Abstract
Coatomer, the coat protein complex of COPI vesicles, is involved in the budding of these vesicles, but the underlying mechanism is unknown. Toward a better understanding of this process, the interaction between coatomer and the cytoplasmic domain of a major transmembrane protein of COPI vesicles, p23, was studied. Interaction of coatomer with this peptide domain results in a conformational change and polymerization of the complex in vitro. This changed conformation also is observed in vivo, i.e., on the surface of authentic, isolated COPI vesicles. An average of four peptides was found associated with one coatomer complex after polymerization. Based on these results, we propose a mechanism by which the induced conformational change of coatomer results in its polymerization, and thus drives formation of the bud on the Golgi membrane during biogenesis of a COPI vesicle.
COPI-coated vesicles mediate protein transport in the early secretory pathway (1–3). The COPI coat consists of a small G protein, ADP-ribosylation factor 1 (ARF1) (4, 5), and coatomer, a heterooligomeric protein complex of seven subunits [(α to ζ coat proteins (COPs)] (6–8). COPI bud formation is initiated by membrane recruitment of ARF1, which in its GTP-bound form (9, 10), together with members of the p24 family, provides membrane-binding sites for coatomer (11, 12). Subsequent coat assembly leads to membrane deformation and the morphological appearance of a bud (13). To gain insight into the mechanism underlying this process, we investigated the interaction with coatomer of p23 (14), a member of the p24 family. p23, a type I transmembrane protein, is highly enriched in COPI vesicles and is present in a ratio to coatomer of approximately 4:1 (14). The cytoplasmic domain of p23 (YLRRFFKAKKLIE) is structurally similar to a classic dilysine motif (KKXX) for retrieval to the endoplasmic reticulum (ER) (15–18), and binds coatomer with the same efficiency as the KKXX motif (14). p23 binding, however, depends on its phenylalanine residues as well as its lysine residues.
The essential components needed for the biogenesis of other coated vesicles are defined as well. Clathrin-coated (19) and COPII-coated vesicles (20) have been generated in vitro from defined constituents. Most recently, a conformational change was shown on binding of GTP to dynamin (21), and this structural change was correlated to the process of pinching off a coated vesicular intermediate. However, two questions remain. (i) How are coat proteins normally bound? (ii) How does this association lead to a mechanical deformation of a membrane?
We describe here a conformational change of the COPI–coat protein complex, coatomer, induced by binding to the cytoplasmic domain of a major transmembrane protein of COPI vesicles, p23. This change in vitro leads to polymerization of the coat complex. The changed conformation of coatomer is also observed in vivo, i.e., on the surface of an authentic isolated COPI vesicle. We propose that the induced conformational change drives polymerization and subsequent deformation of the Golgi membrane during biogenesis of a COPI vesicle.
MATERIALS AND METHODS
Coatomer was isolated as described (22). Purified COPI vesicles were prepared as described in ref. 23 [vesicles were stored in 25 mM Hepes⋅KOH, pH 7.4/250 mM KCl/2.5 mM magnesium acetate/40% (vol/vol) sucrose at −80°C]. For Western blots shown in this study, an anti-γ-COP antibody was used that was generated against the recombinant protein (24).
Synthetic Peptides.
The sequences of the synthetic peptides are shown in Fig. 1A. Dimers were formed by disulfide bridges linking the peptides via N-terminally introduced cysteine residues. For this purpose, newly synthesized monomeric peptides were oxidized in 20% (vol/vol) dimethyl sulfoxide in water for 48 h. Subsequently, the dimers were isolated by using HPLC.
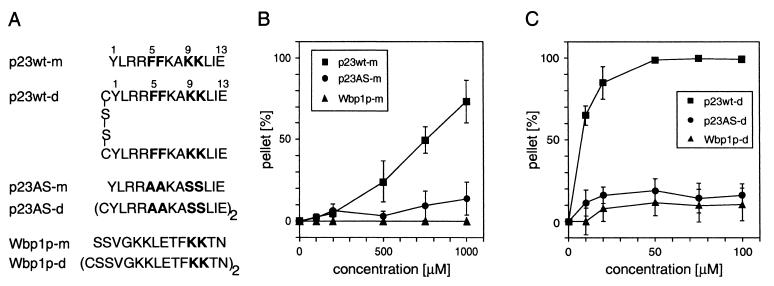
Figure 1.
Precipitation of coatomer with synthetic peptides corresponding to the COOH termini of p23wt, p23AS and Wbp1p. (A) Peptides used in this study. p23wt represents the cytoplasmic domain of p23, which binds coatomer depending on its dilysine and diphenylalanine motifs, whereas p23AS lacks these residues and therefore does not bind coatomer. Wbp1p represents the cytoplasmic domain of a subunit of the yeast N-oligosaccharyl transferase complex bearing a characteristic KKXX ER-retrieval motif known to bind coatomer. (B and C) Precipitation of coatomer (0.09 μM) with monomeric (B) and dimeric (C) peptides. Only p23wt (■) precipitates the complex, whereas mutated p23 peptide (p23AS, ●) and Wbp1p (▴) have no effect. The error bars indicate standard errors (n = 3).
Precipitation of Coatomer.
Pure soluble coatomer [0.09 μM in buffer 1 (25 mM Hepes⋅KOH, pH 7.4/100 mM KCl)] was incubated with increasing concentrations of peptides (monomeric peptides: 100–1,000 μM; dimeric peptides: 10–100 μM) for 1 h at room temperature (RT). Precipitates were pelleted by centrifugation at 40,000 × g for 15 min at 4°C. Pellets and supernatants were analyzed by using SDS/PAGE and immunoblotting using anti-COP antibodies. Quantification of precipitated and soluble coatomer was performed by scanning (in a linear range) the γ-COP signal detected by enhanced chemiluminescence (ECL, Amersham Pharmacia).
Limited Proteolysis of Coatomer and COPI Vesicles.
For experiments with isolated coatomer, pure, soluble protein complex (0.09 μM in buffer 1) was incubated with either 50 μM p23wt-d, p23AS-d, or Wbp1p-d under the conditions described above. Thereafter, thermolysin was added to a final concentration of 0.008 μM, and digestion was performed for the times indicated. Proteolysis was stopped with EDTA (final concentration 40 mM). Proteolytic fragments were separated on 7.5–15% gradient SDS gels and detected with immunoblot analysis.
For experiments with COPI vesicles, 25 μl of a suspension of purified COPI vesicles containing ≈0.5 μg of coatomer (final concentration ≈0.009 μM) were diluted to a final concentration of 10% (wt/vol) sucrose and incubated with 50 μM p23AS-d in a total volume of 100 μl for 30 min at RT. Thereafter, thermolysin was added to a final concentration of 0.008 μM, digestion was performed for 0–2 h, and proteolysis was stopped with EDTA (final concentration 40 mM). Protein was precipitated with chloroform/methanol and separated on 7.5–15% gradient SDS gels under reducing conditions. COPs were analyzed by immunoblot analysis. Coatomer (0, 5 μg, 0.009 μM) was incubated under identical conditions with either 50 μM p23wt-d or p23AS-d in a total volume of 100 μl [25 mM Hepes⋅KOH, pH 7.4/250 mM KCl/2.5 mM magnesium acetate, 10% (wt/vol) sucrose], and digested and analyzed as described for COPI vesicles.
Determination of the Stoichiometry Between Precipitated Coatomer and p23 Peptide.
p23wt-d and p23AS-d (10 nmol in 10 μl of water) were labeled with [125I]iodine [1 mCi (37 GBq)] by using Iodo-Gen iodination reagent (10 μg, Pierce) according to manufactures’ instructions. Iodide was removed by chromatography of soluble peptides on Sep-Pak C18 cartridges (Millipore) and washing with 10 ml of water. Peptides were eluted from the columns with 75% (vol/vol) acetonitrile/0.1% trifluoracetic acid in water. On addition of 0.5% (vol/vol) pyridin, the solvents were evaporated in a Speed Vac. The peptides were dissolved in water and stored at 4°C. Specific radioactivity for p23wt-d was 3–5 × 104 cpm/pmol and for p23AS-d 4–6 × 104 cpm/pmol. The iodinated peptides were used within 4 weeks. p23wt-d (500 pmol) containing from 8 × 105 to 1 × 106 cpm in a total volume of 20 μl of buffer 1 (25 μM p23wt-d, specific activity between 1.6 and 2 × 103 cpm/pmol) was incubated with increasing concentrations of coatomer (3.6–7.2 pmol) for 30 min at RT. Precipitates were pelleted by centrifugation at 40,000 × g for 15 min at 4°C. Pellets were washed three times with buffer 1 containing 500 pmol of nonradioactive p23wt-d and centrifuged at 40,000 × g for 15 min at 4°C. Thereafter, the amount of precipitated peptide was analyzed by counting radioactivity of the pellets (signals detected were in the range of 1–3 × 104 cpm above background). Precipitation of coatomer was determined by analyzing pellets and supernatants on 7.5% SDS gels and immunoblotting. The same experiments were performed with [125I]p23AS-d.
Size Exclusion Chromatography of Monomeric and Dimeric p23wt.
Size exclusion chromatography was performed on a Smart System (Amersham Pharmacia Biotech) by using the Superdex Peptide PC 3.2/30. The column was equilibrated with buffer 1 at a flow rate of 40 μl/min at 8°C. Peptides were dissolved in buffer 1 at a concentration of 1 mg/ml (Fig. 7 A and B) or 0.1 mg/ml (Fig. 7D) and injected as indicated in the figure. Chromatography of the peptides was performed at a flow rate of 20 μl/min at 8°C, and fractions of 40 μl were collected.
RESULTS
p23-tail Peptide-Induced Polymerization of Coatomer.
To study the interaction of p23 with isolated coatomer, we incubated the complex with a synthetic peptide analogous to the cytoplasmic domain of p23 (p23wt-m, Fig. 1A), and after centrifugation observed aggregation of coatomer. At a peptide concentration of about 750 μM, half of the soluble coatomer is precipitated (Fig. 1B). Based on the ≈4:1 stoichiometry between p23 and coatomer in COPI vesicles, we reasoned that the active form of p23 might be an oligomer. Indeed, a dimeric form of p23wt-m, p23wt-d (Fig. 1A), precipitates 50% of coatomer at about 1/100 the concentration of p23wt-m (Fig. 1C). Unrelated proteins like BSA and phosphorylase are not precipitated under identical conditions. Furthermore, a peptide (p23AS; Fig. 1A) that lacks the amino acid residues known to be essential for binding coatomer (14) does not precipitate coatomer in considerable amounts, either in its monomeric or its dimeric form (Fig. 1 B and C). A peptide analogous to the cytoplasmic domain of Wbp1p (25), an ER-resident transmembrane protein that is not part of the transport machinery but rather cargo for retrieval (17), binds coatomer with the same efficiency as the p23-tail peptide. However, both the monomeric and the dimeric form of this peptide (Fig. 1A) did not cause significant precipitation of the complex (Fig. 1 B and C).
Limited Proteolysis of Coatomer.
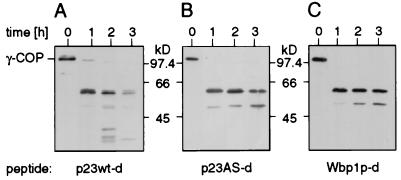
The aggregation of coatomer observed with the p23wt peptides is likely to involve a conformational change of the complex. To probe such a conformational change by limited proteolysis, coatomer was incubated with thermolysin in the presence of the various peptides used in the precipitation. Because γ-COP has been identified as a binding partner of both the ER-retrieval motif (24) and the cytoplasmic domain of p23 (26), proteolysis of this subunit was monitored. γ-COP is partially cleaved to yield a prominent fragment of about 59 kDa (Fig. 2, A–C, 1 h). In the presence of p23 wt-d this 59-kDa fragment is proteolytically degraded into various fragments (Fig. 2A, 2 and 3 h). In contrast, in the presence of p23AS-d, the 59-kDa fragment is clearly more stable (Fig. 2B, 2 and 3 h). Even after 3 h in the presence of the mutated peptide, only one additional fragment (of about 52 kDa) is observed. Partial digestion in the presence of Wbp1p-d resulted in a pattern very similar to that with p23AS-d (Fig. 2C). The increased susceptibility to protease of γ-COP, together with polymerization of coatomer specifically induced by p23wt-d, demonstrates a conformational change of the complex on binding to the peptide.
Figure 2.
Limited proteolysis of coatomer. Proteolysis of coatomer (0.09 μM) with thermolysin (0.008 μM) in the presence of dimerized peptides (50 μM) p23wt-d (A), p23AS-d (B), and Wbp1p-d (C). Western blot analysis of proteolytic fragments of the γ-COP subunit reveals a better susceptibility of this subunit to the protease in the presence of p23wt-d (A, lanes 2 and 3) as compared with p23AS-d (B, lanes 2 and 3 h) and Wbp1p-d (C, lanes 2 and 3 h).
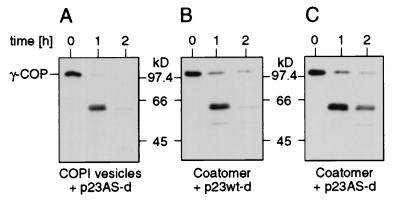
Figure 3.
Limited proteolysis of COPI vesicles. (A) Suspension of purified COPI vesicles containing ≈0.009 μM coatomer treated with thermolysin (0.009 μM) in the presence of p23AS-d (50 μM) as a substrate for the protease. Western blot analysis of proteolytic fragments of γ-COP reveals a prominent fragment of about 59 kDa (lane 1 h) that is highly susceptible to the protease (lane 2 h). (B) Coatomer (0.009 μM) incubated with p23wt-d (50 μM) and treated with thermolysin under the same conditions as used for the COPI vesicles yields a result comparable to that of COPI vesicles (lanes 1 and 2 h). In contrast, after partial digestion in the presence of p23AS-d (50 μM) of coatomer, the 59-kDa fragment is more stable (C, lanes 1 and 2 h).
The increased susceptibility to protease of coatomer observed in its aggregated state in vitro led us to investigate whether this conformational change also occurs under conditions that reflect an in vivo situation, i.e., on the surface of an isolated COPI vesicle. To this end, purified COPI vesicles (23) were subjected to limited proteolysis with thermolysin. The result is shown in Fig. 3. Strikingly, although coatomer is densely packed on the surface of the vesicle, the γ-COP 59-kDa domain is highly susceptible to the protease (Fig. 3A), similar to the precipitated state of coatomer (Fig. 3B) and different to soluble coatomer, where the γ-COP domain again is much more stable (Fig. 3C). (Note that the conditions of proteolysis for technical reasons were different in the experiments shown in Figs. 2 and 3.) These results strongly indicate a physiological role of the observed conformational change in the biogenesis of a COPI vesicle.
Stoichiometry of p23wt-d and Coatomer.
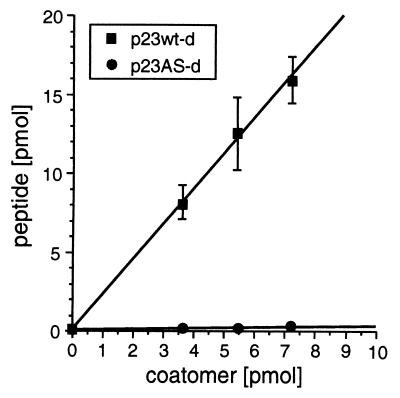
A stoichiometry of p23/coatomer of about 4:1 has been determined in isolated COPI vesicles (14). If the observed conformational change of coatomer with subsequent precipitation reflected a physiological mechanism, then a similar stoichiometry would be expected in the precipitate between the p23 tail peptide and coatomer. To determine the stoichiometry in the precipitates, various amounts of coatomer were incubated with a precipitating amount of 125I-labeled dimerized p23wt peptide, and after centrifugation, the amount of peptide in the precipitates was determined by counting their 125I radioactivity. The result is shown in Fig. 4. A linear increase of radioactivity is observed with increasing amounts of coatomer, and the stoichiometry calculated from the specific radioactivity of the dimerized peptide and coatomer (100% of the input coatomer was precipitated at each concentration) can be determined from the resulting slope. From an average of four experiments, 2.2 moles of dimerized p23wt peptide bind to 1 mole of coatomer under precipitating conditions. This is in good agreement with approximately four molecules of p23 bound to one coatomer in a COPI vesicle. As a control, 125I-labeled p23AS-d peptide did not lead to amounts of radioactivity precipitated above background (Fig. 4).
Figure 4.
Stoichiometry between coatomer and p23wt-d. 125I-labeled p23wt-d (■) or p23AS-d (●) were incubated with increasing concentrations of coatomer. After centrifugation, the amounts of peptide in the precipitates were determined by counting the 125I radioactivity. The error bars indicate standard errors (n = 4).
Size Exclusion Chormatography of Monomeric and Dimeric p23wt.
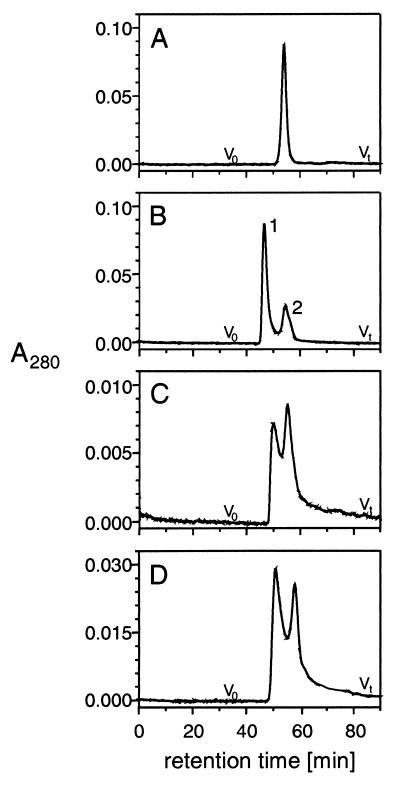
To analyze a possible oligomerization of p23wt-d in solution, size exclusion chromatography was performed. p23wt-m and p23wt-d reduced by DTT elute as a single peak (Fig. 5A). p23wt-d, however, gives rise to two peaks (Fig. 5B): peak 2 elutes with the same retention time as p23wt-m (Fig. 5A), indicating very similar Stokes’ radii of the two species, whereas peak 1, judged from its retention time, is likely to represent a p23wt-d dimer. Thus, an equilibrium seems to exist in solution between two forms of p23wt-d. Accordingly, rechromatography of the fraction corresponding to peak 1 in Fig. 5B results in an elution profile similar to the p23wt-d sample applied in Fig. 5B. However, a different ratio of peak sizes is observed with peak 2 enlarged at the expense of peak 1 (Fig. 5C), again indicating an equilibrium of the two states of the peptide. This equilibrium depends on the concentration of the peptide (Fig. 5D), revealing its bimolecular nature. Thus, peak 1 must be caused by a dimeric state, and peak 2 by the monomeric state, of p23wt-d. A dimeric state of p23wt-d corresponds to four cytoplasmic domains of p23 and is in good agreement with the above stoichiometry between the peptide (2.2 mol p23wt-d, equivalent to 4.4 mol p23wt-m) and 1 mol of polymerized coatomer.
Figure 5.
Size exclusion chromatography of p23-tail peptides. (A) Monomeric p23wt (p23wt-m: 1 mg/ml; sample corresponds to 20 μl). (B) Dimeric p23wt (p23wt-d: 1 mg/ml; sample corresponds to 20 μl). Peak 1 represents the dimerized form of p23wt-d, and peak 2 represents the monomeric form of p23wt-d. (C) Rechromatography of peak 1 of B (40 μl). The appearance of both peaks 1 and 2 demonstrates an equilibrium between two states of p23wt-d. (D) Dimeric p23wt (p23wt-d: 0.1 mg/ml; sample corresponds to 40 μl). The concentration dependence of the equilibrium of peak 1 and 2 indicates a bimolecular event.
DISCUSSION
We have shown here that interaction of coatomer with the cytoplasmic domain of p23 leads to a conformational change in its γ-subunit and concomitant precipitation of the complex. This conformational change is specific in that a Wbp1p peptide, which also binds coatomer via its ER-retrieval motif (24), failed to induce this conformational change. The very same conformation of γ-COP is found in coatomer under conditions that reflect an in vivo situation, namely on the surface of authentic, Golgi-derived COPI vesicles. The structure of p23 that triggers the conformational change is likely to be a tetramer of four peptides, as judged from size exclusion chromatography and determination of its stoichiometry in the precipitated coatomer complex.
In vitro precipitation of coatomer was recently observed in the presence of some bivalent aminoglycoside antibiotics (27) and interpreted in terms of aggregation of coatomer by crosslinking. In light of the data described here, these aminoglycosides may well fit into the binding site for the cytoplasmic domain of p23 that resides in the γ-subunit of the complex (26) and induce its conformational change and therefore precipitate the complex.
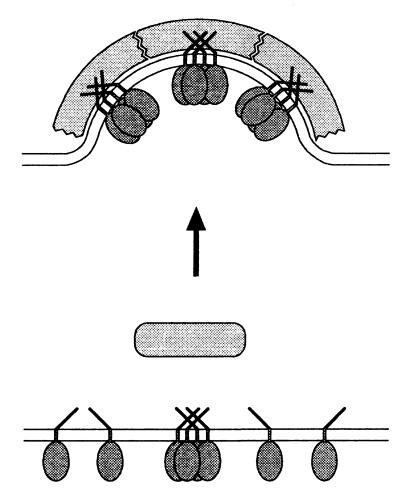
It is of note that a Wbp1p-peptide with a characteristic ER-retrieval motif does bind coatomer but (in contrast to the cytoplasmic domain of p23) does not trigger a conformational change of the complex. Thus, the two classes of domains have distinct and different functions: interaction of the Wbp1p-type of peptide may serve the sorting of cargo to be retrieved to the ER into retrograde COPI vesicles. p23, however, may represent part of the machinery of a COPI vesicle. Not only does it bind to coatomer during the budding reaction, but it also triggers a conformational change that leads to polymerization of the complex (Fig. 6).
Figure 6.
Hypothetical model for bud formation. According to this model, interaction of coatomer with a tetramer of p23 induces a conformational change of the complex. This leads to its polymerization on the surface of a membrane, resulting in the formation of a coated bud. (Note that for simplicity, a role of ARF 1 is not included in this diagram.)
The stoichiometry described here in the precipitate between p23 peptide and coatomer and the stoichiometry of p23 to coatomer in purified COPI vesicles (14) strongly favors a tetramerized state of p23 when it binds to coatomer in vivo. Similarly, other members of the p24 family, known to bind coatomer and bearing a diphenylalanine and dilysine motif, may interact with coatomer in an oligomerized state. Thus, receptor-induced polymerization of coatomer may represent a general mechanism for COPI vesicle formation. However, at present it is not known in which stoichiometries the various p24 family members interact with each other (28) or whether they are able also to form homooligomers. In any case, various oligomers would allow different classes of COPI vesicle to exist, specified by the different p24 members they contain. According to this model, oligomerization of p23 and its relatives must be regulated in vivo. Such regulation may occur via interaction of p24 members with cargo if p24 members serve as cargo receptors (29–31). Another candidate to regulate oligomerization of p24 members is ARF 1, the recruitment of which to Golgi membranes precedes binding of coatomer. However, such a role of ARF 1 in oligomerization of p24 members waits to be determined.
As coatomer is used in many cycles, an important question concerns a reversibility of the conformational change of complex. Because ARF 1 is a component of COPI vesicle and is known to be involved in the uncoating process by hydrolysis of its bound GTP (32), this energy-providing step may well help to reverse the conformational change of coatomer and thus dissociate the coat.
In general, receptor-induced polymerization of coat proteins during their recruitment would strongly increase the efficiency of a coating process, and with the resulting geometry ruled by the nature of the conformationally changed proteins, may well be the driving force to shape a membrane.
Acknowledgments
We thank Dr. Suzanne Pfeffer, Dr. Kai Simons, and Dr. Graham Warren for critically reading the manuscript. This work was supported by grants of the Deutsche Forschungsgemeinschaft (SFB 352 to C.H., J.B.H., and F.W.), the Human Frontier Science Program (to F.W.) and the Fonds der Chemischen Industrie (to F.W.).
ABBREVIATIONS
- ARF1
ADP ribosylation factor 1
- COP
coat protein
- COPI
protein coat composed of ARF1 and coatomer
- ER
endoplasmic reticulum
References
- 1.Rothman J E. Nature (London) 1994;372:55–63. doi: 10.1038/372055a0. [DOI] [PubMed] [Google Scholar]
- 2.Rothman J E, Wieland F T. Science. 1996;272:227–234. doi: 10.1126/science.272.5259.227. [DOI] [PubMed] [Google Scholar]
- 3.Schekman R, Orci L. Science. 1996;271:1526–1533. doi: 10.1126/science.271.5255.1526. [DOI] [PubMed] [Google Scholar]
- 4.Serafini T, Orci L, Amherdt M, Brunner M, Kahn R A, Rothman J E. Cell. 1991;67:239–253. doi: 10.1016/0092-8674(91)90176-y. [DOI] [PubMed] [Google Scholar]
- 5.Taylor T C, Kahn R A, Melancon P. Cell. 1992;70:69–79. doi: 10.1016/0092-8674(92)90534-j. [DOI] [PubMed] [Google Scholar]
- 6.Waters M G, Serafini T, Rothman J E. Nature (London) 1991;349:248–251. doi: 10.1038/349248a0. [DOI] [PubMed] [Google Scholar]
- 7.Stenbeck G, Harter C, Brecht A, Herrmann D, Lottspeich F, Orci L, Wieland F T. EMBO J. 1993;12:2841–2845. doi: 10.1002/j.1460-2075.1993.tb05945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harter C. FEBS Lett. 1995;369:89–92. doi: 10.1016/0014-5793(95)00621-f. [DOI] [PubMed] [Google Scholar]
- 9.Donaldson J G, Cassel D, Kahn R A, Klausner R D. Proc Natl Acad Sci USA. 1992;89:6408–6412. doi: 10.1073/pnas.89.14.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palmer D J, Helms J B, Beckers C J, Orci L, Rothman J E. J Biol Chem. 1993;268:12083–12089. [PubMed] [Google Scholar]
- 11.Stamnes M A, Craighead M W, Hoe M H, Lampen N, Geromanos S, Tempst P, Rothman J E. Proc Natl Acad Sci USA. 1995;92:8011–8015. doi: 10.1073/pnas.92.17.8011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nickel W, Wieland F T. FEBS Lett. 1997;413:395–400. doi: 10.1016/s0014-5793(97)00939-3. [DOI] [PubMed] [Google Scholar]
- 13.Orci L, Palmer D J, Ravazzola M, Perrelet A, Amherdt M, Rothman J E. Nature (London) 1993;362:648–652. doi: 10.1038/362648a0. [DOI] [PubMed] [Google Scholar]
- 14.Sohn K, Orci L, Ravazzola M, Amherdt M, Bremser M, Lottspeich F, Fiedler K, Helms J B, Wieland F T. J Cell Biol. 1996;135:1239–1248. doi: 10.1083/jcb.135.5.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nilsson T, Jackson M, Peterson P A. Cell. 1989;58:707–718. doi: 10.1016/0092-8674(89)90105-0. [DOI] [PubMed] [Google Scholar]
- 16.Jackson M R, Nilsson T, Peterson P A. EMBO J. 1990;9:3153–3162. doi: 10.1002/j.1460-2075.1990.tb07513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackson M R, Nilsson T, Peterson P A. J Cell Biol. 1993;121:317–333. doi: 10.1083/jcb.121.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cosson P, Letourneur F. Science. 1994;263:1629–1631. doi: 10.1126/science.8128252. [DOI] [PubMed] [Google Scholar]
- 19.Takei K, Haucke V, Slepnev V, Farsad K, Salazar M, Chen H, De Camilli P. Cell. 1998;94:131–141. doi: 10.1016/s0092-8674(00)81228-3. [DOI] [PubMed] [Google Scholar]
- 20.Matsuoka K, Orci L, Amherdt M, Bednarek S Y, Hamamoto S, Schekman R, Yeung T. Cell. 1998;93:263–275. doi: 10.1016/s0092-8674(00)81577-9. [DOI] [PubMed] [Google Scholar]
- 21.Sweitzer S M, Hinshaw J E. Cell. 1998;93:1021–1029. doi: 10.1016/s0092-8674(00)81207-6. [DOI] [PubMed] [Google Scholar]
- 22.Pavel J, Harter C, Wieland F T. Proc Natl Acad Sci USA. 1998;95:2140–2145. doi: 10.1073/pnas.95.5.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malhotra V, Serafini T, Orci L, Shepherd J C, Rothman J E. Cell. 1989;58:329–336. doi: 10.1016/0092-8674(89)90847-7. [DOI] [PubMed] [Google Scholar]
- 24.Harter C, Pavel J, Coccia F, Draken E, Wegehingel S, Tschochner H, Wieland F. Proc Natl Acad Sci USA. 1996;93:1902–1906. doi: 10.1073/pnas.93.5.1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.te Heesen S, Janetzky B, Lehle L, Aebi M. EMBO J. 1992;11:2071–2075. doi: 10.1002/j.1460-2075.1992.tb05265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harter C, Wieland F T. Proc Natl Acad Sci USA. 1998;95:11649–11654. doi: 10.1073/pnas.95.20.11649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hudson R T, Draper R K. Mol Biol Cell. 1997;8:1901–1910. doi: 10.1091/mbc.8.10.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dominguez M, Dejgaard K, Füllekrug J, Dahan S, Fazel A, Paccaud J-P, Thomas D Y, Bergeron J J M, Nilsson T. J Cell Biol. 1998;140:751–765. doi: 10.1083/jcb.140.4.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schimmoeller F, Singer K B, Schroeder S, Krueger U, Barlowe C, Riezman H. EMBO J. 1995;14:1329–1339. doi: 10.1002/j.1460-2075.1995.tb07119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fiedler K, Veit M, Stamnes M A, Rothman J E. Science. 1996;273:1396–1399. doi: 10.1126/science.273.5280.1396. [DOI] [PubMed] [Google Scholar]
- 31.Kuehn M J, Herrmann J M, Schekman R. Nature (London) 1998;391:187–190. doi: 10.1038/34438. [DOI] [PubMed] [Google Scholar]
- 32.Tanigawa G, Orci L, Amherdt M, Ravazzola M, Helms J B, Rothman J E. J Cell Biol. 1993;123:1365–1371. doi: 10.1083/jcb.123.6.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]