 ps at constant pressure at 300 K. Finally, 40 ps was performed at constant pressure at 300 K with a lower force constant of 0.1 kcal/mol-Å2. A relaxation time constant of 0.02 ps was used for temperature and pressure coupling. Productive simulation was performed at a temperature of 300 K for 10 ns, using a relaxation time constant of 0.2 ps for the temperature and pressure coupling.
Position restraint was applied to subunit A throughout the mixed-state simulations to aid in maintaining the closed conformation.
In all, we performed 50 ns of simulation on a system comprising ∼70,000 atoms.
RESULTS
Simulations from open domain conformation
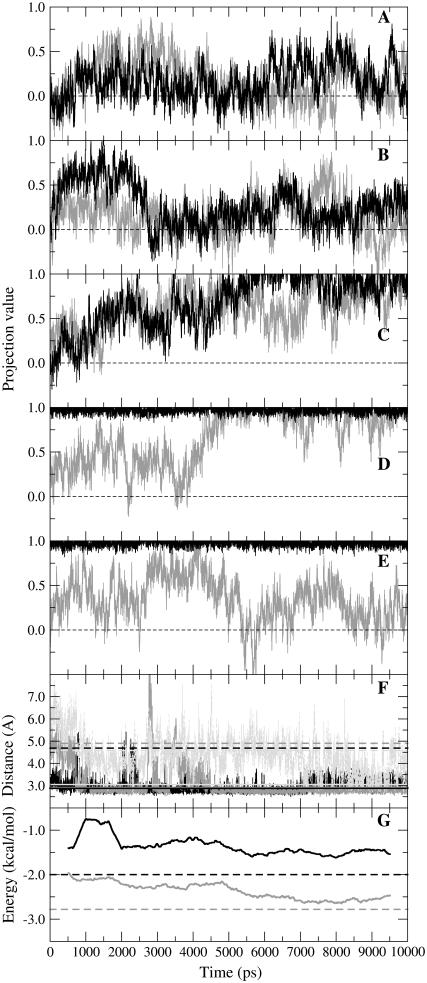
Fig. 2, A–C, shows the projection values for three simulations that start from an open conformation for both subunits. Without NAD+ and both subunits starting from the open structure, only a weak tendency to close is observed (Fig. 2 A). With NAD+ present in subunit A only, and the loop unmodeled in both subunits, an initial tendency to close is observed in subunit A, but full closure is never achieved (Fig. 2 B). However, with NAD+ in subunit A only, and the loop of each subunit modeled to its conformation in the closed structure, both subunits close after ∼7 ns (Fig. 2 C). This will be referred to as the “closing” trajectory. Fig. 2 F shows trajectories of distances between atoms of Arg47, His51, and Arg369 and NAD+ in the closing trajectory. These residues have been identified (apart from Arg47) as closure-inducing residues whose interactions with NAD+ help drive domain closure (5). The movement of these residues from the open to closed x-ray structure is indicated in Fig. 3 A. After 7 ns, these distances correspond almost perfectly with those in the closed x-ray structure. Fig. 2 G shows the trajectories of the backbone hydrogen bond energies between Ala317 and Phe319 and their hydrogen bond partners on the carboxamide group of NAD+. They start off weak and at 10 ns are closer to the energies calculated for the closed x-ray structure. A strengthening of these hydrogen bonds is observed at ∼5 ns, coinciding with increased closure of subunit A as seen in Fig. 2 C. The closing trajectory represents, therefore, a remarkably accurate simulation of the process of NAD+-driven domain closure.
FIGURE 2.
(A–E) Trajectories of rigid-body projection values: 0 is open; 1 is closed. Black is subunit A, and gray is subunit B. (A) No NAD+ present in either subunit, loops as in open x-ray structure. (B) NAD+ present in subunit A, loops as in open structure. (C) NAD+ in subunit A, loops as in closed structure. (D) Subunit A closed on NAD+, subunit B open, loops as in closed structure. (E) Subunit A closed on NAD+, subunit B open, loop as closed for subunit A, open for subunit B. (F) Distances between Arg369 nitrogen atom (NH1) and phosphate oxygen atom (OP1N) (black) (note that this distance has already decreased considerably during the position restraint procedure), Arg47 nitrogen atom (NH2) and phosphate oxygen (OP2A) (dark gray), and His51 nitrogen (NE2) and the nicotinamide ribose oxygen (O2′N) (light gray) from closing trajectory in (C). Broken lines are from open x-ray structure (between 4.5 Å and 5 Å), continuous from closed (∼3 Å). (G) Hydrogen bond energy trajectories for residues 317 (black) and 319 (gray), which form backbone hydrogen bonds with the carboxamide group of NAD+. Because of its fluctuating nature, the running average of the energy using windows 1 ns in length is shown. Broken lines give energies as found in closed x-ray structure. Energies calculated using program DSSP (36).
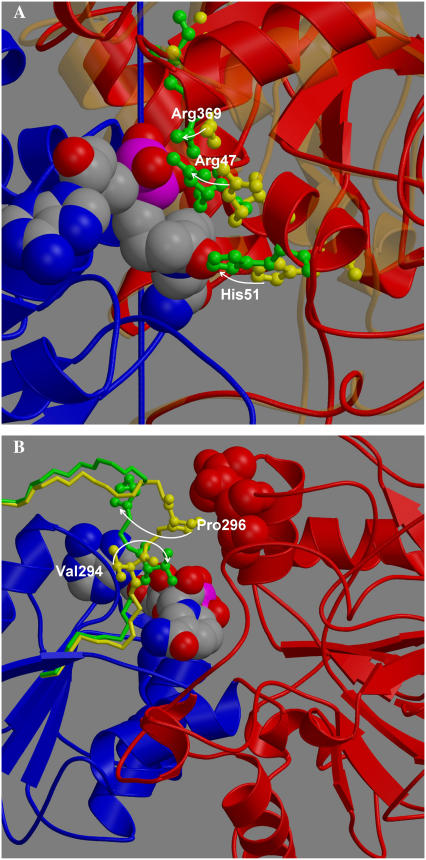
FIGURE 3.
Views on key regions in LADH. (A) The binding domain is in blue, and NAD+ in space-filling model. The movement of Arg47, His51, and Arg369 in going from the open (yellow) to closed x-ray structure (green) is indicated by arrows. The catalytic domain is in bold red (closed) and faint orange (open). (B) Loop in open (yellow) and closed (green) conformation. Rotation of Val294 (ball and stick model) to interact with NAD+ (space filling) by rotation about φ-angle of Gly293 would cause Pro296 (ball and stick model) to move away from contacts 56 and 57 (space filling, red) on catalytic domain, thereby allowing it to close.
Arg369-Diphosphate interaction drives domain closure in initial stages
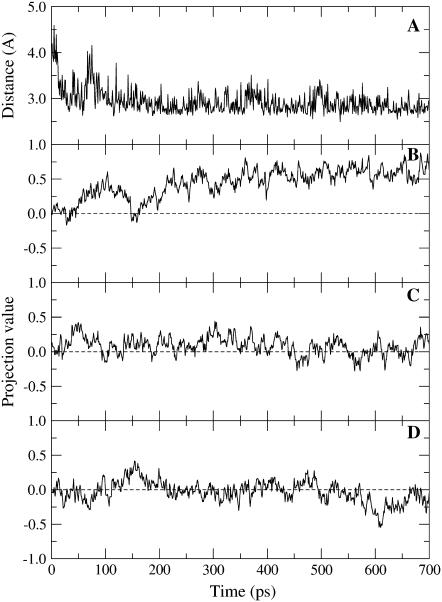
In the closing simulation, the interaction of Arg369 with the diphosphate of NAD+ appears to be particularly strong as it forms mainly during the position restraint procedure (see Fig. 2 F). Fig. 4 B shows the initial 800 ps of the projection trajectory for subunit A where NAD+ is present, but the loop is not modeled (black line in Fig. 2 B). As seen in Fig. 4 A, the Arg369-diphosphate salt bridge also forms very rapidly, and a strong initial tendency for the domains to close is observed. To verify this interaction's role in helping to drive domain closure, Arg369 was mutated to alanine at the point of release of position restraint. Fig. 4 C shows the projection trajectory for this mutant. It shows that the tendency to close is lost. To confirm this finding, another simulation was performed where the charge of the guanidinium group of Arg369 was set close to zero at the point of release of position restraint. Again the tendency to close is lost (see Fig. 4 D). This result supports our assertion that the Arg369-diphosphate interaction helps to drive domain closure in the initial stages. The important role that Arg369 plays in the domain closure process was first recognized in kinetics experiments on isoenzymes of human alcohol dehydrogenases (6) (see Discussion section).
FIGURE 4.
Results from three separate simulations, each with NAD+ in subunit A and loops left unmodeled . The simulations were of the wild-type, an Arg369Ala mutant, and a mutant with the charge of the guanidnium group of Arg369 set close to zero (Arg369nocharge). (A) Distance between the Arg369 nitrogen atom (NH1) and phosphate oxygen atom (OP1N) atom in wild-type. (B) Projection value for subunit A in wild-type (as shown in Fig. 2 B). (C) Projection value for subunit A of Arg369Ala mutant. (D) Projection value for subunit A of Arg369nocharge mutant. Mutations were made at point when position restraint was released for the wild-type. The tendency to close is lost in the mutants, indicating that the Arg369–diphosphate interaction is important in initiating domain closure.
Loop as block to domain closure
The loop comprising residues 290–300 has been referred to as “flexible” (4). At one end of this loop, residues 292–294 contact NAD+ in the closed structure, whereas at the other end, residues 296 and 297 contact residues 51, 56, and 57 on the catalytic domain in the open structure. If residues Pro296 and Asp297 do not move relative to the binding domain, then the domains cannot close. Residue 295 is also a proline. Two consecutive prolines will fix four consecutive main-chain dihedral angles, namely Val (ψ294) Pro (φ295), Pro (ψ295), and Pro(φ296). Analysis of the x-ray structures shows that these dihedral angles do not change appreciably between the open and closed structures and that this region moves as a rigid element. Val292, Gly293, and Val294 are the only residues from the loop that contact NAD+ in the closed structure, and it is noticeable that in going from the open structure to the closed, the side chain of Val294 rotates by 140° to contact the nicotinamide riboside. A DynDom analysis of the movement between the open and closed structures using a window length of three residues and minimum domain size of four residues yielded a “moving” domain comprising residues 293–296 and a “fixed” domain comprising all the other residues, with residues 292–293 and 296–300 assigned as bending. The moving domain rotates 129° relative to the fixed domain about a hinge axis approximately parallel to the region 292–297. This is facilitated primarily by a 105° rotation about the φ-dihedral axis of Gly293 and a 144° rotation about the ψ-dihedral axis of Pro296. Therefore, it is reasonable to regard the region between the φ-dihedral axis of Gly293 and the ψ-dihedral axis of Pro296 as a rigid element. The rotation of the Val294 side chain, which may be induced by the presence of NAD+, is therefore extended out to residue Pro296 through this rigid “arm”. The rigid arm has a characteristic crankshaft form, which means that the contacts between Pro296 and Asp297 and residues 51, 56, and 57 on the catalytic domain are removed in going from the open- to closed-loop conformation (see Fig. 3 B) thus removing the obstacle to closure. This suggests that these contacts exist to prevent domain closure in the absence of NAD+ and that the interaction of Val294 with the nicotinamide riboside helps to stabilize the loop in its closed conformation to allow closure. The results of our simulations presented in Fig. 2, A–C, support this interpretation, as only the simulation with the loop modeled as in the closed structure (Fig. 2 C) was able to close fully. In the simulation corresponding to Fig. 2 B, where the loop is left as in the open structure, the interactions with NAD+ that drive closure are present in subunit A, but the loop remains an obstruction to domain closure. This suggests that domain closure accompanied by the change in conformation of the loop is a much slower process than domain closure without the need for the loop to change conformation. So our hypothesis would be that without NAD, the loop conformation keeps the domains open, but, with NAD, it changes conformation to allow domain closure. This may occur either on binding of NAD to the coenzyme domain or subsequent to binding and concurrent with domain closure.
Domain and loop conformation of x-ray structures
In a study aimed at creating a comprehensive description of domain movements in the PDB (25), 73 LADH protomer structures were assigned to a single family based on sequence similarity. These structures are of horse and human liver alcohol dehydrogenases. Their domain conformations separate into two tight conformational clusters corresponding to 11 open and 62 closed protomers (see http://www.cmp.uea.ac.uk/dyndom/Subgroup.do?subgroupid=1560_m) (25). All open structures have the same open-loop conformation. Likewise all closed conformations have the same closed-loop conformation and are all bound to NAD or an analog that provides the same interactions as the nicotinamide group. Thus, the available structural data are consistent with the hypothesis stated above.
Intersubunit cooperative domain closure
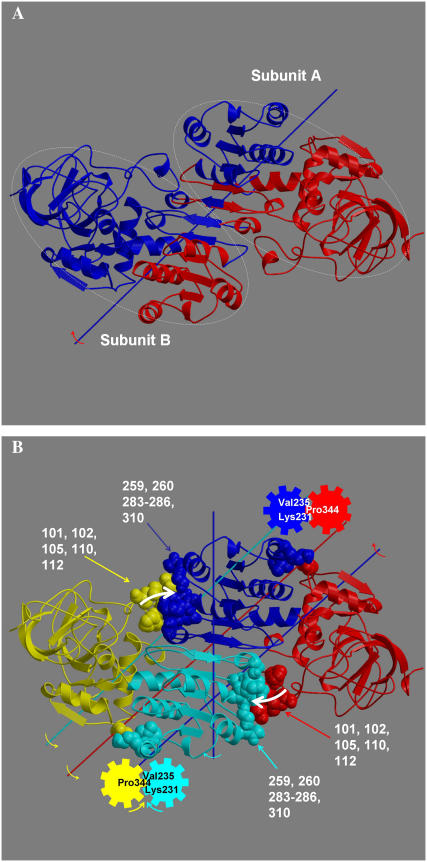
The trajectories in Fig. 2, A–C, hint at cooperative domain closure between subunits. The zero time-lag correlation between subunit A and subunit B projection values for the closing trajectory of Fig. 2 C is 0.38. During the first half of this trajectory when most of the closure occurs, the value is 0.46. To investigate this further, an analysis of the domain movement between the open and closed x-ray structures was undertaken using the DynDom program (14, 15). This analysis was performed on the whole protein, not just the individual subunits (by removing the chain terminators in the PDB files). Fig. 5 A shows a DynDom result of the open and closed x-ray structures. It shows that as the catalytic domain of subunit A closes onto the binding domain of subunit A, the binding domain of subunit B moves with it (as they are assigned as one dynamic domain), closing onto the catalytic domain of subunit B. This suggests that cooperativity acts through contacts between the catalytic domain of one subunit and the binding domain of the other subunit. Fig. 5 B shows a finer-grained analysis. It shows that as the catalytic domain closes, there is a relative twist of the binding domains (2). In closing, the catalytic domain of subunit A pushes on the binding domain of subunit B, causing this twist. This twisting causes residues on the opposite sides of the twist axis to move in the opposite direction. In particular, residues Lys231 and Val235 in the i and i + 4 positions of an α-helix form a cleft into which Pro344 on the catalytic domain of subunit B is wedged. The movement of the helix will move Pro344, causing the catalytic domain of subunit B to rotate in a counterdirection to its binding domain. The overall effect is the closing of subunit B, which in turn will enhance the closing of subunit A through the same mechanism. Although there are many contacting regions between the coenzyme-binding and catalytic domains, focus is drawn to this region as the hinge axis for the relative movement of the catalytic domains (see Fig. 5 B) passes directly between Pro344, Lys231, and Val235. The mechanical equivalent of this is the tooth of one gear between two teeth of another. A rotation of one gear will cause a counterrotation of the other. For a small rotation, fixing one gear will give an axis for rotation of the other that passes between these teeth. Further support for this overall mechanism of cooperativity comes from a principal component analysis of the binding domains (using residues 178–290 and 302–314 from both subunits) from the closing trajectory.
FIGURE 5.
Cooperativity in LADH. (A) DynDom (14) result with a 21-residue window and 250-residue minimum domain size. It shows coenzyme-binding domain of subunit A and catalytic domain of subunit B combined as a single dynamic domain colored blue, and vice-versa to give a dynamic domain colored red. Subunits indicated by ellipses. (B) DynDom result showing relative twist of coenzyme-binding domains. White arrows indicate catalytic domains of one subunit pushing on coenzyme-binding domain of the other subunit. Colored arrows indicate the rotation of the domain of the same color relative to the domain with the color of the shaft that represents that axis of rotation. Coenzyme-binding domains were analyzed separately to give the twist axis. Default DynDom parameters were used.
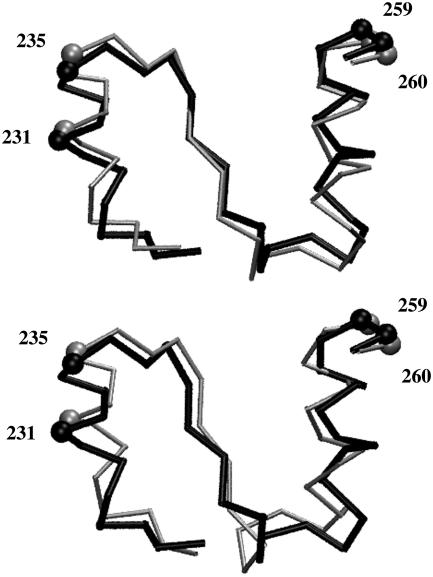
Fluctuation along the first principal component accounts for 61.7% of the total fluctuation. The movement depicted by the first principal component is more complex than the movement between the x-ray structures, which reveals a relative twist of the coenzyme-binding domains as seen in Fig. 5 B. The inner product of the first principal component with the unit vector for the movement between the x-ray structures is 0.42. Despite the differences, there is an indication of a similar mechanism. Most of the movement in the first principal component is located around the αβα motif (residues 224–261) at the base of the Rossman fold. This region links residues 259 and 260, which contact the catalytic domain of the other subunit, with the gearing residues 231 and 235. As can be seen in Fig. 6, in subunit B, the movement in this region in the first principal component is similar to that between the x-ray structures, especially at these key residues. Furthermore, the movement at residues 231 and 235 is consistent with the proposed gearing mechanism.
FIGURE 6.
Results of a principal component analysis (16,17) of the coenzyme-binding domains from the closing trajectory in comparison to x-ray structures. Shown is the αβα motif (residues 224–261) at the base of the Rossman fold, which showed the largest movement in both subunits in the first principal component. At the top is shown the movement from the open x-ray structure in black to the closed x-ray structure in gray (in the frame of reference for which there is no overall external movement of both coenzyme-binding domains). At the bottom is the movement of this region in subunit B from the open x-ray structure in black to the structure corresponding to the maximal projection of the trajectory along the first principal component, in gray. The balls indicate residues 231, 235, 259, and 260, which are implicated in the cooperative mechanism (see Fig. 5 B). The movement of 231 and 235 in going from open to closed is consistent with the proposed gearing mechanism, as this helix would need to move inward during closure in Fig. 5 B, which roughly corresponds to upward in this figure. The movement between the x-ray structures has been exaggerated by a factor of two. Figure created using VMD (37).
Mixed state simulations demonstrate closed subunit drives closure in other subunit
Without the blocking loop, cooperative forces should cause a subunit to close in the absence of NAD+ when the other subunit is closed on NAD+. This would create an unproductive closed subunit and suggests that the purpose of the loop is to prevent this. To test this, two mixed-state simulations were performed, each with subunit A closed on NAD+ and subunit B open without NAD+. For one simulation the loop of subunit B was modeled as in the closed structure, and for the other it was left as in the open form. The projection trajectories of Fig. 2, D and E, show, after some delay, the rapid and persistent full closure of subunit B for the closed-loop structure but a much more open conformation for the open-loop structure. Therefore, this result appears to confirm our conjecture that cooperative forces from one fully closed subunit are able to close the other open subunit but are unable to do so if the loop is in its open conformation.
DISCUSSION
The study combines an analysis of existing structural data with MD simulations. The emerging model has NAD+ bind first to the coenzyme-binding domain before domain closure is induced. Evidence for this is not direct but circumstantial. Kinetics experiments on human β3β3 LADH, which has a cysteine at position 369 instead of an arginine, do support a two-step process in the interaction of LADH with NADH (6). The first process is thought to be the recognition of NADH by LADH and is dependent on the concentration of NADH, whereas the second step is independent of the concentration of NADH, which is consistent with it inducing domain closure from the bound state. A careful analysis that considered the two alternatives of NAD binding first to the coenzyme-binding domain and inducing closure by interacting with the catalytic domain, or the NAD binding first to the catalytic domain and inducing domain closure by interacting with the coenzyme-binding domain, demonstrated that a much more plausible mechanism operates for the former (5). In addition, all open-domain structures with NAD, NAD analogs, or inhibitors have the ligand bound to the coenzyme-binding domain. Thus, our starting point for the MD simulations was with NAD+ bound to the coenzyme-binding domain in the open-domain conformation.
Key interactions that drive domain closure operate at different stages of the domain closure process
Following key interactions between residues on the catalytic domain and NAD+ should allow us to judge their contribution to the domain closure process. The first to form is the Arg369-diphosphate interaction, followed by the Arg47-diphosphate interaction, followed by the hydrogen bonds with Ala317 and Phe319, followed by the His51-ribose interaction much later in the process. This would suggest that the Arg369 interaction is primary in driving domain closure in the initial stages. Our simulations of Arg369 mutants supported this. The strengthening of the hydrogen bonds with Ala317 and Phe319 happens after partial closure occurs and coincides with a further closing. The His51 interaction with the ribose does not appear to contribute greatly in the initial stages of domain closure but forms a tight bond only once the domains are closed. Thus, it appears that many of the key interactions come into play at different stages during domain closure in a sort of relay of interactions. These results provide further evidence that specific interactions help drive domains closed (5) and do not support the general diffusive model proposed for domain closure in proteins (26).
Loop as NAD-sensitive switch that blocks domain closure
The results indicate that the interaction of NAD+ with Val294 stabilizes the loop in its closed conformation. This interaction could occur on binding of NAD+ to the coenzyme-binding domain before domain closure occurs or could occur concurrently with domain closure. With NAD+ present but the loop left unmodeled, domain closure did not occur within the simulation time, as the loop remained a block to closure. This indicates that loop rearrangement could be a much slower process than domain closure. In kinetics experiments on the human isoenzyme, β3β3 LADH, with Cys369 rather than Arg369, the rate of NADH-induced isomerization (a step independent of NADH concentration) was 42 s−1, whereas for the β1β1 isoenzyme with Arg369, it was found to be at least 1200 s−1 (the limit of instrument detection) (6). However, in our simulations, with the loop modeled to its closed conformation, closing occurs at a rate of the order of 108 s−1. As the change of a cysteine to arginine and NADH to NAD+ would not seem to explain a 42 s−1 to 108 s−1 difference if the isomerization were simply domain closure alone, it is likely that the isomerization process measured in these experiments also involves loop rearrangement and that Arg369 is involved in this, possibly as an indirect consequence of its role in helping to drive the domains closed; i.e., as the domains strain to close, residues 51, 56, and 57 on the catalytic domain push on the loop, so helping to change its conformation. It may also be that the difference between the extraordinarily rapid closure found here and the rates derived from kinetics experiments occurs because the NAD+ was already optimally placed in its binding position on the coenzyme-binding domain, whereas the experimentally determined rates may also include initial binding events, which are expected to be much slower.
The structural data also support the hypothesis that the loop acts as an NAD-sensitive switch for domain closure. The role of the interaction between Val294 and NAD in stabilizing the closed conformation of the loop seems crucial. In its absence the loop probably remains a block to closure. The rotation of Val294 is facilitated by a hinge at Gly293 and extended out to the blocking residues via a rigid ProPro motif at residues 295 and 296. This is strongly suggestive of a mechanism that relates closure of the domains to the Val294–NAD interaction. A number of x-ray structures suggest that the formation of this interaction is indeed necessary for domain closure. Two mutants (Val292Ser (27) and Gly293Ala/Pro295Thr double mutant (4)) have been solved in the presence of NAD+. Both have an open-domain conformation, and in both, the nicotinamide riboside was not resolved, although the ADP portion is seen bound to the coenzyme-binding domain in the same way as in the closed structures. Both have loop structures of the open-domain conformation. As Val292, Gly293, and Val294 are the only residues from the loop that contact NAD in the closed domain, it would seem that the interaction of these residues with the nicotinamide riboside stabilizes both the nicotinamide riboside and the closed conformation of the loop. It is likely therefore that theVal292Ser mutant disrupts this stabilizing interaction, leaving the loop in its open conformation. Thus, the loop remains a block to closure, and the domains are kept open. The Gly293Ala/Pro295Thr double mutant is particularly interesting, as these mutations undermine the roles of two important residues in the proposed switch mechanism, the former providing the required flexibility, the latter the required rigidity. In the case of the former, the 105° rotation about the φ-dihedral axis of Gly293 that facilitates the interaction of Val294 with the nicotinamide riboside must be less favorable in an alanine. If so, the loop may be unable to adopt its closed conformation. This would leave the loop as a block to domain closure, and so the domains remain open. In the case of the Pro295Thr mutation, the lack of rigidity would prevent the propagation of the change at Val294 out to the blocking residues 296 and 297. These would remain a block to closure keeping the domains open. The simulations suggest that the interactions between Ala317 and Phe319 and the carboxamide of NAD+ aid in driving domain closure in the later stages. Given that these interactions should require a stable nicotinamide riboside, the loop should therefore also be stabilized in its closed conformation. This is logical, as the domains would already need to have closed partially for this interaction to have an effect.
A few structures exist that have NAD analogs or inhibitors bound and yet have open-domain conformations. The NAD analog 5-β-d-ribofuranosylpicolinamide adenine dinucleotide (CPAD) is known to induce domain closure as supported by a wild-type closed-domain structure bound to CPAD (28). However, a Phe93Trp/Val203Ala double mutant bound to CPAD has an open-domain conformation (29). The explanation is that the pyridine ring (analogous to the nicotinamide ring) has rotated away from the loop to fill space made available by the mutated residues (29). Again the crucial interaction of Val294 with the ligand is unable to form, and the loop remains in its open conformation blocking domain closure. Two structures bound to inhibitors, SAD and thiazole-4-carboxamide adenine-dinucleotide (TAD), also have open structures (12). If Val294 were to rotate to its position that switches the loop to its closed conformation, it would have severe steric overlap with these inhibitors. This may indicate that the Val294 interaction with NAD+ has a role in the specificity of the enzyme–coenzyme interaction. It explains why ATP cannot induce domain closure. In short, all available structures do support our hypothesis that the loop acts as a block to closure, and it is primarily the interaction of Val294 with the nicotinamide riboside that stabilizes it in the closed conformation.
Cooperative domain closure
Our simulation results suggest intersubunit cooperativity in the domain closure process. A careful analysis of the x-ray structures suggests a plausible mechanism. The twist of the coenzyme-binding domains was noted in an early study (2) but was not attributed to cooperativity, although it was noted that it would cause NAD to be buried deeper within the domains. Kinetics experiments on LADH have not presented any clear evidence for cooperative behavior. In the 1970s kinetics experiments using aromatic substrates were interpreted in terms of an asymmetric model whereby the products were required to dissociate in the first subunit to react before the other was able to react (30,31). However, evidence for this anticooperativity between subunits came to be disputed (32), and the simpler model involving functionally independent subunits was established.
Summary of mechanism
The emerging picture has NAD+ bind to the coenzyme-binding domain of one subunit to release the blocking loop for domain closure. Interactions between NAD+ and specific residues on the catalytic domain drive closure. Cooperative forces act to close the other subunit, but it remains open because of the blocking loop. When NAD+ binds to the open subunit, it releases the loop, and domain closure occurs due to forces from NAD+ directly and those from cooperativity.
With all the evidence from the simulations and the existing x-ray structures taken together, a convincing case for the mechanism described above has been presented. The case for this overall mechanism is strengthened by the mutual dependency of the three submechanisms involved. The first submechanism is the binding of NAD+ to the coenzyme-binding domain and its influence on the loop conformation. The second is the domain closure process, which appears to be driven by specific interactions and would result in a release of free energy. The third is the cooperative domain closure, which is dependent on the second, as it must harness the released energy to drive the other subunit closed. The first is a result of the third, as it is required to prevent closure of a subunit in the absence of NAD+. Individual residues crucial to the operation of all three of these submechanisms have been identified, which will allow them to be investigated further using experimental techniques.
The overall mechanism may relate to product release on formation of the aldehyde and NADH. It is feasible that energy stored in the twisted coenzyme-binding domains is used to help drive the domains open.
This result taken together with related results for the domain enzyme citrate synthase (11,33) and maltose-binding protein (34,35) also confirms that some domain proteins have mechanisms that keep their domains open so that their binding sites remain accessible to the functional ligand.
Acknowledgments
We thank Nobuhiro Gō, Hisashi Ishida, Mariko Higuchi, Yasumasa Joti, and Guoying Qi for their contributions.
This work was support by the Japan Ministry of Education, Culture, Sports, Science and Technology; Japan Atomic Energy Research Institute; Wellcome Trust (Joint Infrastructure Fund); and the Biotechnology and Biological Sciences Research Council.
References
- 1.Eklund, H., J. P. Samama, L. Wallen, C. I. Branden, A. Akeson, and T. A. Jones. 1981. Structure of a triclinic ternary complex of horse liver alcohol-dehydrogenase at 2.9 a resolution. J. Mol. Biol. 146:561–587. [DOI] [PubMed] [Google Scholar]
- 2.Eklund, H., J. P. Samama, and T. A. Jones. 1984. Crystallographic investigations of nicotinamide adenine-dinucleotide binding to horse liver alcohol-dehydrogenase. Biochemistry. 23:5982–5996. [DOI] [PubMed] [Google Scholar]
- 3.Colonna-Cesari, F., D. Perahia, M. Karplus, H. Eklund, C. I. Branden, and O. Tapia. 1986. Interdomain motion in liver alcohol dehydrogenase: Structural and energetic analysis of the hinge bending mode. J. Biol. Chem. 261:15273–15280. [PubMed] [Google Scholar]
- 4.Ramaswamy, S., D. H. Park, and B. V. Plapp. 1999. Substitutions in a flexible loop of horse liver alcohol dehydrogenase hinder the conformational change and unmask hydrogen transfer. Biochemistry. 38:13951–13959. [DOI] [PubMed] [Google Scholar]
- 5.Hayward, S. 2004. Identification of specific interactions that drive ligand-induced closure in five enzymes with classic domain movements. J. Mol. Biol. 339:1001–1021. [DOI] [PubMed] [Google Scholar]
- 6.Stone C. L., M. B. Jipping, K. Owusu-Dekyi, T. D. Hurley, T. K. Li, and W. F. Bosron. 1999. The pH-dependent binding of NADH and subsequent enzyme isomerization of human liver beta(3)beta(3) alcohol dehydrogenase. Biochemistry. 38:5829–5835. [DOI] [PubMed] [Google Scholar]
- 7.Luo, J., and T. C. Bruice. 2004. Anticorrelated motions as a driving force in enzyme catalysis: The dehydrogenase reaction. Proc. Natl. Acad. Sci. U S A. 101:13152–13156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo, J., and T. C. Bruice. 2002. Ten-nanosecond molecular dynamics simulation of the motions of the horse liver alcohol dehydrogenase.PhCH2O- complex. Proc. Natl. Acad. Sci. U S A. 99:16597–16600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo, J., and T. C. Bruice. 2001. Dynamic structures of horse liver alcohol dehydrogenase (HLADH): Results of molecular dynamics simulations of HLADH-NAD(+)-PhCH2OH, HLADH-NAD(+)-PhCH2O-, and HLADH-NADH-PhCHO. J. Am. Chem. Soc. 123:11952–11959. [DOI] [PubMed] [Google Scholar]
- 10.de Groot, B. L., S. Hayward, D. M. F. van Aalten, A. Amadei, and H. J. C. Berendsen. 1998. Domain motions in bacteriophage T4 lysozyme: A comparison between molecular dynamics and crystallographic data. Proteins. 31:116–127. [DOI] [PubMed] [Google Scholar]
- 11.Roccatano, D., A. E. Mark, and S. Hayward. 2001. Investigation of the mechanism of domain closure in citrate synthase by molecular dynamics simulation. J. Mol. Biol. 310:1039–1053. [DOI] [PubMed] [Google Scholar]
- 12.Li, H., W. H. Hallows, J. S. Punzi, V. E. Marquez, H. L. Carrell, K. W. Pankiewicz, K. W. Watanabe, and B. M. Goldstein. 1994. Crystallographic studies of 2 alcohol dehydrogenase-bound analogs of thiazole-4-carboxamide adenine-dinucleotide (tad), the active anabolite of the antitumor agent tiazofurin. Biochemistry. 33:23–32. [DOI] [PubMed] [Google Scholar]
- 13.Al-Karadaghi, S., E. S. Cedergren-Zeppezauer, S. Hovmoller, K. Petratos, H. Terry, and K. S. Wilson. 1994. Refined crystal-structure of liver alcohol-dehydrogenase NADH complex at 1.8-Angstrom resolution. Acta Crystallogr. D. Biol. Crystallogr. 50:793–807. [DOI] [PubMed] [Google Scholar]
- 14.Hayward, S., and H. J. C. Berendsen. 1998. Systematic analysis of domain motions in proteins from conformational change: New results on citrate synthase and T4 lysozyme. Proteins. 30:144–154. [PubMed] [Google Scholar]
- 15.Hayward, S., and R. A. Lee. 2002. Improvements in the analysis of domain motions in proteins from conformational change: DynDom version 1.50. J. Mol. Graph. Model. 21:181–183. [DOI] [PubMed] [Google Scholar]
- 16.Kitao, A., F. Hirata, and N. Go. 1991. The effects of solvent on the conformation and the collective motions of protein: normal mode analysis and molecular dynamics simulation of melittin in water and in vacuum. Chem. Phys. 158:447–472. [Google Scholar]
- 17.Amadei, A., A. B. M. Linssen, and H. J. C. Berendsen. 1993. Essential dynamics of proteins. Proteins. 17:412–425. [DOI] [PubMed] [Google Scholar]
- 18.Pearlman, D. A., D. A. Case, J. W. Caldwell, W. S. Ross, T. E. Cheatham, S. Debolt, D. Ferguson, G. Seibel, and P. Kollman. 1995. Amber, a package of computer-programs for applying molecular mechanics, normal-mode analysis, molecular-dynamics and free- energy calculations to simulate the structural and energetic properties of molecules. Comput. Phys. Commun. 91:1–41. [Google Scholar]
- 19.Jorgensen, W. L., J. Chandrasekhar, J. D. Madura, R. W. Impey, and M. L. Klein. 1983. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 79:926–935. [Google Scholar]
- 20.Cummins, P. L., K. Ramnarayan, U. C. Singh, and J. E. Gready. 1991. Molecular-Dynamics Free-Energy Perturbation Study on the Relative Affinities of the Binding of Reduced and Oxidized Nadp to Dihydrofolate-Reductase. J. Am. Chem. Soc. 113:8247–8256. [Google Scholar]
- 21.Pavelites, J. J., J. L. Gao, P. A. Bash, and A. D. Mackerell. 1997. A molecular mechanics force field for NAD(+), NADH, and the pyrophosphate groups of nucleotides. J. Comput. Chem. 18:221–239. [Google Scholar]
- 22.Hoops, S. C., K. W. Anderson, and K. M. Merz. 1991. Force-Field Design for Metalloproteins. J. Am. Chem. Soc. 113:8262–8270. [Google Scholar]
- 23.Ryckaert, J. P., G. Ciccotti, and H. J. C. Berendsen. 1977. Numerical-Integration of Cartesian Equations of Motion of a System with Constraints - Molecular-Dynamics of N-Alkanes. J. Comput. Phys. 23:327–341. [Google Scholar]
- 24.Berendsen, H. J. C., J. P. M. Postma, W. F. v. Gunsteren, and A. D. Nola. 1984. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81:3684–3690. [Google Scholar]
- 25.Qi, G., R. A. Lee, and S. Hayward. 2005. A comprehensive and non-redundant database of protein domain movements. Bioinformatics. 21:2832–2838. [DOI] [PubMed] [Google Scholar]
- 26.Gerstein, M., A. M. Lesk, and C. Chothia. 1994. Structural mechanisms for domain movements in proteins. Biochemistry. 33:6739–6749. [DOI] [PubMed] [Google Scholar]
- 27.Rubach, J. K., S. Ramaswamy, and B. V. Plapp. 2001. Contributions of Valine-292 in the nicotinamide binding site of liver alcohol dehydrogenase and dynamics to catalysis. Biochemistry. 40:12686–12694. [DOI] [PubMed] [Google Scholar]
- 28.Li, H., W. H. Hallows, J. S. Punzi, K. W. Pankiewicz, K. A. Watanabe, and B. M. Goldstein. 1994. Crystallographic studies of isosteric NAD analogs bound to alcohol-dehydrogenase - specificity and substrate-binding in 2 ternary complexes. Biochemistry. 33:11734–11744. [DOI] [PubMed] [Google Scholar]
- 29.Colby, T. D., B. J. Bahnson, J. K. Chin, J. P. Klinman, and B. M. Goldstein. 1998. Active site modifications in a double mutant of liver alcohol dehydrogenase: Structural studies of two enzyme-ligand complexes. Biochemistry. 37:9295–9304. [DOI] [PubMed] [Google Scholar]
- 30.Bernhard, S. A., M. F. Dunn, P. L. Luisi, and P. Schack. 1970. Mechanistic studies on equine liver alcohol dehydrogenase.1. stoichiometry relationship of coenzyme binding sites to catalytic sites active in reduction of aromatic aldehydes in transient state. Biochemistry. 9:185–192. [DOI] [PubMed] [Google Scholar]
- 31.Dunn, M. F. 1974. Comparison of kinetics and stoichiometry of proton uptake with aldehyde reduction for liver alcohol-dehydrogenase under single turnover conditions. Biochemistry. 13:1146–1151. [DOI] [PubMed] [Google Scholar]
- 32.Weidig, C. F., H. R. Halvorson, and J. D. Shore. 1977. Evidence for site equivalence in reaction-mechanism of horse liver alcohol-dehydrogenase with aromatic substrates at alkaline-pH. Biochemistry. 16:2916–2922. [DOI] [PubMed] [Google Scholar]
- 33.Daidone, I., D. Roccatano, and S. Hayward. 2004. Investigating the accessibility of the closed domain conformation of citrate synthase using essential dynamics sampling. J. Mol. Biol. 339:515–525. [DOI] [PubMed] [Google Scholar]
- 34.Millet, O., R. P. Hudson, and L. E. Kay. 2003. The energetic cost of domain reorientation in maltose-binding protein as studied by NMR and fluorescence spectroscopy. Proc. Natl. Acad. Sci. USA. 100:12700–12705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stockner, T., H. J. Vogel, and D. P. Tieleman. 2005. A salt-bridge motif involved in ligand binding and large-scale domain motions of the maltose-binding protein. Biophys. J. 89:3362–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kabsch, W., and C. Sander. 1983. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 22:2577–2637. [DOI] [PubMed] [Google Scholar]
- 37.Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD: Visual molecular dynamics. J. Mol. Graph. 14:33–38. [DOI] [PubMed] [Google Scholar]