Abstract
Steroidogenic factor 1 (SF-1/Nr5a1) is an orphan nuclear receptor encoded by the Ftz-F1 gene and is required for gonad and adrenal development and regulation of hormone production within the reproductive and adrenal axes. To extend our understanding of Ftz-F1 and its role in SF-1 expression, we identified and characterized a yeast artificial chromosome (YAC) containing Ftz-F1. Within this YAC, Ftz-F1 is centrally located and flanked by genes encoding a second orphan nuclear receptor, germ cell nuclear factor, and proteasome (prosome, macropain) subunit β type 7. Three lines of transgenic mice carrying the YAC were generated and in two lines (lines 7 and 14), RT-PCR and ribonuclease protection analysis showed that expression of transgenic SF-1 mimicked that of endogenous SF-1, both spatially and quantitatively. In the third line (line 15), pituitary and hypothalamic expression were absent. Comparison of the integrated transgenes revealed that line 15 was truncated at the end of intron 4 and revealed a region within the locus that is responsible for SF-1 expression in the pituitary and hypothalamus. The line 14 transgene was introduced into a mouse strain lacking functional SF-1. Examination of SF-1-deficient, transgene-positive mice revealed that the YAC was able to rescue adrenal and gonad development, which normally arrests in the SF-1-null embryos and showed that the 153-kb transgene integrated in line 14 is sufficient to properly direct SF-1 expression and support its biological activity. Thus, the study defines a region of Ftz-F1 that contains the requisite set of regulatory elements to direct SF-1 cell-specific expression and all temporal and quantitative changes need for its biological activity.
Abbreviations: Gcnf, Germ cell nuclear factor; GFP, green fluorescent protein; HR, homologous recombination; PFG, pulse-field gel; PFGE, pulse-field gel electrophoresis; Psmb7, proteasome (prosome, macropain) subunit β type 7; RNase, ribonuclease; RPA, RNase protection assay; SF-1, steroidogenic factor 1; tg, transgenic; VMH, ventromedial hypothalamus; YAC, yeast artificial chromosome; YIP, yeast integrating plasmid; ySLH1, yac SLH1
Steroidogenic factor 1 (SF-1, Ad4BP, Nr5a1) is an orphan nuclear receptor and one of four known transcripts, SF-1, ELP1, ELP2, and ELP3, derived from the Ftz-F1 gene through differential splicing and promoter usage (1). Of the Ftz-F1 products, SF-1 has received the greatest interest due to its pivotal roles in endocrine regulation and development of adrenal glands and gonads (1–5). As a key regulator of endocrine homeostasis, SF-1 controls transcription of many genes that are essential for the regulation or production of steroid hormones. This includes genes for cholesterol side-chain cleavage enzyme, cytochrome P450 aromatase, steroidogenic acute regulatory protein, α-subunit of glycoprotein hormones, LHβ, inhibinα, GnRH, GnRH receptor, and FSH receptor (2, 6–18). Within the promoters of these genes are specific response elements, typically PyCAAG-GTCA, to which SF-1 binds and stimulates transcriptional activity. Thus, SF-1 is thought contribute to the transcriptional activity in endocrine and reproductive tissues and, consequently, through activation of its target genes to steroid hormone production. In support of its role as an endocrine regulator, SF-1 is expressed in hormone producing cells of the adrenal glands, gonads, and anterior pituitary (5, 19–21). In the testes and ovaries, SF-1 is expressed in supporting cells (Sertoli and granulosa cells) and steroidogenic cells (Leydig and theca cells), whereas in the adrenal, SF-1 is localized to the steroid-producing adrenalcortical cells (22). Pituitary SF-1 is limited to gonadotrope cells (23).
SF-1 was initially recognized as a key regulator of adrenal and gonad development, when mice lacking the orphan receptor were generated and characterized with severe adrenal insufficiency and XY sex reversal, due to the absence of adrenal glands and gonads (3–5). A lack of corticosterone in these animals resulted in their death by postnatal d 8 and the absence of testicular androgens resulted in genetic males with female genitalia (3). Examination of SF-1-null embryos revealed that adrenal and gonad development initiated but was soon followed by apoptosis of the organs’ precursor cells (3). In addition, SF-1 null mice lacked the ventromedial hypothalamic nucleus and exhibited impaired expression of several pituitary genes, including LHβ, FSHβ, glycoprotein α-subunit, and the GnRH receptor (5, 23, 24). Lastly, mice lacking SF-1 presented with structural and functional abnormalities of the splenic vascular system and in humans, XY sex reversal and adrenal failure were observed in a patient harboring a mutation in the FTZ-F1 gene (25, 26).
In contrast to the information available on SF-1-target genes, its functions and expression patterns, the transcriptional mechanisms that direct SF-1 to its designated tissues remains poorly understood. Transient transfection approaches have identified some of the cis-elements and trans-factors that control basal transcription of the promoter region and include an E box, a CCAAT box, and three Sp1-binding sites (27–30). Such studies also revealed that similar promoter elements are employed for SF-1 transcription in testis, pituitary, and adrenal cells, suggesting that a common mechanism is used for promoter activity across different tissues (27–29). Proteins binding these elements have also been characterized, with some elements showing multiple binding complexes and differences between cell types (27, 28). However, despite their contributions to our understanding of SF-1’s promoter, these studies have not elucidated any mechanisms that explain how SF-1 expression is restricted to various target tissues or how its transcriptional regulation contributes to its developmental and endocrine functions.
More direct approaches using transgenic mice have been used to help uncover regions of the gene necessary for correct temporal and spatial expression of SF-1. Transgenic mouse studies using a 674-bp SF-1 promoter demonstrated expression of a LacZ reporter within the gonad (31). Yet, despite showing that some elements needed for gonad expression are present in this fragment, the studies revealed that a majority of key regulatory sequences required for full SF-1 production are missing. A second transgenic mouse study employed a 50-kb region of the mouse Ftz-F1 locus that demonstrated correct expression in most but not all SF-1-positive tissues (32). These studies have defined an important region that contributes to SF-1’s tissue-specific expression but also revealed that the region is insufficient to recapitulate all patterns of SF-1 expression. Furthermore, neither transgenic study was able to provide data on the transgene’s ability to support correct quantitative expression of SF-1. Consequently, new models are required to help define the Ftz-F1 locus and identify the region that fully recapitulates spatial, temporal, and quantitative expression of SF-1 and provide a comprehensive starting point from which studies can emerge to identify important control sequences.
To explore the mechanisms that regulate SF-1 expression in vivo, we have isolated a 500-kb yeast artificial chromosome (YAC) that contains the entire rat Ftz-F1 locus plus substantial amounts of 5′ and 3′ sequence. Three lines of transgenic mice carrying the Ftz-F1 YAC were developed, and studies revealed that SF-1 expression from two transgenes mimicked that of the endogenous gene, both spatially and quantitatively, whereas a third lacked expression in the pituitary and hypothalamus. Mice containing one of the Ftz-F1 transgene but lacking endogenous SF-1 were created by crossing transgenic mice with mice containing a targeted deletion for SF-1. Characterization of these mice demonstrated that the transgene directs SF-1 expression to an extent that rescues all known defects due to SF-1 deficiency and therefore has within its sequence all the regulatory elements required for SF-1 biological activity.
RESULTS
Identification and Characterization of a Ftz-F1-Containing YAC
A complete understanding of SF-1 transcriptional regulation requires defining the genomic region that independently and accurately controls its expression. Because data on SF-1 indicated that distal regulatory elements are essential for its proper expression, it was postulated that the required region encompasses considerable DNA sequence and therefore, to ensure sufficient sequence was obtained, a rat Ftz-F1-containing YAC was employed for expression studies. Screening of a YAC library identified three Ftz-F1-positive clones (y277D3, y376D1, y385G2). Restriction enzyme digestion followed by pulse field gel electrophoresis (PFGE) and Southern blot analysis identified a 500-kb Ftz-F1-containing NotI fragment present in two of the YAC clones (y376D1 and y385G2). To facilitate characterization of the locus and to reduce the cloned segment to a size compatible for transgenic analysis, the NotI fragment was subsequently isolated from y385G2 and ligated to pYAC4L vector arms to yield the smaller Ftz-F1-containing YAC, ySLH1 (Fig. 1). PFGE and Southern blot analysis with probes to either the promoter region of SF-1 (5′ probe) or the 3′ end of Ftz-F1 (exon 7) confirmed that uncut ySLH1 migrated similar to the NotI fragment from y385G2 and contained Ftz-F1 (Fig. 1A and data not shown). ySLH1 was further characterized by PFGE and Southern blot analysis after digestion with several rare-cutting restriction endonucleases. Hybridization with Ftz-F1 5′ and 3′ probes, as well as probes to the vector arms (selectable yeast marker gene URA3 and TRP1), revealed the positions of the restriction endonuclease recognition sites and indicated that Ftz-F1 is centrally located within ySLH1 (Fig. 1B). Assembly of three overlapping rat contigs obtained from high throughput genome sequence entries in GenBank (AC108247, AC129763, and AC119301) confirmed the location of the restriction endonuclease sites, indicating that the YAC had not undergone any significant rearrangement, and identified additional genes located within ySLH1; germ cell nuclear factor (Gcnf), a second orphan nuclear receptor, and proteasome (prosome, macropain) subunit β type 7, Psmb7 (Fig. 1B). In addition, the gene encoding Gpr144, a member of the G protein-coupled receptor family, resides between Psmb7 and Ftz-F1 (data not shown).
Fig. 1. Characterization of the Ftz-F1-Containing YAC, ySLH1.
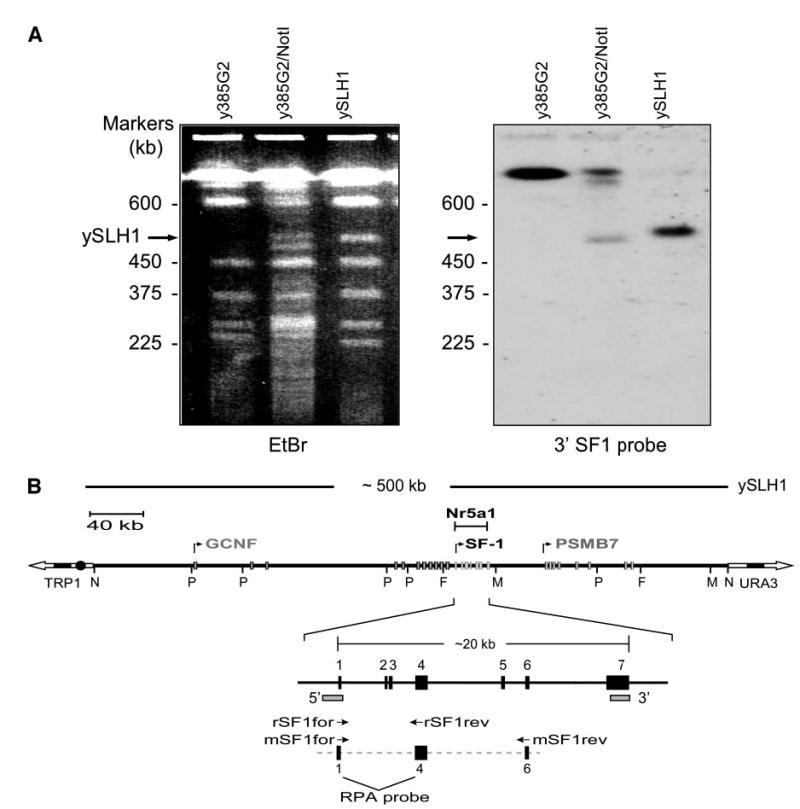
A, Pulse field electrophoresis of y385G2, y385G2 digested with NotI, and ySLH1, a 500-kb NotI subclone of y385G2. The gel was stained with ethidium bromide (EtBr, left) and characterized by Southern blot analysis (right) using the 3’SF1 probe (shown in B as 3′-marked rectangle). B, Schematic depiction of ySLH1. ySLH1-containing yeast were subject to digestion with restriction endonucleases and analyzed by PFGE and Southern blot analysis (not shown). Positions of the restriction endonuclease sites for PmeI (P), NotI (N), FseI (F), and MluI (M) were determined using 5′ and 3′ probes (marked as rectangles) and TRP1 and URA3 probes corresponding to the vector arms. Three different contigs (AC108347, AC119301, AC129763) from the working draft sequence of the rat genome project were assembled to generate a map of the rat Ftz-F1 locus and the positions of the above restriction endonuclease sites were confirmed. Annotated are the Ftz-F1 gene and its adjacent genes encoding a second orphan nuclear receptor GCNF (NR6A1) and a proteosome protein PSMB7. Boxes represent the different genes’ exons and arrows the direction and start of transcription. Primers used for RT-PCR analysis of rat SF-1 (rSF1for and rSF1rev) and mouse SF-1 (mSF1for and mSF1rev) are indicated as are exons within the probe employed for RNAse protection analysis.
SF-1 Expression from ySLH1 Mimics that of Endogenous SF-1
To determine whether genomic sequences residing within ySLH1 confer proper SF-1 expression, transgenic mice were produced using the ySLH1 YAC and the expression profile of rat SF-1 was compared with that of endogenous mouse SF-1. Three YAC-containing transgenic lines (lines 7, 14, and 15) were obtained, and transgene expression was measured by RT-PCR and RPA [ribonuclease (RNase) protection analysis], with RNA isolated from a variety of tissues from both positive and negative transgenic mice. Primers (rSF1for and rSF1rev) used to detect rat SF-1 were specific for exons 1 and 4 and did not cross-react with mouse SF-1, as evidenced by the lack of amplified product in SF-1-expressing tissues of negative littermates (Fig. 2A;+, transgenic tissue;–, negative tissue). In all three transgenic lines, a 954-bp product was detected in testis, ovary, adrenal, and spleen of transgenic animals, whereas no product was observed in tissues of negative littermates (Fig. 2A). Southern blot analysis using a radiolabeled primer that hybridizes to sequences between the amplification primers confirmed that the products were derived from SF-1 mRNA and analysis of mouse SF-1 mRNA demonstrated that transgene expression mimicked that of the endogenous mouse gene (data not shown). Surprisingly, transgene expression in the pituitary and hypothalamus was detected only in mice derived from lines 7 and 14, revealing an interesting expression deficiency in line 15 (Fig. 2A, bottom). RPA was used to compare SF-1 mRNA levels derived from the rat transgene and mouse endogenous gene. In this assay, the RNA probe differentiated between rat and mouse SF-1 mRNA because different protected sizes were obtained for the two species (Fig. 2B, compare lanes 2 and 3). Both rat and mouse transcripts were detected in the testis, ovary, and adrenal of transgenic lines 7 and 14, whereas only mouse transcripts were observed in tissues from negative littermates (Fig. 2B and data not shown). Evaluation of mRNA levels revealed that expression of SF-1 from the YAC closely resembled that of the endogenous gene, suggesting that ySLH1 contains not only the regulatory information for correct spatial expression of SF-1 but also that needed to control the gene’s expression level and shield it from the potential regulatory influence of surrounding chromatin.
Fig. 2. Analysis of Transgene Expression in ySLH1-Lys Transgenic Mice.
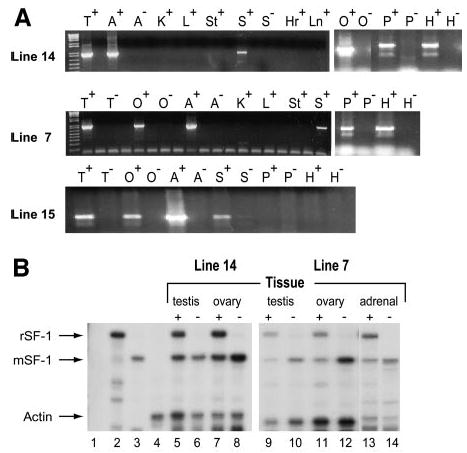
RT-PCR was used to examine expression of transgenic rat SF-1 in lines 7, 14, and 15 (A). For each line, the rat transcript was amplified using RNA from tissues isolated from trans-genic (indicated as +) or negative littermate (indicated as –) animals. Samples are heart (Hr), kidney (K), liver (L), lung (Ln), spleen (S), stomach (St), testis (T), ovary (O), adrenal (A), pituitary (P), and hypothalamus (H). RNase protection analysis was used to measure mRNA levels in transgenic lines 7 and 14 (B). SF-1 and actin transcript levels were measured using their respective, specific RNA probes. The SF-1 probe distinguished rat and mouse transcripts by protecting distinct sized mRNAs. Controls are shown in lanes 1–4 and are as follows: 1, tRNA with probes to both SF-1 and actin; 2, rat testis RNA with SF-1 probe; 3, mouse testis RNA with SF-1 probe alone; 4, mouse testis RNA with actin probe. RNA isolated from tissues noted above the lanes of transgenic mice (indicated as +) or negative littermates (indicated as –) was assayed for both SF-1 and actin mRNAs. The lanes are as follows: 5, line 14 transgenic mouse testis; 6, negative mouse testis; 7, line 14 transgenic mouse ovary; 8, negative mouse ovary; 9, line 7 transgenic mouse testis; 10, line 7 negative mouse testis; 11, line 7 transgenic mouse ovary; 12 line 7 negative mouse ovary; 13, line 7 transgenic mouse adrenal; 14, line 7 negative mouse adrenal. Arrows show protected fragments for transgenic rat SF-1 (rSF-1), endogenous mouse SF-1 (mSF-1), and actin.
Transgene Mapping Reveals Region Required for Pituitary and Hypothalamic Expression of SF-1
Mapping of the integrated ySLH1 transgenes was undertaken to confirm the regions required for accurate SF-1 expression and to explore potential differences in the integrated sequences that might explain the inability of line 15 to express within the hypothalamus and pituitary. High-molecular-weight DNA was prepared from liver cells isolated from mice of each transgenic line and the DNA analyzed by Southern blot analysis, after digestion with PmeI and separation by PFGE. Multiple hybridization probes, which span the Ftz-F1 locus, were employed (Fig. 3, top, denoted with arrows along the locus) and revealed that, within line 14, there were two transgene inserts that contained complete copies of Ftz-F1 (Fig. 3). The largest insert contained one intact PmeI fragment of the YAC (~153 kb) and a smaller insert fragmented between the 3′ SF1 and SPH10 probes (Fig. 3, A and B). Both line 14 inserts terminate just 5′ to the PmeI site, but the exact breakpoint has not been determined (data not shown). Similarly, the 3′ terminus of the largest insert was located near the downstream PmeI site to its 3′ side (data not shown). Several smaller fragments were also detected within line 14, but these do not have a complete Ftz-F1 gene and therefore do not generate relevant transcripts. Line 7 contained a single complete PmeI fragment with the 5′ end terminating close to the upstream PmeI site and the 3′ end terminating approximately 40 kb 3′ to the downstream PmeI site (data not shown). Interestingly, mapping of line 15 revealed a significant loss of DNA sequence at the 3′ end of ySLH1, resulting in a transgene that terminated just 5′ to exon 5 (Fig. 3, A and B). Notably, the transgene’s 5′ end extended beyond that determined for either line 7 or 14 (data not shown). Thus, the mapping data identified a region between the 3′ breakpoints of lines 14 and 15 that is essential for pituitary and hypothalamic expression of SF-1.
Fig. 3. Mapping of Transgenes Integrated in Lines 14 and 15.
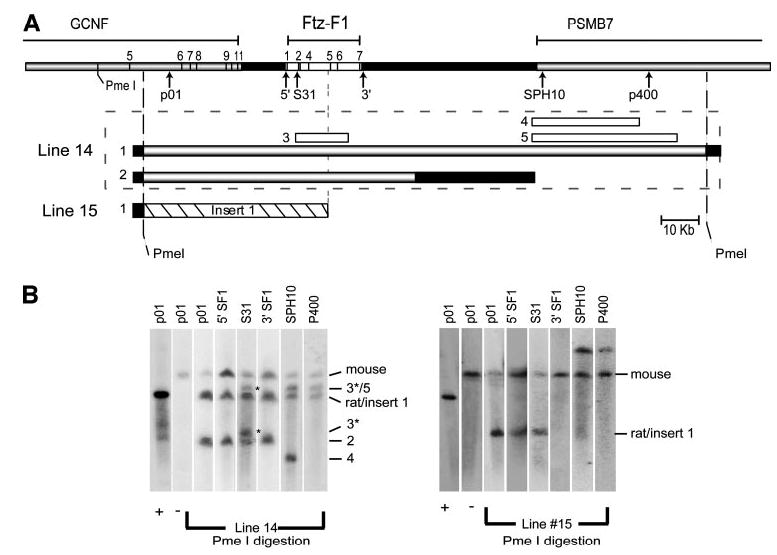
A, Schematic diagram of the genomic regions within transgenic lines 14 and 15. The Ftz-F1 locus is depicted as a rectangle (top) with the Ftz-F1, GCNF, and PSMB7 genes annotated. Exons are numbers for GCNF and Ftz-F1 indicated by lines. The positions of various probes used in the analysis are noted by arrows. Transgenic inserts are numbered and represented as bars below the locus (line 14, gray and white bars; line 15, dashed bar). Inserts for line 14 are denoted within the dashed box. Positions of the recognition sequences for the restriction endonuclease PmeI are also noted. B, Southern blot analysis of high-molecular-weight DNA isolated from lines 14 and 15. Liver plugs were digested with PmeI, resolved by PFGE, and analyzed by Southern blot analysis using the indicated probes (top of the blot and marked by arrows in A). Signals corresponding to the endogenous mouse gene (mouse) and rat transgene inserts (numbers correspond to those in A) are noted. Asterisks mark smaller inserts within Ftz-F1 that also appear in Fig. 4.
ySLH1 Rescues Defects Observed in SF-1-Null Mice
Whereas the above studies suggested that SF-1 expression from transgenes 14 and 7 follow that of endogenous SF-1, the data were limited to a narrow temporal window and, therefore, could not address many of the complexities of SF-1 expression. To provide a more sensitive measure of transgene function, studies were undertaken to determine whether transgenic SF-1 could rescue the developmental and endocrine abnormalities of SF-1-null mice (3). Accordingly, line 14 transgenic mice were mated to mice carrying a targeted mutation for SF-1 (Nr5a1tm1Kl), which effectively disrupts function of the orphan receptor, to generate Ftz-F1+/− mice carrying the transgene (3). Mating of the resulting Ftz-F1+/−; transgenic (tg) + mice produced animals carrying the YAC transgene and lacking a functional endogenous allele (Fig. 4, shaded box). In addition to a band corresponding to the intact transgene, two slower migrating products were observed that represent small fragments of the YAC within the BglII digestion product. The fragments span only a portion of the coding region (from intron one to three) and therefore should not contribute to the transgenic SF-1 mRNA pool (asterisk in Figs. 3 and 4).
Fig. 4. Southern Blot Analysis for Rescue Genotype Determination.
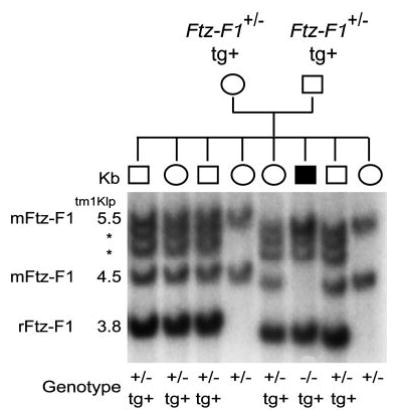
Tail DNA was isolated from the resulting progeny of SF1 +/−; tg/+ matings, digested with BglII, and analyzed using a probe directed against intron 3 of the rat Ftz-F1 gene. Bands for the endogenous mouse allele (4.5 kb), the rat transgene (3.8 kb), the mouse knockout allele (5.5 kb), and two truncated transgene products (*) are indicated. The determined genotypes are indicated at the bottom.
All transgenic mice lacking endogenous SF-1 (Ftz-F1−/−; tg+) survived past the first week of life, and their general health was not noticeably different from wild-type mice and body weights were similar between rescued and control mice (Table 1). Because SF-1-null mice die within 1 wk of birth from adrenal insufficiency caused by agenesis of the gland, survival of the rescued animals indicated that adrenal gland development was sufficient to produce the required steroid hormones. Indeed, comparison of rescued and wild-type mice revealed that adrenal glands in the rescued animals were similar in body position, size, and shape to that of wild-type littermates, and histological analysis showed normal structure and zonation of the medulla and cortex (top, Fig. 5, A and B).
Table 1.
Body Weights of Rescue and Control Mice
| Genotype | Sex | n | 1 mo (g) | 2 mo (g) | 3 mo (g) |
|---|---|---|---|---|---|
| Ftz-F1−/−;tg+ | Male | 15 | 23.8 ± 1.8 | 31.3 ± 2.0 | 32 ± 2.2 |
| Ftz-F1+/−;tg+ | Male | 14 | 22.3 ± 1.9 | 29.8 ± 1.8 | 30.9 ± 1.9 |
| Ftz-F1−/−;tg+ | Female | 12 | 20.2 ± 1.7 | 23.8 ± 2.5 | 27.5 ± 2.4 |
| Ftz-F1+/−;tg+ | Female | 17 | 19.7 ± 1.2 | 23.6 ± 2.2 | 28.3 ± 2.2 |
mo, Months.
Fig. 5. Morphology of Adrenal Glands and Gonads in SF-1-Null Mice Rescued with a Ftz-F1-Containing YAC Transgene (Line 14).
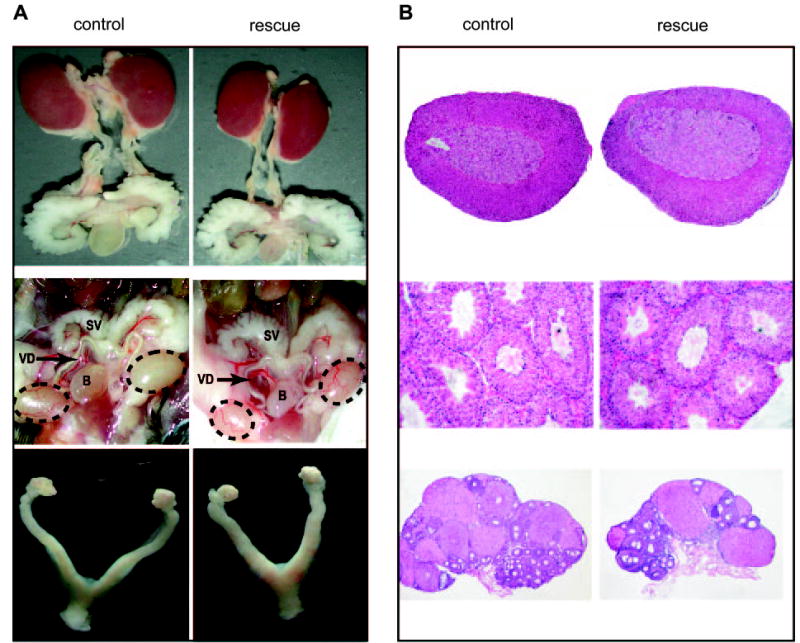
A, Comparison of organ structures from control (left) and rescued (right) animals. Top, Adrenal glands, kidneys, and components of the urogenital system from wild-type and SF-1-null mice rescued with the YAC transgene. Middle, Testes and internal genitalia of wild-type and rescue adult male mice. Testes are indicated within dotted ovals. SV, Seminal vesicle; B, bladder; VD, vas deferens. Bottom, Ovaries and internal genitalia of control and rescue adult female mice. B, Histology of adrenal glands and gonads from control (left) and rescued (right) animals. Five-micrometer sections of Bouin’s fixed, paraffin-embedded tissues were stained with hematoxylin and eosin as described in the Materials and Methods. Top, Adrenal glands from wild-type and rescued mice. Middle, Testes from adult wild-type and rescued mice. Bottom, Ovaries from control and rescued 4-month-old diestrous mice.
Because SF-1-deficient mice present with XY sex reversal, genotypic sex of the rescue mice was determined by assaying for the presence of the Y chromosome-specific gene Sry. This confirmed that all genotypes corresponded to the observed genders, indicating that the embryonic endocrine environment supported proper development of the external genitalia (data not shown). With respect to male rescue mice, this signified that testicular development progressed to the extent that androgen levels were sufficient to masculinize the external genitalia. Further examination revealed that male reproductive tracts from rescued animals were indistinguishable from control male littermates (middle, Fig. 5A). Notably, the androgen-dependent Wolffian duct derivatives, i.e. the seminal vesicles, vas deferens, and epididymis, were well developed, and testes from the rescued animals were appropriately descended and of similar size and shape to that of normal littermates. In addition, male rescue mice were fertile, with fertility and litter sizes equivalent to controls and histological analysis of the testes showed normal morphology of the seminiferous epithelium and similar germ cell numbers between rescue and wild-type animals (Table 2 and middle, Fig. 5B).
Table 2.
Mating Data for Ftz-F1−/−;tg+ mice
| Genotype | Sex | n | Mating Successa | Fertilityb | Litters | Average Litter Sizec |
|---|---|---|---|---|---|---|
| Ftz-F1+/+ | Male | 3 | 3/3 | 3/3 | 3 | 10.0 ± 2.0 |
| Ftz-F1+/− | Male | 4 | 6/6 | 4/4 | 6 | 9.8 ± 2.0 |
| Ftz-F1−/−;tg+ | Male | 19 | 25/25 | 19/19 | 25 | 8.6 ± 1.6 |
| Ftz-Fl+/−;tg+ | Male | 13 | 19/19 | 13/13 | 19 | 9.5 ± 2.0 |
| Ftz-F1+/+;tg+ | Male | 6 | 7/7 | 6/6 | 7 | 10.2 ± 1.6 |
| Ftz-F1+/− | Female | 11 | 13/15 | 11/13 | 13 | 8.3 ± 1.9 |
| Ftz-F1+/+ | Female | 12 | 17/19 | 12/12 | 17 | 7.6 ± 2.0 |
| Ftz-F1−/−;tg+ | Female | 22 | 25/28 | 19/22 | 25 | 8.2 ± 1.6 |
| Ftz-F1+/−;tg+ | Female | 23 | 28/30 | 22/23 | 28 | 10.1 ± 2.1 |
| Ftz-F1+/−;tg+ | Female | 10 | 14/15 | 9/10 | 14 | 10.7 ± 1.6 |
Number of litters produced/number of matings.
Number fertile animals/number tested.
Values are expressed as average ± SEM.
Female rescue mice were also fertile, with mating success and fertility not different from controls (Table 2). In addition, the average litter size between rescued females was indistinguishable from wild-type and Ftz-F1+/− animals (Table 2). However, comparison of the rescued, Ftz-F1+/−, or wild-type mice to YAC transgenic mice (Ftz-F1+/+;tg+) revealed a statistically significant increase in the litter size of transgenic females (P ≤ 0.005), with an approximate increase of two pups per litter. Examination of reproductive tract structures from rescued mice revealed them similar to those of control littermates, whereas the ovaries were found to be slightly smaller to normal in size (bottom, Fig. 5A). Histological analysis of ovaries from rescued females showed normal follicular development and presence of corpora luteum (bottom, Fig. 5B). Overall, the studies demonstrate that expression of SF-1 from the transgene can direct development of the gonads and later fulfill the functional requirements of SF-1 in the adult gonad and that the transgene also directed proper expression of SF-1 within the pituitary, where it is required for the production of the gonadotropin hormones (33).
Immunolocalization of Transgenic SF-1 Expression
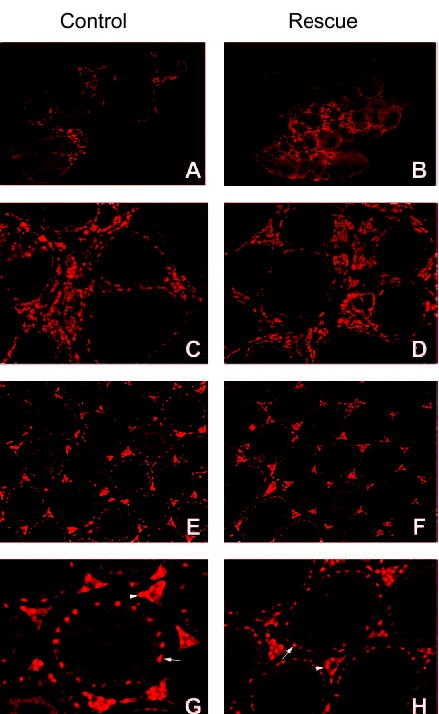
With endogenous SF-1 protein absent, cellular expression of transgenic SF-1 was evaluated by immunohistochemistry using an SF-1-specific antibody and tissues isolated from YAC-rescued mice. Comparison of SF-1 expression between rescued and control animals revealed that cellular expression of transgenic SF-1 was indistinguishable from that of endogenous SF-1. In the ovaries of control and rescued mice, expression of SF-1 was predominantly located in the theca/interstitial cells (Fig. 6, A–D). In the testis, SF-1 expression was observed within the Leydig cells located in the interstitial region and Sertoli cells in the seminiferous tubules (Fig. 6, E–H, in G and H; arrowheads mark expressing Leydig cells and arrows expressing Sertoli cells). Notably, no difference was observed between rescue and control groups. In the adrenal gland and hypothalamus, SF-1 expression was similar between animal groups, with staining limited to cells in the cortex of the adrenal and ventral medial hypothalamus (Fig. 7A). Pituitary SF-1 in control and rescue mice was localized to gonadotrope cells, as indicated by its colocalization with LH β subunit (Fig. 7B). The lack of SF-1 staining in the pituitary of newborn knockout mice confirmed the specificity of the antibody (Fig. 7A, bottom). The above studies demonstrate that SF-1 expressed from the transgene follows the same cellular profile as that of the endogenous SF-1 and that the sequence information within transgenic line 14 is sufficient to express SF-1 to a level of accuracy that fulfills its functional obligations in development and endocrine regulation.
Fig. 6. Immunohistochemical Evaluation of SF-1 in Gonads of Control and Rescued Mice.
Tissues were harvested and SF-1 visualized using a bovine SF-1 antibody (described in the Materials and Methods). A–D, Sections from ovaries isolated from control and rescue mice, respectively. C and D, Higher magnifications of panels A and B, respectively. E and G and F and H, Sections from testes isolated from control and rescue mice, respectively. G and H, Higher magnifications of panels E and F, respectively. Arrowheads in G and H mark expressing Leydig cells, whereas arrows mark expressing Sertoli cells.
Fig. 7. Immunohistochemical Evaluation of SF-1 in Adrenal, vmh, and Pituitary of Control and Rescued.
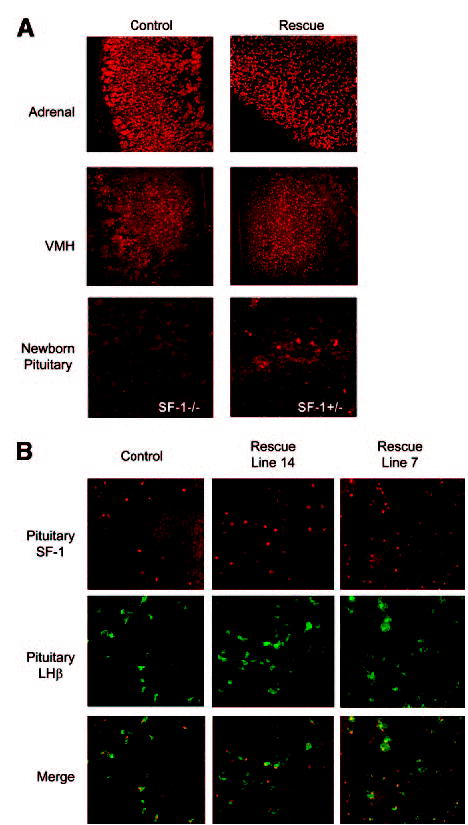
Mice Tissues were harvested from adult animals (except where noted) and visualized for SF-1 as described in the legend of Fig. 6. Adult pituitary sections were also stained for LHβ, to marked gonadotrope cells. A, SF-1 staining in the adrenal gland and vmh of control and rescued animals and pituitary of SF-1-null (SF-l−/−) and heterozygous (SF-1−/+) newborns mice. B, Pituitary staining for SF-1 (red, top) and LHβ (green, middle) in control, line 14 rescued, and line 7 rescued mice. Merged images (bottom) revealed colocalization of SF-1- and LHβ-expressing cells.
Comparative Genomics Reveals Potential Regulatory Regions within the Ftz-F1 Locus
Characterization of mice carrying the ySLH1 transgene demonstrated that a 153-kb region of the Ftz-F1 locus fully recapitulated expression of the endogenous gene to an extent that it rescued all known defects in the Ftz-F1-null mice. In addition, evaluation of one transgenic line in which 3′ sequences downstream of intron 4 were deleted revealed a region that contains important regulatory elements for SF-1 expression in the pituitary and hypothalamus. To help identify elements that control SF-1 expression, genomic sequence data from human, rat, and chicken were compared across the 153-kb region, using the web-based sequence analysis tool ECR Browser to identify and annotate evolutionary conserved regions (ECRs; Ref. 34). The analysis revealed over 40 noncoding ECRs in the comparison between rat and human and nine ECRs in the comparison of human and chicken (Fig. 8, black peaks). Because the evolutionary distance from chicken to human is much greater than from rat to human, the ECRs identified in the chicken/human analysis represent more highly conserved noncoding sequences and therefore ones most likely to contain important functional information. Hence, these regions represent good candidate sites for transcriptional control of Ftz-F1 and are marked as ECRs 1–9 (Fig. 8). Comparison of the genomic segments integrated in transgenic lines 14 and 15 revealed several potential ECRs involved in tissue-specific expression. Lines 7, 14, and 15, each expressed in the adrenal gland and gonads and also contain ECRs 1–3. Thus, these sites represent good candidates for contributing to expression in these tissues (Fig. 8). Furthermore, sequences between the 3′ ends of YACs 14 and 15 were identified as necessary for expression in the pituitary and hypothalamus, thereby implicating ECRs 4–9 in this expression program (Fig. 8).
Fig. 8. Whole Genome Alignment of the Rat Sequence within ySLH1 with the Corresponding Sequences from Human and Chicken.
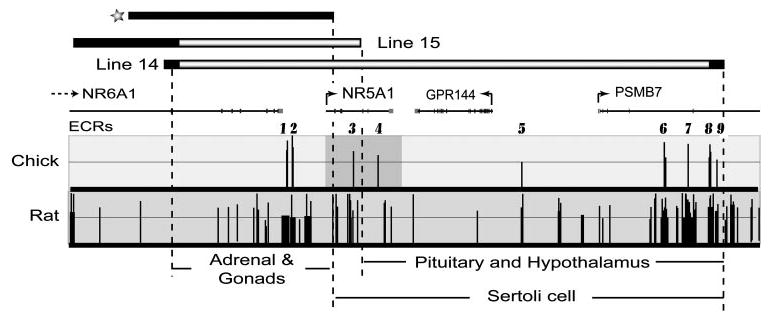
With human sequence as the base genome, alignments were performed with rat and chicken sequences within the NR5A1 locus and the conserved regions visualized using the ECR browser navigation tool (http://ecrbrowser.dcode.org). Genes within the region are marked as lines above the depicted alignments and correspond to NR5A1, PSMB7, NR6A1, and GPR144. Arrows mark the direction of transcription and boxes, exons. Dark peaks in the graphed area depict noncoding, evolutionarily conserved sequences (ECRs), which were identified as sequences having a minimal identity threshold of 75% over a minimal length of 100 bp and do not correspond to repetitive sequences. The x-axis represents positions in the base genome and the y-axis represents percent identity between the base (human) and the aligned genome specified. Transcriptional orientation of the genes are indicated by arrows. Bars above the graph indicate the genomic segment integrated in transgenic lines 14 and 15. Black regions within the bar indicate the breakpoint area. The top black bar (marked with a star) indicates the approximate position of a previously reported GFP transgene (32).
DISCUSSION
Using a transgenic rescue strategy, we have established that sequences within a 153-kb region of the Ftz-F1 locus are sufficient to direct SF-1 expression to an extent that fully supports its biological activity, and consequently identified a genomic region that contains the complete repertoire of elements required for cell-specific expression and all requisite temporal and quantitative changes in SF-1. Thus, the sequence information defined within the YAC provides an important reference point for identifying transcriptional elements regulating SF-1. Currently, our knowledge of SF-1 transcriptional control is primarily limited to data derived from studies on its promoter, which, as noted previously, have not provided any substantial mechanistic insight into SF-1’s cell-specific expression or other features of its regulation. In part, progress in this area has been hampered by the inability to identify the genomic region that correctly directs SF-1 transcription in vivo and has led to conclusions that SF-1 expression is largely directed by regulatory elements located at significant distances from the transcriptional start site (32). Fortunately, more recent reports using transgenic mice have, to some extent, overcome this obstacle and confirm the requirement for distal regulatory elements, as well as proximal promoter elements, in directing SF-1 expression in vivo. In one study, a transgene containing 676 bp of the SF-1 promoter expressed in the embryonic gonad, revealing an important functional contribution by this promoter to embryonic gonad expression (31, 35). However, data from this study also revealed that many important regulatory sequences are located outside the promoter region, as expression from the transgene was poorly restricted and absent from other sites of SF-1 expression.
A second transgenic mouse study avoided the limitations imposed by small promoter fragments by using a 50-kb region of the mouse Ftz-F1 gene, which included 45 kb of 5′-flanking sequence, exon 1, and part of exon 2, to direct expression of the green fluorescent protein (GFP) (32). In these animals, GFP expression was observed in the adrenal cortex, interstitial cells of the testis and ovary, and the ventromedial hypothalamus (VMH). However, transgene expression was notably absent in adult pituitary gonadotropes and Sertoli cells, when compared with endogenous SF-1. Thus, this GFP transgene was able to direct many but not all aspects of SF-1 tissue-specific expression. Importantly, neither study was able provide sufficient detail to assess all qualities of SF-1 expression, such as transcription levels or dynamic changes, that are necessary for proper SF-1 function.
By evaluating transgene activity via its ability to rescue the biological deficiencies in SF-1-null mice, the studies reported here offer a more sensitive measure of SF-1 expression and extend evaluation to include embryonic and postnatal development by virtue of recovered SF-1 function. Importantly, the studies demonstrated that SF-1 produced from the transgene imparts the developmental and reproductive functions attributed to SF-1 and therefore reflect accurate quantitative and qualitative regulation by the sequences within the 153-kb transgene. Endogenous SF-1 expression begins early in development in a population of cells that subsequently contribute to the adrenal glands and gonads (36). In these cells, SF-1 contributes to cell survival, as without it, the cells regress by programmed cell death and the gonads and adrenals fail to form (3). Therefore, the presence of normal adrenal glands and gonads in rescued mice show that transgenic SF-1 was expressed in these cells at the necessary time point in development and at sufficient levels to induce organ formation, with the additional implication that any requisite changes in expression levels were properly orchestrated by the transgene. Because SF-1 expression is strongly attenuated in the developing ovary, normal ovarian development in the rescue animals suggests either that the expression change occurred or that it is unnecessary for its development (22). Future studies will help elucidate the validity of these options.
In contrast to the direct genetic evidence that defines SF-1 as a developmental regulator, the lack of major steroidogenic organs in SF-1-null mice has circumvented in vivo evaluation of many SF-1 target genes and support for its role in steroid hormone production and endocrine homeostasis remains largely circumstantial. There are, however, in vivo data demonstrating that SF-1 is required for expression of the gonadotropin genes. Using a cre/lox strategy, Zhao et al. (33) specifically deleted SF-1 in mouse pituitary gonadotropes and showed that LH and FSH levels were markedly diminished and the animals infertile. In relation to our studies, where fertility was normal in male and female rescued mice, pituitary transgene function must have been sufficient to support proper expression of the gonadotropin genes. Notably, colocalization studies showed transgenic SF-1 properly expressed within gonadotrope cells of the pituitary. Also, despite the lack of direct in vivo evidence for SF-1’s role in steroid synthesis, the expression profile of ySLH1 and the normal functioning adrenal glands and gonads in rescued mice strongly suggests that the YAC transgene fulfilled any biological directive necessary for SF-1-regulated steroid biosynthesis, but establishment of this awaits direct in vivo confirmation of SF-1’s role in steroid production.
Importantly, proper transgene expression was supported by RT-PCR and RPA studies in transgenic mice and immunohistochemical analysis in rescued mice, which confirmed SF-1 expression in the appropriate cells and tissues. Because the SF-1 knockout allele disrupted nearly all of the protein coding sequence and no evidence of SF-1 was detected in the pituitary of newborn SF-1-null mice (Fig. 7A), immunodetection of SF-1 in the rescue tissues was specific to the protein produced from the transgene. Western blot analysis supported this conclusion as well because there were no bands observed at sizes other than that for SF-1 (data not shown). Furthermore, no significant signal was observed by Western blot analysis using liver protein samples, indicating that employed SF-1 antibody did not detect LRH-1, a closely related orphan nuclear receptor that is expressed in liver, as well as SF-1-positive tissues (37).
Lastly, because the 153-kb transgene contains all necessary sequences for proper SF-1 expression, it provides an important reference point from which we can delineate the functional elements controlling SF-1 expression. However, effective investigation of the regulatory mechanism requires a strategy to sort through this large amount of sequence to identify regions of greatest potential for containing transcriptional control elements. To facilitate the discovery of such elements, areas of evolutionary conservation were identified by examining the human, rat, and chicken whole genome alignments within the region encompassed by the 15-kb region and revealed over 40 noncoding ECRs in the human/rat comparison and nine ECRs in the human/chicken comparison (Fig. 8 and Ref. 38). The high probability that such sequences are functional, particularly the more distally related human to chicken ECRs, identifies them as strong candidates for SF-1 regulatory elements that can serve as a foundation for future evaluation of the locus.
The data presented here together with previous transgenic studies offer important insight into potential roles of the identified ECRs and regions defined by the transgenes. Comparison of the current study to the one previously, which employed 50 kb of mouse Ftz-F1 fused to GFP, revealed both similarities and differences that serve to solidify the earlier data and extend it to provide insight on SF-1 expression in additional cell types (32). In the earlier study, the trans-gene contained approximately 17 kb additional 5′ sequence, relative to inserts in lines 14 and 7, and 8.5 kb less 3′ sequence than line 15 (Fig. 8, black rectangle and asterisk). Data from these studies showed GFP expression in ovarian theca and testicular Leydig cells, as well as cells within the hypothalamus and spleen. GFP was also observed within the developing gonads and adrenal, but cellular morphology precluded accurate identification of the expressing cells. In support of these findings, transgenes evaluated in the present study also demonstrated expression within the gonads, adrenals, spleen, and hypothalamus. Confirmation of the earlier findings is especially relevant given that only a single transgene was evaluated previously; leaving the noted caveat that integration site might have influenced the study’s outcome. It is also noteworthy that the earlier study did not report the size of the integrated transgene, so it is unclear if the data reflect results from a fully intact transgene or a smaller integrated one. Together, however, the studies convincingly place ECRs 1and 2 at the top of the candidate list for regions housing elements that control expression in adrenal cortical cells, ovarian theca cells, and testicular Leydig cells (Fig. 8).
In contrast to this earlier study, transgenes evaluated here expressed within gonadotropes and Sertoli cells, indicating that sequences defined by the transgenes’ differences house regulatory elements required for expression within these two cell types. Characterization of line 15 further eliminated sequences between the start of exon 2 through intron 4, and notably ECR 3, as a potential location for elements that direct SF-1 pituitary expression. Thus, several ECRs emerge as good candidate regulatory regions that control gonadotrope and Sertoli cell expression and, based on proximity to Ftz-F1, ECRs 4 and 5 appear to be the most promising for gonadotrope expression and ECRs 3–5 for Sertoli cell expression (Fig. 8). Characterization of these, as well as ECRs 1 and 2, should provide important insight to our understanding of SF-1 transcription.
A second and conflicting difference between the studies stems from the determined requirements for hypothalamic expression. In the current study, SF-1 mRNA was not detected in the hypothalamus of line 15, whereas in the previous study, hypothalamic expression of GFP was observed, despite the use of a smaller transgene (Fig. 8). The reason for the discrepancy is currently unknown but may reflect differences in the detection methods, influences from sites of integration, divergent regulatory regions in mouse and rat, or, more interestingly, a transcriptional mechanism involving interactions between activating and silencing elements. Studies herein evaluated SF-1 transcript levels by RT-PCR and, despite detectable transcripts for endogenous SF-1 and L7, transcripts for transgene-derived SF-1 were not observed (Fig. 2 and data not shown). In the earlier study, GFP fluorescence and immunohistochemical techniques were used to detect transgene expression. Thus, perhaps use of this reporter induced vmh expression, either from ectopic induction or artifacts from the employed antibodies. Alternatively, the RT-PCR data may reflect differential splicing in the vmh within line 15, which precluded detection of transcripts when using the exon1/4 primer set. Or, because the conflicting data in each case derived from only a single transgenic line, it is equally possible that integration site effects influenced hypothalamic expression in one of the studies. Notably, the two transgenes are derived from different species (mouse vs. rat) and thus, there may be species differences in the location of elements required for hypothalamic SF-1 expression. Lastly, if the data from each study accurately reflect the transcriptional mechanism, the data suggest that transgenic line 15 contains a potent silencing element located between exons 2 and 5, which suppressed upstream elements inducing hypothalamic expression of the SF-1. Such a mechanism further implies that sequences downstream of exon 5 are able to overcome the silencing activity. Thus, hypothalamic expression would require cooperative interaction of two positive regulatory regions to overcome the effects of a silencing region. Future characterization of additional transgenic lines will resolve the apparent conflict and identify mechanisms not only for hypothalamic expression but other tissues as well. Importantly, the studies have set the stage for deciphering regulatory elements required for accurate SF-1 expression and reveal the transcriptional mechanisms involved in the unique cellular expression patterns of this gene.
MATERIALS AND METHODS
Yeast Strain and Propagation
Saccharomyces cerevisiae yeast strain AB1380 (MATa, ura3-52, trp1, ade2–1, his5, lys2–1, can1–100) was grown in YPD medium and used to propagate the 500-kb YAC containing the rat Ftz-F1 gene. YAC-containing yeast were grown in complete medium, lacking at least one selective nutrient required for YAC maintenance. The original YACs purchased from Invitrogen Life Technologies (Carlsbad, CA) were provide in yeast strain J57D(MATα, ura3, trp1, ade2–101, can1–100, leu2–3, 112 his3–6) and grown in complete medium lacking tryptophan and uracil. All media were prepared as described elsewhere and all yeast cell cultures were carried out overnight at 30 C under vigorous agitation (39, 40). Transformation of spheroplasted yeast cells was performed as described except yeast cells were treated with 80 μg/ml of Arthrobacter luteus lyticase (Sigma, St. Louis, MO) for approximately 90 min at 37 C (41).
YAC Clones
DNA pools of a rat YAC library constructed by the Whitehead Institute for Biomedical Research and the Massachusetts Institute of Technology Center for Genome Research (Cambridge, MA) (average insert size of 830 kb; Ref. 42) were purchased from Invitrogen Life Technologies and screened for the presence of the rat Ftz-F1 gene using the PCR (primary screen primers: 5′-CCAACCTCGAGCCCCCATAAAG-ATAGGAATA-3′ and 5′-GCCCAAGCTTTTACTGAAGGGAA-GGAGAAT-3′; secondary screen primers 5′-GGGGGCGGC-CGCGACTATTCGTACGACGAG-3′ and 5′-GCGGGAGCTC-ACTGCTCGGCTTTCCCCTCAG-3′). DNA was amplified using Taq DNA polymerase, reaction conditions recommended by the manufacturer, and 30 amplification cycles (95 C for 1 min, 33 C for 1 min, 72 C for 30 sec; Invitrogen). Three Ftz-F1-containing YACs, 277D3, 376D1, and 385G2, were identified and purchased from Invitrogen Life Technologies. YAC clone ySLH1 was generated by subcloning a 500-kb Ftz-F1-containing NotI fragment from clone 385G2 by in-gel ligation of the isolated fragment with NotI/BamHI digested pYAC4L vector arms (39). pYAC4L was constructed by inserting a polylinker (EcoRI-SfiI-AscI-NotI-PmeI-SacI) having EcoRI compatible overhangs into the unique EcoRI site of pYAC4 (Stratagene, La Jolla, CA). Ligated DNA was transformed into yeast strain AB1380 and transformants were selected for growth in the absence of uracil and tryptophan. Ura+/trp+ clones were characterized by PFGE and Southern blot analysis. The 500-kb subclone ySLH1 was identified. ySLH1Lys was generated by retrofitting ySLH1 with a pLysNeo vector, resulting in the replacement of the ura3 marker with lys2 (43). Retrofitting was performed by transformation of ySLH1-containing yeast with a 7.8-kb NotI/SalI fragment of pLysNeo and selection for growth on minimal medium lacking lysine and tryptophan. Positive colonies were screened by PFGE and Southern blot analysis utilizing a probe to the lys2 cassette (data not shown).
Pulse-Field Gel Electrophoresis and Restriction Endonuclease Mapping
High-molecular weight DNA was analyzed and mapped by PFGE and Southern blot analysis, as described (40). Yeast cells were embedded in 1% Seaplaque low-melting point agarose [BioWhittaker Molecular Applications (BMA) Rockland, ME] plugs at 109 cells/ml, and unused plugs were stored at 4 C in 10 mM Tris-HCl, 50 mM EDTA (pH 8.0). The agarose-embedded DNA was washed with 10 mM Tris-HCl, 1 mM EDTA (pH 8.0) (once for 30 min on ice) and equilibrated (two times for 30 min on ice) in 200 μl of the restriction enzyme buffer before digestion with restriction enzymes (New England Biolabs, Beverly, MA). The DNA was fractionated by PFGE (CHEF-DR II, Bio-Rad, Hercules, CA) using the following conditions: 0.5× TBE, 6 V/cm, 40 sec switch time, 24 h, at 14 C. Under these conditions, separation of DNAs up to 610 kb was routinely achieved. The resolved genomic DNA was analyzed by Southern blot hybridization according to standard procedures. Hybridization probes included 5′SF-1, 3′SF-1, URA, TRP, and LYS and were prepared using the random primer labeling system according to the vendor’s recommendations (NEN-New England Nuclear, Boston, MA). DNA for each probe, except LYS, were generated by PCR using YAC DNA or the appropriate plasmid as template and the following primer sets. 5′ SF1, which spans the major transcription start site for SF-1, was generated with the following primers: 5′-CGCGCTCGAGCCCCCATAAAGATAG-GAATA-3′, and 5′-GGGGAAGCTTCTATCGGGCTGTCAG-GAACT-3′. 3′ SF1, which binds to the last exon (exon 7) of SF1, was generated by PCR using primers 5′-GTGAAATTC-CTGAACAACCA-3′ and 5′-GGGGAAGCTTAGTCTGCTTGG CCTGCAGC-3′. URA DNA, to detect the acentric pYAC4 vector arm, was amplified with 5′-GTACCACCAAGGAAT-TACTGG-3′ and 5′-CGGGTGTATACAGAATAGCAG-3′, and TRP DNA, to detect the centric pYAC4 vector arm, was amplified with primer pair 5′-AAATAGTTCAGGCACTCCG-3′ and 5′-TCTGTGAAGCTGCACTGAG-3′. LYS DNA was isolated from the pCH37 by digestion with HindIII and EcoRI (43).
YAC Purification and Screening of Transgenic Mice
For the generation of transgenic mice, YAC DNA was purified as reported (44). In short, ySLH1-Lys was grown and preparative agarose plugs containing high-mass DNA were prepared as described above. The plugs were loaded on a 1% (wt/vol) Seaplaque (BMA) agarose gel and the DNA fractionated by PFGE using the following conditions: 0.5× TBE (44.5 mM Tris, 44.5 mM boric acid, 1 mM EDTA), 6 V/cm, 40-sec switch time, 24 h, at 14 C. A portion of the gel was stained with ethidium bromide to determine the location of ySLH1 and the gel region containing the YAC was cut from the unstained portion of the gel. The YAC-containing gel slice was rotated 90 degrees relative to the original direction of mobility and electrophoresed through a 4% (wt/vol) low melting point agarose gel (Nusieve agarose GTG, BMA) in 0.5× TBE at 50 V for 18 h at 4 C to concentrate the DNA. A gel slice containing the concentrated YAC DNA was excised and equilibrated in 50 ml of injection buffer [10 mM Tris-HCl (pH 7.5), 0.25 mM EDTA, 100 mM NaCl] for 2 h on ice. The gel slice was heated to 68 C for 10 min and digested with β-agarose (4 U/100 mg gel slice) overnight at 42.5 C. YAC DNA was recovered by filtration through a 0.2 μm syringe filter (13-mm Acrodisc; PALL Gelman Sciences, Pawtucket, RI). Isolated DNA, at a final concentration of 1–2 ng/μl, was used to generate transgenic mice (C57B6/SJL) through the Transgenic and Gene-Targeting Institutional Facility at the University of Kansas Medical Center.
The genotypes of all founders and their offspring were determined by PCR of mouse tail DNA. Tail snips were digested in 500 μl lysis buffer [100 mM EDTA, 50 mM Tris (pH 8.0), and 1% SDS] in the presence of 400μl/ml proteinase K (Invitrogen) at 55 C overnight. Three hundred microliters of saturated NaCl (concentration > 6 M) was added and the digests were mixed by inversion, followed by incubation on ice for 10 min. The sample was then centrifuged at 6000 rpm for 10 min and 500 μl of supernatant was precipitated by adding 1 ml of 100% ethanol and mixing by inversion. The precipitated DNA was spooled and rinsed by dipping in a solution of 70% ethanol. DNA was resuspended in 150 μl of TE buffer [10 mM Tris, 1 mM EDTA (pH 8.0)]. Transgene amplification was performed using two pairs of primers specific for rat Ftz-F1: 5′-CTGCACAACCAGCTGGCCC-3′ and 5′-CGGGCTGTCAGGAACTTCTTCC-3′ amplified a 301-bp region located upstream of exon 1, whereas 5′-ACATCTC-CTTCCTGATCATCTGGTC-3′ and 5′-GCTTCAAGGGTT-GTGACAAGGGAC-3′ amplified a 242-bp region positioned downstream of exon 7. Amplification was performed using a touchdown PCR procedure (95 C for 2 min, followed by 94 C for 30 sec, 70 C to 55 C for 30 sec with a 0.5 C/cycle ramp, 72 C for 30 sec, and then 10 cycles of 94 C for 30 sec, 55 C for 30 sec, 72 C for 30 sec, and finally 72 C for 10 min) and Biolase DNA polymerase, according to manufacturer’s recommendation (Bioline USA Inc., Randolph, MA). These primers specifically recognize rat Ftz-F1 and not endogenous mouse Ftz-F1. Animals were bred to the outbred CD1 strain and manipulations were conducted according to the Guide for Care and Use of Experimental Animals.
RNase Protection and RT-PCR
Total RNA was isolated from cells as described (45). For detection of SF-1 by RPA, an RNA probe was generated from rSF1-rev, which was constructed by PCR amplification of exons 1 and 3c [based on the mouse Ftz-F1 gene structure (1)] of rat Ftz-F1 using rat testis cDNA as template, primers SF3 (5′-GGGGTCTAGAAGTTTGCAGTCCGCCGCT-3′) and SF1.7 (5′-CTTGCAGCTCTCGCACGT-3′), and HF Taq polymerase (Invitrogen). The cDNA was cloned into the XbaI site of pGEM4Z (Promega Madison, WI). Antisense RNA probe was generated by linearizing the plasmid with HindIII followed by in vitro transcription using SP6 polymerase, according to the manufacturer’s recommendations (Promega). This produced a 394-base probe that protects 268 bases of rat SF-1 mRNA. This same probe protects a smaller portion of mouse SF-1 mRNA. The actin RNA probe was generated as described previously, resulting in a 188-base antisense transcript that protects 126 bases of actin mRNA (45). Note, that the actin probe was produced to have approximately 50-fold lower specific activity than the SF-1 probe. RNase protection assays were preformed as described elsewhere (45, 46).
RT-PCR was performed, with slight modification, as previously described (47). Briefly, F1 transgenic mice positive for the YAC transgene were killed and total RNA was isolated from various tissues. cDNA was generated using Superscript reverse transcriptase (Invitrogen Life Technologies). The cDNA was used as a template in PCR with intron-spanning primers specific for either rat or mouse SF-1 transcripts or the ribosomal protein L7. Primers used in each reaction are given in Table 3. Amplification of ribosomal protein L7 was used to confirm cDNA synthesis for each sample. Amplified products were examined by agarose gel electrophoresis and Southern blot analysis.
Table 3.
RT-PCR Primers for Expression Analysis
| Gene | Upstream Primer | Sequence | Downstream Primer | Sequence |
|---|---|---|---|---|
| Rat SF-1 | rSF-1For (exon 1) | AGAAGTTCCTGACAGCCCGA | rSF-1Rev (exon 4) | ATAAAGGTCTGGTCCGCCATC |
| Mouse SF-1 | mSF-1For (exon 1) | GAAGTTTCTGAGAGCCCGC | mSF-1Rev (exon 7) | AACTGGAGCACTAACTCTTGG |
| Mouse L7 | L7.1 | GGAAAGGCAAGGAGGAAGCA | L7.2 | TCCTCCATGCAGATGATGC |
YAC Rescue of the SF-1 Knockout Mice
Heterozygous mating pairs containing a targeted mutation that disrupts function of SF-1 [Nr5a1tm1Klp (formerly Ftzf1tm1Klp)] were purchased from The Jackson Laboratory (Bar Harbor, ME; Ref. 3). Heterozygous Nr5a1tm1Klpmice were mated to the line 14 YAC transgenic founder mouse (tg) to generate heterozygous transgenic animals (Ftz-F1+/−; tg+). These animals were bred to generate mice homozygous for the SF-1 null allele and positive for the YAC transgene (Ftz-F1 −/−; tg+). All rescued animals were on a mixture of background strains (129p; C57B/6; DBA/2 from Nr5a1tm1Klp and C57Bl/6; CD1 from transgenics). Animals were genotyped by Southern blot analysis as described (3). Tail DNA was isolated, digested with BglII, and analyzed by Southern blot analysis using a probe generated from an 836-bp DNA fragment just within a BglII boundary of the rat Ftz-F1fourth intron. This probe hybridized to 4300- and 5000-bp bands for the mouse wild-type and mutant alleles, respectively, and to a 3780-bp band for the rat transgene. Wild-type (Ftz-F1+/+), homozygous mutant (Ftz-F1−/−), transgenic (Ftz-F1+/+; tg+), heterozygous transgenic (Ftz-Fl +/−; tg+), and rescued (Ftz-F1−/−; tg+) mice were examined for the presence of adrenals and gonads, tissue morphology, and fertility status. All animals were cared for in accordance with National Institutes of Health guidelines and experimental procedures were approved by the Laboratory Animal Research Committee at the University of Kansas Medical Center. Mice were maintained on a 12-h light, 12-h dark cycle and given food and water ad libitum.
Histology and Immunohistochemistry
Testes, ovaries, hypothalamus, pituitary, and adrenal glands from adult (2- to 4-month-old) mice were rapidly dissected. One testis, ovary, and adrenal from each animal were immersion fixed in Bouin’s solution at 24 C for 16–24 h (depending on tissue size). Tissues were dehydrated with ethanol and embedded in paraffin according to standard procedures. Five-micrometer sections were stained with hematoxylin and eosin. The remaining ovary and adrenal and the pituitary and hypothalamus were frozen immediately after dissection. Ten-micrometer sections were cut and immunofluorescently labeled for SF-1, using a 1:2000 dilution of a polyclonal rabbit antibody directed against bovine SF-1 (generously provided by Dr. K. Morohashi, Division for Sex Differentiation, National Institute for Basic Biology, National Institutes of Natural Sciences, Myodaiji-Cho, Okazaki 444-8787, Japan). For pituitary analysis, SF-1 and LHβ double immunostaining was performed using guinea pig, antirat LHβ antiserum (National Hormone and Pituitary Program) at a 1:50 dilution in combination with 1:2000 dilution of anti-SF-1 (described above). Tissue sections were fixed in methanol-acetone (1:1) at −20 C for 10 min, rehydrated, incubated with blocking solution for 1 h, and then incubated with primary antibody overnight. Tetramethyl rhodamine isothiocyanate-conjugated goat anti-rabbit secondary antibody (The Jackson Laboratory) was used to visualize the SF-1 signal. Cy2-conjugated donkey antiguinea pig secondary antibody (The Jackson Laboratory) was used to detect LHβ expression. The combined secondary antibodies were applied at a dilution of 1:200 for 1 h at room temperature.
For immunofluorescent analysis of the testes, tissues were immersion fixed for 16 h at +4 C in freshly prepared 4% paraformaldehyde buffered with PBS (pH 7.4), ethanol dehydrated, and embedded in paraffin according to standard procedures. Five-micrometer sections were generated and, after paraffin removal, were subjected to antigen retrieval by submerging sections in 0.01 M citrate buffer (pH 6) and microwaving (full power) for 8 min. Sections were washed in PBS-T and immunofluorescence for SF-1 performed as described above.
Whole Genome Alignment Analysis
Genome sequence alignment for rat, human and chicken Nr5a1 was performed and visualized using the ECR browser navigation tool (http://ecrbrowser.dcode.org), which provides a dynamic conservation profile based on a series of pairwise genome alignments (34). With the human genome as the base sequence, the parameters were set at 75% identity over a length of 100 bp. Conserved, noncoding regions are noted by colored peaks.
Mapping of Integrated Transgenes
High-molecular-weight mouse genomic DNA was isolated from livers of wild-type, line 14, line 7, and line 15 YAC transgenic animals and analyzed by PFGE and Southern blot hybridization. Single cell liver suspension was prepared as described (48). Livers were cut into small pieces that were sheared mechanically by sequential, repeated passage through 16-, 18-, and 20-gauge needles. The cells were washed twice with Dulbecco’s PBS and resuspended at a concentration of 3 × 107 cells/ml and embedded in 1% (wt/vol) low melting temperature agarose (SeaPlaque agarose (BMA) in PBS was added to the liver suspensions and these suspensions were cast into plugs in prechilled molds (Bio-Rad, Hercules, CA). The plugs were incubated in 50 ml of LDS solution [1%(wt/vol) lithium dodecyl sulfate, 100 mM EDTA, 10 mM Tris-HCl (pH 8.0)] at 37 C for 1 h, the LDS solution was changed and plugs were incubated overnight at 37 C. Plugs were then washed twice for 30 min in 0.2 × NDS [0.2% (wt/vol) lauryl sarcosine, 100 mM EDTA, 2 mM Tris base (pH 9.5)], followed by three 30-min washes in TE (pH 8.0) [10 mM Tris-HCl (pH 8.0), 1 mM EDTA]. The plugs were stored in TE at +4 C. Before digestion with restriction enzymes eight 4-× 2-mm slices of agarose plug were washed in TE, pH 8.0 (once for 30 min on ice) equilibrated in 200 μl of appropriate enzyme buffer (two times for 30 min on ice) followed by o/n incubation in the presence of 40 U of the enzyme (New England Biolabs) at +4 C. Plugs were then digested at the appropriate enzymatic temperature for 24 h with an additional 50 U of enzyme. The digested DNA was fractionated by PFGE using the following conditions: 0.5× TBE, 6 V/cm, initial switching time 1 sec, final switching time 25 sec, 22 h, at +14 C. DNA fragments of 9–400 kb were resolved under these conditions. Southern blot analysis was performed using various hybridization probes spanning ySLH1. Probe positions are indicated in Fig. 8 and were prepared by radiola-beling PCR amplified DNA. Specifics of each probe and the employed primers can be obtained by contacting the authors.
Acknowledgments
We would like to thank Dr. Paul Terranova (Department of Molecular and Integrative Physiology, University of Kansas Medical Center, Kansas City, KS) for his insightful suggestions and generous support and the National Hormone and Pituitary Program for antirat LHβ antisera.
Footnotes
This work was supported by National Institutes of Health Grant R01 HD38498 (to L.L.H.) and a Specialized Cooperative Center Program in Reproduction Research (U54 HD 33994 to Paul Terranova) from the National Institute of Child Health and Human Development.
Current address for S.P.S.: Affymetrix UK, Ltd., Voyager, Mercury Park, Wycombe Lane, Wooburn Green, High Wycombe HP100HH, United Kingdom.
References
- 1.Ninomiya Y, Okada M, Kotomura N, Suzuki K, Tsukiyama T, Niwa O. Genomic organization and isoforms of the mouse ELP gene. J Biochem (Tokyo) 1995;118:380–389. doi: 10.1093/oxfordjournals.jbchem.a124918. [DOI] [PubMed] [Google Scholar]
- 2.Parker KL, Schimmer BP. Steroidogenic factor 1: a key determinant of endocrine development and function. Endocr Rev. 1997;18:361–377. doi: 10.1210/edrv.18.3.0301. [DOI] [PubMed] [Google Scholar]
- 3.Luo X, Ikeda Y, Parker KL. A cell-specific nuclear receptor is essential for adrenal and gonadal development and sexual differentiation. Cell. 1994;77:481–490. doi: 10.1016/0092-8674(94)90211-9. [DOI] [PubMed] [Google Scholar]
- 4.Sadovsky Y, Crawford PA, Woodson KG, Polish JA, Clements MA, Tourtellotte LM, Simburger K, Milbrandt J. Mice deficient in the orphan receptor steroidogenic factor 1 lack adrenal glands and gonads but express P450 side-chain-cleavage enzyme in the placenta and have normal embryonic serum levels of corticosteroids. Proc Natl Acad Sci USA. 1995;92:10939–10943. doi: 10.1073/pnas.92.24.10939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shinoda K, Lei H, Yoshii H, Nomura M, Nagano M, Shiba H, Sasaki H, Osawa Y, Ninomiya Y, Niwa O. Developmental defects of the ventromedial hypothalamic nucleus and pituitary gonadotroph in the Ftz-F1 disrupted mice. Dev Dyn. 1995;204:22–29. doi: 10.1002/aja.1002040104. [DOI] [PubMed] [Google Scholar]
- 6.Sugawara T, Kiriakidou M, McAllister JM, Holt JA, Arakane F, Strauss JF. Regulation of expression of the steroidogenic acute regulatory protein (StAR) gene: a central role for steroidogenic factor 1. Steroids. 1997;62:5–9. doi: 10.1016/s0039-128x(96)00152-3. [Erratum (1997) 62:395] [DOI] [PubMed] [Google Scholar]
- 7.Sugawara T, Kiriakidou M, McAllister JM, Kallen CB, Strauss JF. Multiple steroidogenic factor 1 binding elements in the human steroidogenic acute regulatory protein gene 5′-flanking region are required for maximal promoter activity and cyclic AMP responsiveness. Biochemistry. 1997;36:7249–7255. doi: 10.1021/bi9628984. [DOI] [PubMed] [Google Scholar]
- 8.Wolfe MW. The equine luteinizing hormone β-subunit promoter contains two functional steroidogenic factor-1 response elements. Mol Endocrinol. 1999;13:1497–1510. doi: 10.1210/mend.13.9.0345. [DOI] [PubMed] [Google Scholar]
- 9.Keri RA, Nilson JH. A steroidogenic factor-1 binding site is required for activity of the luteinizing hormone B subunit promoter in gonadotropes of transgenic mice. J Biol Chem. 1996;271:10782–10785. doi: 10.1074/jbc.271.18.10782. [DOI] [PubMed] [Google Scholar]
- 10.Dorn C, Ou Q, Svaren J, Crawford PA, Sadovsky Y. Activation of luteinizing hormone β gene by gonadotropin-releasing hormone requires the synergy of early growth response-1 and steroidogenic factor-1. J Biol Chem. 1999;274:13870–13876. doi: 10.1074/jbc.274.20.13870. [DOI] [PubMed] [Google Scholar]
- 11.Tremblay JJ, Drouin J. Egr-1 is a downstream effector of GnRH and synergizes by direct interaction with Ptx1 and SF-1 to enhance luteinizing hormone β gene transcription. Mol Cell Biol. 1999;19:2567–2576. doi: 10.1128/mcb.19.4.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halvorson LM, Ito M, Jameson JL, Chin WW. Steroidogenic factor-1 and early growth response protein 1 act through two composite DNA binding sites to regulate luteinizing hormone β-subunit gene expression. J Biol Chem. 1998;273:14712–14720. doi: 10.1074/jbc.273.24.14712. [DOI] [PubMed] [Google Scholar]
- 13.Ito M, Park Y, Weck J, Mayo KE, Jameson JL. Synergistic activation of the inhibin α-promoter by steroidogenic factor-1 and cyclic adenosine 3′,5′-mono-phosphate. Mol Endocrinol. 2000;14:66–81. doi: 10.1210/mend.14.1.0410. [DOI] [PubMed] [Google Scholar]
- 14.Barnhart KM, Mellon PL. The orphan nuclear receptor, steroidogenic factor-1, regulates the glycoprotein hormone α-subunit gene in pituitary gonadotropes. Mol Endocrinol. 1994;8:878–885. doi: 10.1210/mend.8.7.7527122. [DOI] [PubMed] [Google Scholar]
- 15.Heckert LL. Activation of the rat follicle-stimulating hormone receptor promoter by steroidogenic factor 1 is blocked by protein kinase a and requires upstream stimulatory factor binding to a proximal E box element. Mol Endocrinol. 2001;15:704–715. doi: 10.1210/mend.15.5.0632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levallet J, Koskimies P, Rahman N, Huhtaniemi I. The promoter of murine follicle-stimulating hormone receptor: functional characterization and regulation by transcription factor steroidogenic factor 1. Mol Endocrinol. 2001;15:80–92. doi: 10.1210/mend.15.1.0583. [DOI] [PubMed] [Google Scholar]
- 17.Ngan ES, Cheng PK, Leung PC, Chow BK. Steroidogenic factor-1 interacts with a gonadotrope-specific element within the first exon of the human gonadotropin-releasing hormone receptor gene to mediate gonadotrope-specific expression. Endocrinology. 1999;140:2452–2462. doi: 10.1210/endo.140.6.6759. [DOI] [PubMed] [Google Scholar]
- 18.Duval DL, Nelson SE, Clay CM. A binding site for steroidogenic factor-1 is part of a complex enhancer that mediates expression of the murine gonadotropin-releasing hormone receptor gene. Biol Reprod. 1997;56:160–168. doi: 10.1095/biolreprod56.1.160. [DOI] [PubMed] [Google Scholar]
- 19.Morohashi KI, Omura T. Ad4BP/SF-1, a transcription factor essential for the transcription of steroidogenic cytochrome P450 genes and for the establishment of the reproductive function. FASEB J. 1996;10:1569–1577. doi: 10.1096/fasebj.10.14.9002548. [DOI] [PubMed] [Google Scholar]
- 20.Sadovsky Y, Crawford PA. Developmental and physiologic roles of the nuclear receptor steroidogenic factor-1 in the reproductive system. J Soc Gynecol Investig. 1998;5:6–12. doi: 10.1016/s1071-5576(97)00096-8. [DOI] [PubMed] [Google Scholar]
- 21.Hatano O, Takayama K, Imai T, Waterman MR, Takakusu A, Omura T, Morohashi K. Sex-dependent expression of a transcription factor, Ad4BP, regulating steroidogenic P-450 genes in the gonads during prenatal and postnatal rat development. Development. 1994;120:2787–2797. doi: 10.1242/dev.120.10.2787. [DOI] [PubMed] [Google Scholar]
- 22.Ikeda Y, Shen W-H, Ingraham HA, Parker KL. Developmental expression of mouse steroidogenic factor-1, an essential regulator of the steroid hydroxylases. Mol Endocrinol. 1994;8:654–662. doi: 10.1210/mend.8.5.8058073. [DOI] [PubMed] [Google Scholar]
- 23.Ingraham HA, Lala S, Ikeda Y, Luo X, Shen W-H, Nachtigal MW, Abbud R, Nilson JH, Parker KL. The nuclear receptor steroidogenic factor 1 acts at multiple levels of the reproductive axis. Genes Dev. 1994;8:2303–2312. doi: 10.1101/gad.8.19.2302. [DOI] [PubMed] [Google Scholar]
- 24.Ikeda Y, Luo X, Abbud R, Nilson JH, Parker KL. The nuclear receptor steroidogenic factor 1 is essential for the formation of the ventromedial hypothalamic nucleus. Mol Endocrinol. 1995;9:478–486. doi: 10.1210/mend.9.4.7659091. [DOI] [PubMed] [Google Scholar]
- 25.Morohashi K, Tsuboi-Asai H, Matsushita S, Suda M, Nakashima M, Sasano H, Hataba Y, Li CL, Fukata J, Irie J, Watanabe T, Nagura H, Li E. Structural and functional abnormalities in the spleen of an mFtz-F1 gene-disrupted mouse. Blood. 1999;93:1586–1594. [PubMed] [Google Scholar]
- 26.Achermann JC, Ito M, Hindmarsh PC, Jameson JL. A mutation in the gene encoding steroidogenic factor-1 causes XY sex reversal and adrenal failure in humans. Nat Genet. 1999;22:125–126. doi: 10.1038/9629. (Letter) [DOI] [PubMed] [Google Scholar]
- 27.Scherrer SP, Rice DA, Heckert LL. Expression of steroidogenic factor 1 in the testis requires an interactive array of elements within its proximal promoter. Biol Reprod. 2002;67:1509–1521. doi: 10.1095/biolreprod.102.006932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daggett MA, Rice DA, Heckert LL. Expression of steroidogenic factor 1 in the testis requires an E box and CCAAT box in its promoter proximal region. Biol Reprod. 2000;62:670–679. doi: 10.1095/biolreprod62.3.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woodson KG, Crawford PA, Sadovsky Y, Milbrandt J. Characterization of the promoter of SF-1, an orphan nuclear receptor required for adrenal and gonadal development. Mol Endocrinol. 1997;11:117–126. doi: 10.1210/mend.11.2.9881. [DOI] [PubMed] [Google Scholar]
- 30.Nomura M, Bartsch S, Nawata H, Omura T, Morohashi K. An E box element is required for the expression of the ad4bp gene, a mammalian homologue of ftz-f1 gene, which is essential for adrenal and gonadal development. J Biol Chem. 1995;270:7453–7461. doi: 10.1074/jbc.270.13.7453. [DOI] [PubMed] [Google Scholar]
- 31.Wilhelm D, Englert C. The Wilms tumor suppressor WT1 regulates early gonad development by activation of Sf1. Genes Dev. 2002;16:1839–1851. doi: 10.1101/gad.220102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stallings NR, Hanley NA, Majdic G, Zhao L, Bakke M, Parker KL. Development of a transgenic green fluorescent protein lineage marker for steroidogenic factor 1. Mol Endocrinol. 2002;16:2360–2370. doi: 10.1210/me.2002-0003. [DOI] [PubMed] [Google Scholar]
- 33.Zhao L, Bakke M, Krimkevich Y, Cushman LJ, Parlow AF, Camper SA, Parker KL. Steroidogenic factor 1 (SF1) is essential for pituitary gonadotrope function. Development. 2001;128:147–154. doi: 10.1242/dev.128.2.147. [DOI] [PubMed] [Google Scholar]
- 34.Ovcharenko I, Nobrega MA, Loots GG, Stubbs L. ECR Browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res. 2004;32:W280–W286. doi: 10.1093/nar/gkh355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaboissier MC, Kobayashi A, Vidal VI, Lutzkendorf S, Van De Kant HJ, Wegner M, de Rooij DG, Behringer RR, Schedl A. Functional analysis of Sox8 and Sox9 during sex determination in the mouse. Development. 2004;131:1891–1901. doi: 10.1242/dev.01087. [DOI] [PubMed] [Google Scholar]
- 36.Hatano O, Takakusu A, Nomura M, Morohashi K. Identical origin of adrenal cortex and gonad revealed by expression profiles of Ad4BP/SF-1. Genes Cells. 1996;1:663–671. doi: 10.1046/j.1365-2443.1996.00254.x. [DOI] [PubMed] [Google Scholar]
- 37.Falender AE, Lanz R, Malenfant D, Belanger L, Richards JS. Differential expression of steroidogenic factor-1 and FTF/LRH-1 in the rodent ovary. Endocrinology. 2003;144:3598–3610. doi: 10.1210/en.2002-0137. [DOI] [PubMed] [Google Scholar]
- 38.Couronne O, Poliakov A, Bray N, Ishkhanov T, Ryaboy D, Rubin E, Pachter L, Dubchak I. Strategies and tools for whole-genome alignments. Genome Res. 2003;13:73–80. doi: 10.1101/gr.762503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Green ED, Hieter P, Spencer FA 1997 Yeast artificial chromosomes. In: Birren B, Green ED, Klapholz S, Myers RM, Roskams J, eds. Genome analysis; a laboratory manual. 1st ed. Plainview, NY: Cold Spring Harbor Laboratory Press; 297–565
- 40.Riethman H, Birren B, Gnirke A 1997 Preparation, manipulation, and mapping of HMW DNA. In: Birren B, Green ED, Klapholz S, Myers RM, Roskams J, eds. Genome analysis; a laboratory manual. 1st ed. Plainview, NY: Cold Spring Harbor Laboratory Press; 83–248
- 41.Gnirke A, Huxley C, Peterson K, Olson MV. Micro-injection of intact 200- to 500-kb fragments of YAC DNA into mammalian cells. Genomics. 1993;15:659–667. doi: 10.1006/geno.1993.1121. [DOI] [PubMed] [Google Scholar]
- 42.Haldi ML, Lim P, Kaphingst K, Akella U, Whang J, Lander ES. Construction of a large-insert yeast artificial chromosome library of the rat genome. Mamm Genome. 1997;8:284. doi: 10.1007/s003359900412. [DOI] [PubMed] [Google Scholar]
- 43.Poorkaj P, Peterson KR, Schellenberg GD. Single-step conversion of P1 and P1 artificial chromosome clones into yeast artificial chromosomes. Genomics. 2000;68:106–110. doi: 10.1006/geno.2000.6267. [DOI] [PubMed] [Google Scholar]
- 44.Peterson KR, Li QL, Clegg CH, Furukawa T, Navas PA, Norton EJ, Kimbrough TG, Stamatoyannopoulos G. Use of yeast artificial chromosomes (YACs) in studies of mammalian development: production of β-globin locus YAC mice carrying human globin developmental mutants. Proc Natl Acad Sci USA. 1995;92:5655–5659. doi: 10.1073/pnas.92.12.5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen JK, Heckert LL. Dmrt1 expression is regulated by follicle-stimulating hormone and phorbol esters in postnatal Sertoli cells. Endocrinology. 2001;142:1167–1178. doi: 10.1210/endo.142.3.8021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harju S, Peterson KR. Sensitive ribonuclease protection assay employing glycogen as a carrier and a single inactivation/precipitation step. Biotechniques. 2001;30:1198–1200. doi: 10.2144/01306bm02. 1202–1204. [DOI] [PubMed] [Google Scholar]
- 47.Heckert LL, Sawadogo M, Daggett MA, Chen JK. The USF proteins regulate transcription of the follicle-stimulating hormone receptor but are insufficient for cell-specific expression. Mol Endocrinol. 2000;14:1836–1848. doi: 10.1210/mend.14.11.0557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Navas PA, Peterson KR, Li Q, McArthur M, Stamatoyannopoulos G. The 5′HS4 core element of the human β-globin locus control region is required for high-level globin gene expression in definitive but not in primitive erythropoiesis. J Mol Biol. 2001;312:17–26. doi: 10.1006/jmbi.2001.4939. [DOI] [PubMed] [Google Scholar]


