Abstract
Steroidogenic factor 1 (SF-1, Nr5a1, and Ad4bp) is an orphan nuclear receptor required for adrenal and gonad development and endocrine regulation. To extend our understanding of SF-1 function and the mechanisms controlling its expression, a transgenic rescue strategy was employed to locate important transcriptional control regions and to reveal functional roles of the protein. A rat yeast artificial chromosome containing Ftz-F1, the gene encoding SF-1, was used to generate mice with different transgenes that varied in size. Rat SF-1 mRNA expression was assayed to assess each transgene’s targeting ability. SF-1-deficient/transgene-positive (SF-1−/−; tg/+) “rescue” mice were then generated and the animals’ developmental and reproductive status was evaluated. The results identified differences in expression patterns and rescue abilities that provided insight into SF-1 transcriptional control and function. Comparing transgene maps and mRNA profiles placed critical transcriptional elements for pituitary and hypothalamic expression to a region 3′ to intron 4, whereas examination of rescued mice revealed that an ~153-kb region of the Ftz-F1 locus recapitulates most or all activity ascribed to the endogenous allele. A second line of rescued mice was hypomorphic, with males showing defects in androgen-dependent tissues due to abnormal Leydig cell differentiation. Histological analysis of embryonic (e14.5) and adult testes from these mice implicated SF-1 in roles that are distinct in fetal and adult Leydig cells.
Keywords: SF-1, testes, adrenal, YAC transgenic mice, steroidogenesis
INTRODUCTION
SF-1 is an orphan nuclear receptor encoded by the Ftz-F1, which is evolutionarily conserved among vertebrates and invertebrates. Structural analysis of the mouse Ftz-F1 gene indicated four distinct transcripts, ELP1, ELP2, ELP3, and SF-1, which differ by their inclusion of alternative exons and use of different promoters.1 The most investigated transcript in humans and rodents is the SF-1, which regulates endocrine homeostasis via its role as a transcription factor for genes encoding various steroid-producing enzymes (P450SCC, P450arom, StAR, and αGSU) as well as steroidogenesis-regulating hormones (LHβ, GnRH, and inhibin α) and their cognate receptors (GnRHR and FSHR).2 An essential role for SF1 in embryonic development was uncovered after generations of SF-1 knockout mice showed deficiencies in adrenal and gonadal development.3 Absence of adrenal steroids in SF-1 null mice resulted in their death shortly after birth, and the lack of gonadal steroids during embryonic development resulted in male-to-female sex reversal. The mice were also characterized by the absence of a ventromedial nucleus of the hypothalamus and impaired expression of genes within gonadotropes of the pituitary.
SF-1 expression sites correlate with their steroidogenic function. SF-1 is expressed by all primary steroidogenic cells, such as cortical cells of the adrenal and Leydig and theca cells of the gonads, and by tissues such as the ventromedial nucleus of the hypothalamus and gonadotropes of the pituitary.4 Its expression is also age dependent and demonstrates developmental regulation.5 Onset of SF-1 expression occurs at e9.5 within the cells of the urogenital ridge that gives rise to gonads and adrenal glands. After sex determination is initiated at e12.5, SF-1 expression is downregulated in the ovary and persists within the testes, where it is thought to contribute to sexual differentiation in the male.
However, because the gonads fail to develop past e10 in knockout mice, it has been difficult to evaluate SF-1’s role in the later events of testes development and function. In order to make a correlation between physiological processes and molecular pathways regulating SF-1 function, we developed a transgenic approach that employs a yeast artificial chromosome (YAC) containing the Ftz-F1 together with a significant amount of the 5′ and 3′ flanking sequence. Several lines of transgenic mice, varying in transgene size, were generated and characterized for expression profiles. Subsequently, selected transgenes were introduced into the SF-1 null mouse background to obtain transgene positive (tg/+), SF-1 negative (SF-1−/−) rescue mice (SF-l−/−;tg/+). Characterization of the rescue mice revealed differences in SF-1 expression profiles and rescue abilities between different lines. The studies established a region encompassing Ftz-F1 that is required for proper expression of SF-1 and revealed a role for SF-1 in Leydig cell differentiation.
METHODS
Rat Ftz-F1 Containing YAC
DNA pools of a rat YAC library constructed by the Whitehead Institute for Biomedical Research and MIT Center for Genome Research were screened for the presence of rat Ftz-F1. One of the identified Ftz-F1 -containing YACs, clone 385G2, was purchased from Invitrogen Life Technology. YAC clone ySLH1 was generated by subcloning 500-kb Ftz-F1 containing NotI fragment, as described earlier.6
Generation and Screening of Transgenic Mice
YAC DNA was purified for pro-nuclear microinjection as reported.7 Isolated DNA, at a concentration of 1–2 ng/L, was used to generate transgenic mice (C57B6/SJL mouse strain) through the Transgenic and Gene-Targeting Institutional Facility at the University of Kansas Medical Center. Three separated YAC transgenic mouse lines, 7, 14, and 15, were produced. Animals were genotyped for the presence of the YAC transgene by PCR of tail DNA, as described.6 All animal manipulations were conducted according to the Guide for Care and Use of Experimental Animals.
YAC Rescue of the SF1 Knockout Mice
Heterozygous breeder pairs containing a targeted mutation that disrupts the function of SF-1 (Nr5a1tm1K1p, formerly Ftzf1tm1K1p)3 were purchased from The Jackson Laboratory (Bar Harbor, Maine). SF-1+/− mice were mated to transgenic mice to generate SF-1 heterozygous transgene-positive animals (SF-1+/−;tg/+). These animals were bred to generate rescue mice that were homozygous null for SF-1 and positive for the YAC transgene (SF-1−/−; tg/+). Rescue animals containing only one YAC transgene were selected for characterization. Animals were genotyped using DNA isolated from either tail biopsy (postnatal) or isolated yolk sac (embryo), as described.6
Generation of Liver Plugs from Transgenic Mice and Mapping of Endogenous Transgenes
High molecular weight mouse genomic DNA was isolated from transgenic mice and analyzed by pulse field gel electrophoresis and Southern blot hybridization, as described.8 Agarose plugs containing high molecular weight DNA were prepared from single-cell liver suspensions at a concentration of 3 × 107 cells/ml and digested with restriction enzyme Pme I. Digested DNA was fractionated by PFGE and analyzed by Southern blot analysis with different probes spanning ySLH1 (P .01; 5′SF1; S31#3; 3′SF1; SPH10; P400) (Fig. 1B).
RT-PCR
Transgenic mice and negative littermates were sacrificed and total RNA isolated from various tissues, as described elsewhere.9 Complementary DNA was generated and used as a template in PCR with primers specific for either rat or mouse SF-1 cDNAs or ribosomal protein L7, as a control for cDNA synthesis. Specifics of the methods are described elsewhere.10
Immunohistochemistry
Whole embryos or testes from adult animals were rapidly dissected and fixed in freshly prepared 4% paraformaldehyde solution prepared with phosphate-buffered saline (pH 7.4), followed by dehydration and paraffin embedding, according to standard procedures. Sections 5 μ were immunofluorescently labeled for SF-1, using a 1:2000 dilution of polyclonal rabbit primary antibody (generously provided by K. Morohashi), or for P450SCC (generously provided by M.J. Soares), using 1:200 dilution of rabbit antiserum. TRITC-conjugated goat anti-rabbit secondary antibody (The Jackson Laboratory, Bar Harbor, Maine) was used to visualize SF-1 and P450SCC signals. For SF-1 analysis, hypothalamus, pituitary, adrenal glands, and gonads were also collected and processed as just described.
RESULTS
To evaluate transcriptional regulation of SF-1 in vivo, we employed YACs as transgenes and monitored transgene expression in the embryo and postnatal animals. Several Ftz-F1 -containing rat YACs were identified, and one was chosen for further characterization (ySLH1). Within ySLH1, Ftz-F1 is centrally located and flanked by genes encoding a second orphan nuclear receptor, germ-cell nuclear factor (Gcnf), and proteasome (prosome, macropain) subunit beta type 7 (Psmb7, Fig. 1 A). Three lines of transgenic mice carrying the YAC were generated, and structural characterization of the integrated transgenes was performed prior to functional analysis. Notably, fragmentation of YAC DNA during integration provided several lines with different transgene sizes (Fig. 1B). Lines 7 and 14 contained a fragment of approximately 153 kb, with breakpoints not fully defined but located near the outer regions specified by the two Pme I sites, whereas line 15 contained a transgene that lost a significant amount of Ftz-F1 encoding sequence terminated at the end of intron 4 (Fig. 1B). Two transgene segments containing a full copy of Ftz-F1 were integrated in line 14, whereas only one full copy of the gene was found in line 7.
For each line the expression profile for SF-1 was compared with that for endogenous SF-1. RT-PCR analysis was performed using rat-specific primers that detect only transgenic, not endogenous, SF-1 transcripts. RT-PCR and RPA analysis in lines 7 and 14 revealed expression of the rat transgene in all SF-1–positive tissues such as testes, ovaries, adrenal, pituitary, hypothalamus, and spleen, and no expression was observed in SF-1–negative tissues (Fig. 2 and data not shown). By contrast, no transgene expression was observed in the VMH or pituitary in line 15 (Fig. 2), suggesting that sequences lost from the transgene’s 3′ are needed to fulfill this expression pattern.
Although RT-PCR and RPA assays indicated correct tissue-specific expression of the transgenes in lines 7 and 14, temporal and cellular regulatory profiles and physiological changes of SF-1 could not be accurately measured in these mice. Therefore, to extend characterization of transgene function, lines 7 and 14 transgenes were introduced into a SF-1 null background and rescue mice, which lacked endogenous SF-1 but contained the transgene, were obtained. The line 14 transgene was able to completely rescue the null phenotype, as SF1−/−;tg/+mice were healthy with normally developed adrenal glands and gonads (Fig. 3A and data not shown). Reproductive performance of both sexes among line 14 rescue mice was indistinguishable from that of WT animals, including similar litter size and litter numbers within the reproductive lifetime period (Table 1). Further examination of transgene function was performed using immunodetection of transgenic SF-1 protein within steroidogenic tissues in rescue mice lacking endogenous protein production. Comparison of SF-1 expression between rescued and control animals revealed that SF-1 expressed from the transgene follows the same cellular profile as that of the endogenous SF-1 (Fig. 3C and data not shown).6
In contrast to line 14, the line 7 transgene was unable to rescue all defects of the characterized SF-1 null mice, demonstrating reduced reproductive success in females and complete infertility in males. Line 7 rescue male mice presented with reduced testis size and hypoplastic or completely undeveloped accessory sexual organs, and some were characterized with feminized external genitalia and cryptorchid testes (Fig. 3B and data not shown). Evaluation of testes at different stages of development, beginning at e14.5, revealed a delay of fetal Leydig cell differentiation. Immunohistochemical analysis of P450SCC at e14.5 in rescued animals showed a significant deficiency in its expression, indicating a lack of functional Leydig cells at this time point (Fig. 4A and B). Interestingly, by postnatal day 0, P450SCC expression was similar to that of the control animals, indicating a delay rather than a block in development of the fetal Leydig cells (Fig. 4C and D). Surprisingly, the Leydig cell population in adult rescue testes was markedly reduced compared to that of control animals, yet P450SCC expression in the remaining Leydig cells appeared normal (Fig. 4E and F). Thus, insufficient expression of SF-1 from the line 7 transgene created a hypomorphic phenotype with defects in both fetal and adult Leydig cells and revealed distinct roles for SF-1 in the establishment of the fetal and adult Leydig cell population.
DISCUSSION
We have demonstrated the successful use of YAC transgenesis to evaluate structural sequence requirements for SF-1 transcription. Our study established that ~153-kb sequence is sufficient for accurate SF-1 expression in all SF-1–positive sites. Previous transgenic studies devoted to the identification of such sequence requirements were unable to direct transgenic SF-1 expression to all appropriate steroidogenic tissues. One transgenic mouse study, using a 674-bp SF-1 promoter directing a Lac-Z reporter, demonstrated an expression profile limited to the gonad.11 Another mouse study employed a 50-kb region of the Ftz-F1 locus and demonstrated correct expression in most, but not all, SF-1–positive tissues,12 further supporting a role for distal regulatory regions in the multilevel complex profile of SF-1 expression. In an attempt to include all required regulatory sequences, we employed a YAC transgenic system, which easily accommodates large amounts of genomic sequence. The study revealed a genomic region that recapitulates SF-1 expression to all appropriate steroidogenic tissues, resolving a limitation of the previous transgenic studies. Interestingly, analysis of line 15 revealed a loss of transgene sequence that correlated to expression deficiencies. Further analysis to identify the sequences lost in line 15 determined a regulatory region responsible for hypothalamic and pituitary expression of SF-1. Comparison of the integrated transgenes in lines 7, 14, and 15 placed these elements in a region downstream of the 3′ end in intron 4.
Transfer of the YAC transgene into the SF-1 null mouse background extended analysis of SF-1 into a more physiological context and opened new possibilities to perform in vivo functional analyses that are limited in the knockout mice, as they lack steroidogenic organs and die shortly after birth. Thus, in the transgenic rescue model, the developmental and physiological ramifications of altered SF-1 levels can be evaluated. In the case of line 7 rescue mice, the hypomorphic phenotype allowed for in vivo assessment of SF-1 function in Leydig cell development. Notably, the phenotype is due to altered SF-1 activity and not to the effects of additional genes within the YAC, as no phenotypic changes were associated with a second YAC rescue model and neither were they present in transgenic mice in the wild-type background. Examination of line 7 hypomorphic mice allowed evaluation of Leydig cell development, which indicated distinct roles for SF-1 in the establishment of fetal and adult Leydig cell populations. Thus, in the SF-1 hypomorphic mice, fetal Leydig cells were developmentally delayed, whereas adult Leydig cells were either blocked at a required proliferation stage or absent altogether. The different Leydig cell defects suggest several interesting hypotheses regarding SF-1 function in Leydig cells and the relation between fetal and adult populations. Thus, the phenotype may reflect a single Leydig precursor cell that forms both adult and fetal populations, and SF-1’s role in the cell’s development depends on the animal’s developmental stage. Alternatively, the two Leydig populations may derive from distinct precursors in which SF-1 acts differently. Additional studies are ongoing to distinguish these possibilities and further our understanding of SF-1 in Leydig cell development.
Although it is tempting to speculate that the phenotype observed is due to the intrinsic effects of SF-1 in Leydig cells, the potential role of Sertoli cells, which also express SF-1, in this process cannot be overlooked. Thus, Sertoli cells, through production of desert hedgehog, are known to direct development of fetal Leydig cells.13 Currently, it is not known if a similar Sertoli cell/Leydig cell relationship exists in the development of adult Leydig cells. It is now critical to extend these studies and distinguish between the intrinsic (Leydig) and extrinsic (Sertoli) roles of SF-1 in Leydig cell development. Further evaluation of hypomorphic mice will likely provide the opportunity to do this.
FIGURE 1.
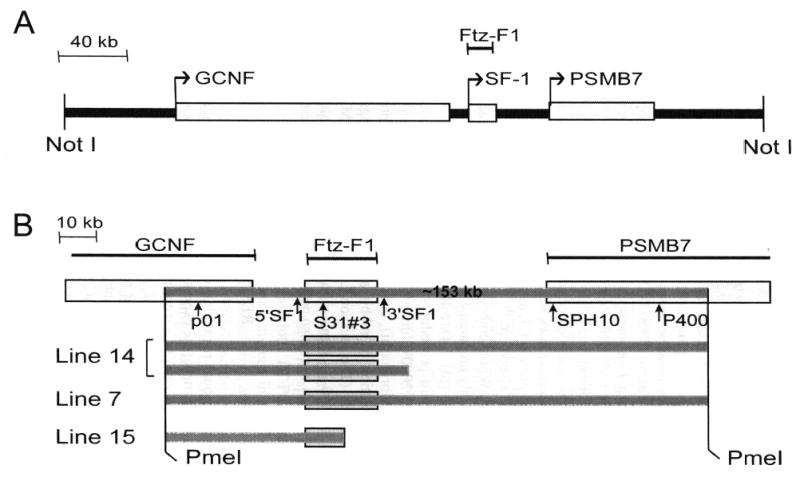
Structural characterization of the Ftz-F1-containing YAC, ySLH1. (A) Schematic depiction of ySLH1. The gene encoding SF-1, Ftz-F1, is centrally located and flanked by genes for proteosome protein PSMB7 and the orphan nuclear receptor GCNF. Genes are represented by rectangles and the NotI restriction endonuclease used in cloning ySHL1 noted. (B) Schematic representation of the YAC transgenes mapped within transgenic lines 7, 14, and 15. Positions of genes (white bars), Pme I restriction sites, and hybridization probes used in mapping analysis (p01, 5′SF1, S31#3, 3′SF1, SPH10, P400) are indicated on top. Gray bars denote the sequences integrated into each line, with the top one marking the ~153-kb fragment established as sufficient for SF-1 expression.
FIGURE 2.
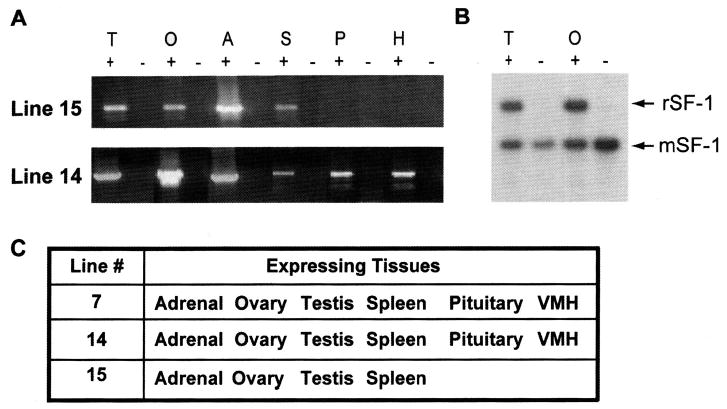
Transgene expression profile for lines 7, 14, and 15. (A) RNA isolated from transgene positive (+) and negative (−) tissues was analyzed for rat SF-1 mRNA in lines 14 and 15, by RT-PCR. Tissues are: testis (T), ovary (O), adrenal (A), spleen (S), pituitary (P), and hypothalamus (H). (B) Ribonuclease protection analysis from line 14 mice, showing levels of transgenic SF-1 (rSF-1) and endogenous SF-1 (mSF-1) mRNAs in testes and ovaries transgenic mice (+) and their negative (−) littermates. (C) Summary table for transgene expression in lines 7, 14, and 15. Tissues positive for transgene expression are indicated in the column next to their respective lines.
FIGURE 3.
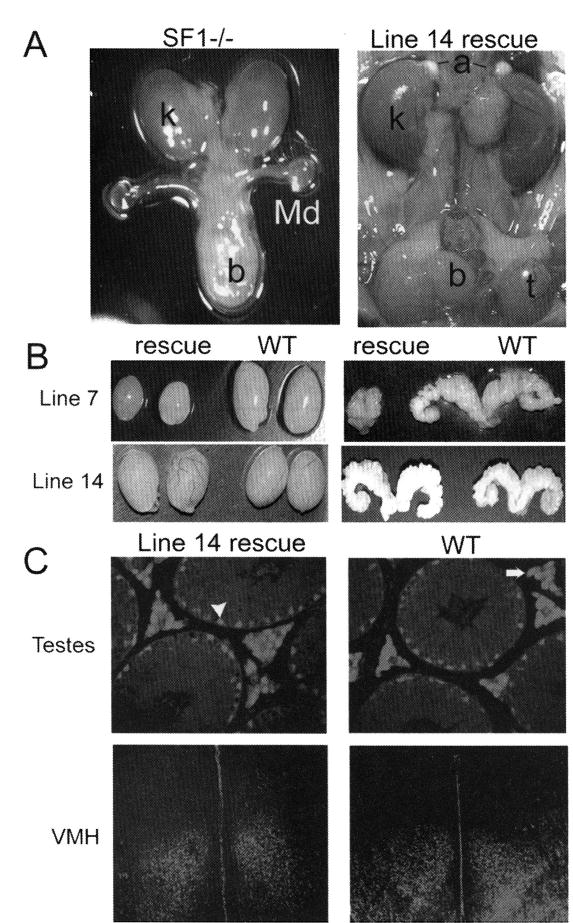
SF-1 function in rescue mice. (A) Normal development of adrenal glands (a) and testes (t) in line 14 rescue mice (right) compared with absence of adrenal glands and gonads and persistence of müllerian duct in SF-1 null mice (left). (B) Anatomical evaluation of lines 7 and 14 rescued mice revealed differences in the ability of their transgenes to rescue SF-1 function in the null mice. Significantly smaller testes and seminal vesicles size were registered in line 7 rescue mice compared with wild-type (WT) littermates, whereas testes and seminal vesicle weights were equivalent in WT and line 14 rescue animals. (C) Immunodetection of transgenic SF-1 in line 14 rescue mice and endogenous SF-1 in WT mice. Similar SF-1 expression was observed in Sertoli (arrowhead) and Leydig cells (arrow) of rescue and WT testes as well as similar expression pattern indicated in VMH of rescue and WT animals.
FIGURE 4.
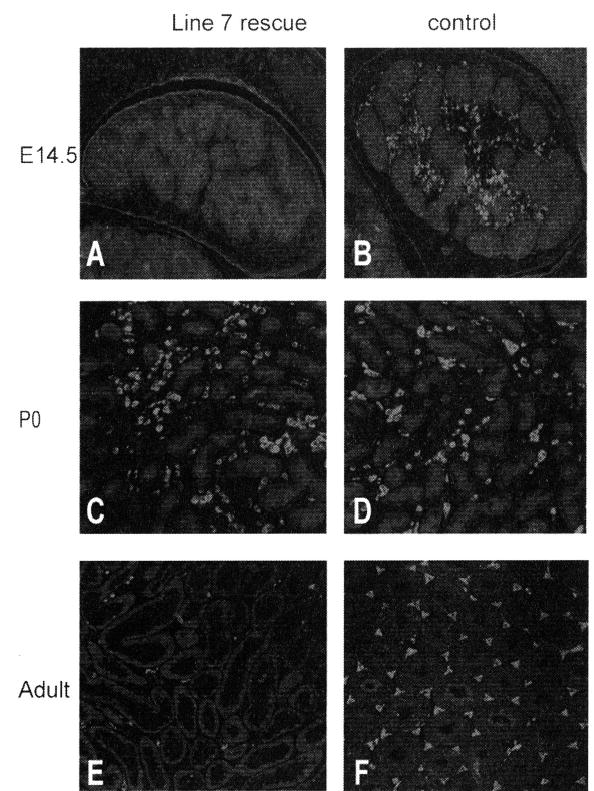
P450SCC expression in Leydig cells of line 7 rescue mice. Immunohistochemistry was used to evaluate P450SCC expression in line 7 (A,C,E) and control mice (B,D,F) at embryonic day 14.5 (A,B), postnatal day 0 (C,D), and adult ages (E,F). P450SCC was used as a marker for the Leydig cell population.
TABLE 1.
Reproductive performance of rescue mice in line 14
| Genotype | Sex | n | Litter number, observed in the group | Average litter size |
|---|---|---|---|---|
| Rescue | Male | 19 | 25 | 8.6 ± 1.6 |
| WT | Male | 10 | 12 | 8.9 ± 2.1 |
| Rescue | Female | 22 | 25 | 8.2 ± 1.6 |
| WT | Female | 12 | 17 | 7.6 ± 2.0 |
Acknowledgments
This work was supported by National Institutes of Health Grant R01 HD38498 (to L.L.H.) and a Specialized Cooperative Center Program in Reproduction Research (U54 HD 33994 to Paul Terranova) from the National Institute of Child Health and Human Development.
References
- 1.Ninomiya Y, et al. Genomic organization and isoforms of the mouse ELP gene. J Biochem (Tokyo) 1995;118:380–389. doi: 10.1093/oxfordjournals.jbchem.a124918. [DOI] [PubMed] [Google Scholar]
- 2.Parker KL, Schimmer BP. Steroidogenic factor 1. A key determinant of endocrine development and function. Endocr Rev. 1997;18:361–377. doi: 10.1210/edrv.18.3.0301. [DOI] [PubMed] [Google Scholar]
- 3.Luo X, Ikeda Y, Parker KL. A cell-specific nuclear receptor is essential for adrenal and gonadal development and sexual differentiation. Cell. 1994;77:481–490. doi: 10.1016/0092-8674(94)90211-9. [DOI] [PubMed] [Google Scholar]
- 4.Morohashi K, et al. Functional difference between Ad4BP and ELP, and their distributions in steroidogenic tissues. Mol Endocrinol. 1994;8:643–653. doi: 10.1210/mend.8.5.8058072. [DOI] [PubMed] [Google Scholar]
- 5.Ikeda Y, Ingraham WH, Parker KL. Developmental expression of mouse steroidogenic factor 1, an essential regulation of the steroid hydroxylases. Mol Endocrinol. 1994;8:654–662. doi: 10.1210/mend.8.5.8058073. [DOI] [PubMed] [Google Scholar]
- 6.Karpova T, et al. A Ftz-F1 -containing yeast artificial chromosome recapitulates expression of steroidogenic factor 1 in vivo. Mol Endocrinol. 2005;19:2549–2563. doi: 10.1210/me.2004-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peterson KR, et al. Use of yeast artificial chromosomes (YACs) in studies of mammalian development: production of beta-globin locus YAC mice carrying human globin developmental mutants. Proc Natl Acad Sci USA. 1995;92:5655–5659. doi: 10.1073/pnas.92.12.5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riethman, H. et al. 1997. Preparation, manipulation, and mapping of HMW DNA. In Genome Analysis: A Laboratory Manual. 1st Ed. B. Birren & E.D. Green, Eds. :83–248. Plainview. Cold Spring Harbor Laboratory Press.
- 9.Chen J, Heckert L. Dmrt1 expression is regulated by follicle-stimulating hormone and phorbol esters in postnatal Sertoli cells. Endocrinology. 2001;142:1167–1178. doi: 10.1210/endo.142.3.8021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harju S, Peterson K. Sensitive ribonuclease protection assay employing glycogen as a carrier and a single inactivation/precipitation step. Biotechniques JID -8306785. 2001;30:1198–200. doi: 10.2144/01306bm02. 1202, 1204. [DOI] [PubMed] [Google Scholar]
- 11.Wilhelm D, Englert C. The Wilms tumor suppressor WT1 regulates early gonad development by activation of Sf1. Genes Dev. 2002;16:1839–1851. doi: 10.1101/gad.220102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stallings NR, et al. Development of a transgenic green fluorescent protein lineage marker for steroidogenic factor 1. Mol Endocrinol. 2002;16:2360–2370. doi: 10.1210/me.2002-0003. [DOI] [PubMed] [Google Scholar]
- 13.Park SY, et al. Nuclear receptors Sf1 and Dax1 function cooperatively to mediate somatic cell differentiation during testis development. Development. 2005;132:2415–2423. doi: 10.1242/dev.01826. [DOI] [PubMed] [Google Scholar]


