Abstract
After a primary infection, herpes simplex virus is maintained in a latent state in neurons of sensory ganglia until complex stimuli reactivate viral lytic replication. Although the mechanisms governing reactivation from the latent state remain unknown, the regulated expression of the viral immediate early genes represents a critical point in this process. These genes are controlled by transcription enhancer complexes whose assembly requires and is coordinated by the cellular C1 factor (host cell factor). In contrast to other tissues, the C1 factor is not detected in the nuclei of sensory neurons. Experimental conditions that induce the reactivation of herpes simplex virus in mouse model systems result in rapid nuclear localization of the protein, indicating that the C1 factor is sequestered in these cells until reactivation signals induce a redistribution of the protein. The regulated localization suggests that C1 is a critical switch determinant of the viral lytic–latent cycle.
As a human pathogen, herpes simplex virus (HSV) causes disease ranging from relatively mild recurrent lesions to more significant disseminated illness such as encephalitis (1, 2). After an initial infection, the virus is maintained in a latent state in neurons of sensory ganglia until stimuli (i.e., stress, UV exposure, infection, hormonal alterations, or tissue damage) results in the reactivation of viral lytic replication (1). However, little is known concerning the biochemical mechanisms that govern the reactivation process, primarily due to the inability to establish a latent infection in tissue culture that accurately reproduces in vivo latency characteristics (1).
Previous studies have focused primarily on the role of viral-encoded proteins in the establishment and reactivation from the latent state (1). However, viral gene expression in the latent state is severely limited (1). Furthermore, the cell specificity of the site of viral latency and the sensitivity of reactivation to host stimuli suggest that cellular proteins play the primary role in the establishment of the latent state and the recurrent reactivation process. As the viral immediate early gene (IE) products are critical for lytic replication, transcription of these genes is also likely to be an important event in the reactivation from the latent state.
During lytic replication, the coordinated expression of these IE genes is controlled by the assembly of multiprotein enhancer complexes containing viral [αTIF (VP16)] and cellular {Oct-1, GA binding protein (GABP), C1 factor [host cell factor (HCF)]} proteins (3–14). However, in a latent infection of sensory neurons, Oct-1 and GABP are present at relatively low levels (15) and the viral transactivator α-trans-induction-factor (αTIF) is not expressed, suggesting that reactivation may proceed by means of an αTIF-independent mechanism. On induction from latency, the steady-state level of Oct-1 increases (16), while the transcriptional activity of GABP is regulated by stress-induced protein kinase pathways (17, 18). Little, however, is known concerning the activity and regulation of the C1 factor under these conditions. This protein is the critical coordinator/mediator of the enhancer activation assemblies, specifically interacting with each component of the regulatory complex as well as with other associated cellular transcription factors. Furthermore, recent studies have determined that the transcriptional activity of GABP is mediated through direct interaction with C1 (J.L.V. and T.M.K., unpublished work). This unique coactivator consists of a family of polypeptides that are derived from a common precursor by site-specific proteolysis (9–11). In addition to its role in the activation of transcription, the protein has also been implicated in the control of cell-cycle progression (19). As such, C1-dependent modulation of the viral lytic–latent cycle is an attractive model.
In these studies, it is demonstrated that the C1 factor is uniquely sequestered in sensory neurons until reactivation signals induce a redistribution of the protein. The regulated localization suggests that C1 may be a critical switch determinant for the initiation of reactivation of HSV from the latent state.
MATERIALS AND METHODS
Tissue Preparation and Immunohistochemistry.
Trigeminal ganglia and control tissues were isolated from 7-week-old BalbC mice and immediately placed in fixative at 4°C or were incubated in DMEM/5.0% fetal bovine serum at 37°C in a CO2 incubator for the appropriate time before fixation at 4°C for 24 hr. For reactivation by scarification, ganglia were immediately placed in fixative at the appropriate time points after scarification, as for the induction of HSV from latently infected mice. Perfusion of mice with fixative before removal of ganglia was done according to standard protocols. In all experiments, ganglia or tissue(s) were removed within a maximum of 2 min. Tissues were fixed in 4.0% paraformaldehyde in PBS for 20–22 hr at 4°C, embedded in paraffin blocks, and sectioned on silated slides. The paraffin was removed and the sections were rehydrated according to standard procedures. After preblocking, the slides were incubated with the appropriate primary antibody (5 μg/ml anti-C1 antibody/3% BSA in PBS) for 2 hr at 4°C in a humidified chamber, washed, and developed as recommended by the manufacturer (Zymed). In some cases, slides were counterstained with hematoxylin. Ganglia and tissue sections were photographed using a Zeiss Axiophot fluorescence microscope fitted with a Princeton Instruments CCD digital camera and a Sony UP-5500 video printer.
Tissue Extracts and Western Blots.
Extracts of trigeminal ganglia from 7-week-old balbC mice were prepared as described (20) by homogenization of 0.25 g tissue in 1 ml t-ext buffer [50 mM Hepes, pH 7.4/150 mM NaCl/1% Nonidet P-40/0.5 mM EDTA/20 μg/ml−1 RNase A/“complete” protease inhibitor (aprotinin, leupeptin, Pefabloc, EDTA (Boehringer Mannheim)] in a tissue grinder using a Teflon pestle attached to a variable speed drill. The homogenates were incubated on ice for 15 min and subsequently cleared by centrifugation at 20,000 × g for 20 min. Replicate sets of extract from HeLa cells (15 μg) and mouse trigeminal ganglia (50 μg) were resolved by SDS/PAGE, transferred to Immobilon-P (Millipore) and probed with affinity-purified antibodies directed against the human C1 factor. The blots were developed for chemiluminescent detection (Pierce) or for chemifluorescent quantitation according to the manufacturers’ recommendations (Molecular Dynamics).
RESULTS AND DISCUSSION
Distinct Localization of the C1 Factor in Sensory Neurons.
Because HSV does not establish latent infections in tissue culture, expression and localization of the C1 factor was analyzed in a mouse model system where HSV latency is uniquely established in the neurons of the sensory ganglia (i.e., trigeminal ganglia). Experimentally, the virus can be induced from the latent state by a number of procedures, including excision and incubation of the ganglia in tissue culture medium (1).
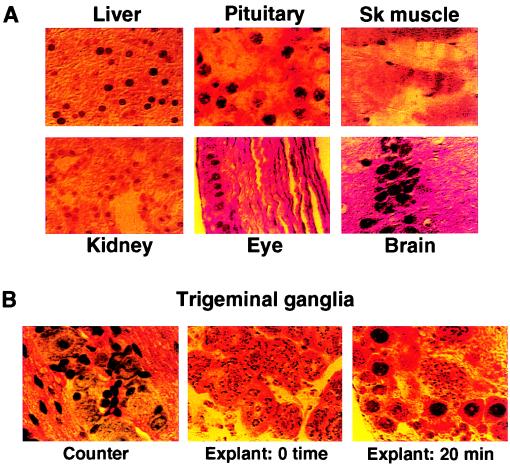
As shown in Fig. 1A, tissue sections from various organs were stained with affinity-purified anti-C1 antibody. In each case, the protein was expressed and localized in the nucleus, consistent with previous studies indicating the ubiquitous expression of the factor (10, 20–21). In direct contrast, staining of trigeminal ganglia exhibits a distinct pattern of C1 localization (Fig. 1B, Center). In these tissues, the neurons exhibit cytoplasmic granulation without nuclear staining, suggesting that C1 is uniquely sequestered in the cytoplasm of these cells. Strikingly, under conditions used to reactivate HSV, ganglia that were incubated in tissue culture medium for 20 min exhibited a clearance of the cytoplasmic granules and appearance of strong nuclear staining (Fig. 1B, Right) in 10–30% of the neurons. As tissues and ganglia can be subject to induced changes during tissue removal and preparation, animals were perfused with fixative before dissection. In these animals, explanted trigeminal ganglia exhibited cytoplasmic localization of the C1 factor while control tissues exhibited nuclear staining, demonstrating that the distinct localization patterns were not a function of the experimental preparations (Fig. 2A). Nuclear localization of the C1 factor, as detected by immunohistochemistry, thereby directly correlated with the reactivation of HSV. Because this protein is critical to the assembly of the HSV IE enhancer and the transcriptional activation of these viral genes, the unique cytoplasmic localization of this coactivator in sensory neurons may contribute to the establishment of HSV latency in these cells. Furthermore, nuclear localization of the C1 factor on stimulation indicates that the protein could play a significant role in the viral reactivation process.
Figure 1.
Distinct localization of the C1 factor in sensory neurons. (A) Sections of representative mouse tissues were stained with purified anti-C1 sera. In each case, the protein is detected in the cell nucleus. (B) Mouse trigeminal ganglia were excised and immediately fixed (0 time) or were explanted into media for 20 min before fixation (20 min). Representative sections from the ganglia were counterstained with hematoxylin to visualize the clustered neurons and neuronal nuclei (Counter) or were stained with purified anti-C1 sera. Although the protein is detected in cytoplasmic punctate structures at 0 time, it is primarily localized to the nucleus of explanted ganglia neurons. Control antibodies directed against neurofilaments demonstrated that the tissue was intact and permeable to antibody staining (data not shown).
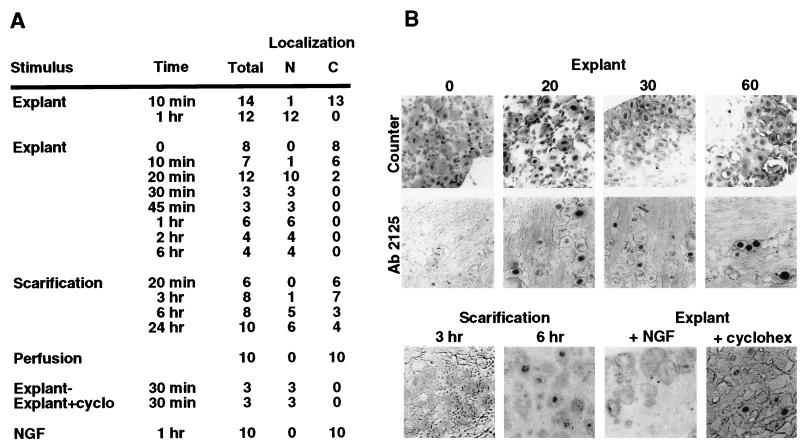
Figure 2.
Correlation of C1 localization and HSV reactivation. (A) Trigeminal ganglia were explanted and incubated in media for various times before fixation (Explant), excised at various times after eye scarification (Scarification), excised after fixation-perfusion (Perfusion), incubated in the presence of NGF (7.5S NGF, 10–100 ng/ml), or incubated in the presence of cyclohexamide (+cyclo, 100 μg/ml). The treated ganglia were fixed, embedded, and stained with anti-C1 sera or control sera as described. The total number of ganglia in each experiment is shown with the number that exhibit nuclear (N) or cytoplasmic (C) phenotypic localization of the C1 factor. Each ganglion contains 200–400 neurons per section. Designation as nuclear indicates that 10–30% of the neurons in a ganglion exhibited nuclear staining, whereas cytoplasmic exhibit 0.2–0.8% nuclear staining. Three to five sections from each embedded ganglion were stained to determine an experimental point. (B) Representative sections from the experimental sets listed above. Sections were counterstained to visualize intact neuronal nuclei (Counter) or were stained with anti-C1 Ab2125 serum.
Nuclear Localization of C1 Correlates with the Initiation of Viral Reactivation.
As described above, explant of latently infected trigeminal ganglia into tissue culture medium will result in viral lytic replication. The results of a series of explant experiments are summarized in Fig. 2A and represented in Fig. 2B. After a 10-min incubation, 1 of 14 ganglia exhibited a significant number of neurons in which C1 was localized to the nucleus. After 1 hr of incubation, 100% of the ganglia showed nuclear staining with anti-C1 sera.
To determine the time course of the localization relative to the stimulus, ganglia were explanted for various times before fixation. The localization of the C1 factor to the neuronal nucleus occurred within 20 min of ganglia explant and incubation. This indicates that localization of C1 to the nucleus is a very early phenomenon relative to the induced expression of other cellular transcription factors (16) and suggests that C1 is a primary target in the signaling that results in the initiation of reactivation of HSV from the latent state.
Nuclear Localization of the C1 Factor Directly Correlates with Stimuli for Viral Reactivation.
Explantation of ganglia is a primary means of experimentally reactivating HSV from the latent state. Other stimuli, such as scarification of the infected animals’ eyes, will also induce viral replication, although at a significantly lower frequency (22) and an altered reactivation time course. To explore further the correlation between C1 localization and HSV reactivation, ganglia were removed and immediately fixed at various times after eye scarification. Under these conditions, C1 was detected in the neuronal nuclei between 3 and 6 hr after stimulation. As with reactivation by explant, the localization of the factor correlated directly with stimuli that induce reactivation from the latent state and occurred at an initial point in the process time course.
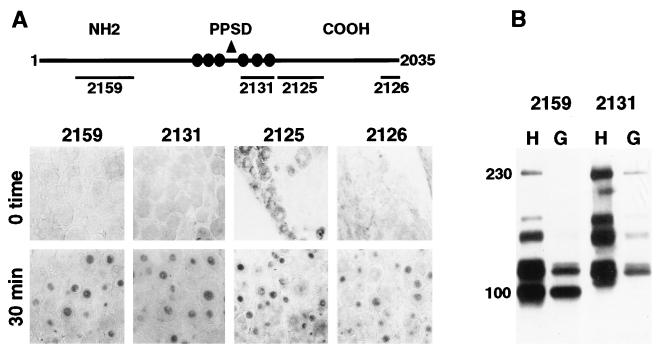
As illustrated in Fig. 3A, antisera directed against different domains of the C1 factor result in similar staining patterns in rapidly fixed or incubated trigeminal ganglia. In all cases, the sera show strong nuclear staining in ganglia that were explanted and incubated for 30 min. Although all sera do exhibit some cytoplasmic staining in rapidly excised and fixed ganglia (0 time), Ab2125 most strongly detects this pool of the C1 factor, suggesting that these determinants may be readily exposed. In addition, clearance of the cytoplasmic staining on stimulation of the ganglia (Figs. 1B and 3A) strongly indicates that this reactivity is C1 factor specific.
Figure 3.
Antisera directed against domains of the C1 factor. (A) Schematic representation of the 2,035-aa C1 factor indicating the protein domains recognized by the various anti-C1 sera [2159, 2131, 2125, 2126 (10)]. NH2, amino terminus; PPSD, proteolytic processing domain; COOH, carboxyl terminus. The circles represent the sites of specific proteolytic processing that generate the family of polypeptides. Representative sections of trigeminal ganglia were immediately fixed or were explanted into media for 30 min. Sections were stained with the indicated sera. (B) Protein extracts of HeLa cells (H) and trigeminal ganglia (G) were subjected to Western blot with the indicated sera. The family of C1 polypeptides ranges from 65 to 230 kDa (8–10).
Inhibition of C1 Nuclear Localization by Nerve Growth Factor (NGF).
A number of studies have demonstrated that the reactivation of HSV from the latent state may be reduced or inhibited in the presence of exogenously added NGF (23–25). Therefore, ganglia were explanted and incubated in the presence of high levels of 7.5S NGF for 1 hr. As illustrated (Fig. 2 A and B), nuclear localization of the C1 factor was completely prevented by NGF, directly correlating with observations concerning HSV reactivation. In addition, the inhibition of C1 localization by NGF suggests a significant signaling pathway by which the localization of this protein may be regulated.
In support of the results obtained with NGF, explanting ganglia directly into PBS in the absence of additional serum factors results in nuclear localization of the protein. This indicates that nuclear localization of C1 is a function of the depletion of specific factors that maintain the cytoplasmic state of the protein (data not shown).
Nuclear Localization of the C1 Factor Occurs in the Absence of de Novo Protein Synthesis.
Explant into culture containing high levels of cycloheximide had no effect on the nuclear localization of the protein, demonstrating that the transport does not require de novo protein synthesis (Fig. 2 A and B). This observation is supported by quantitative Western blots of protein extracts prepared from both rapidly explanted ganglia and from incubated ganglia, which illustrate that the levels of C1 protein do not change on incubation (data not shown). In addition, in comparison to extracts prepared from tissue culture cells (HeLa), extracts from ganglia exhibit a similar pattern of C1 polypeptides, suggesting that the protein undergoes its characteristic proteolytic processing in these cells (Fig. 3B). Quantitation of the product polypeptides indicates that the processed forms are present in the same relative ratios to one another as are found in tissue-culture cells.
Activation of the C1 Factor as an Initial Event in the Reactivation of HSV from the Latent State.
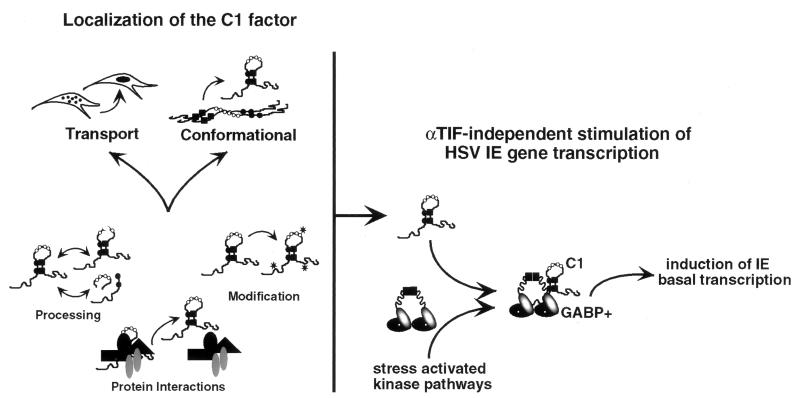
Two models that can account for the localization of the C1 factor are illustrated in Fig. 4 Left; (i) signal-induced transport from a cytoplasmic location and (ii) significant conformational alterations of nuclear pools that may be initially inaccessible to antisera. Either of these mechanisms may depend on phosphorylation of the C1 factor, altered proteolytic processing of the protein, or specific protein–protein interactions that would be regulated by signals that initiate the induction of HSV from the latent state. As the pattern of proteolysis does not differ on explant, it is unlikely that processing plays a significant role in the activation of the protein. Rather, interactions with neuron-specific proteins (J.L.V. and T.M.K., unpublished observations) may be the critical regulatory mechanism that controls the localization of the protein.
Figure 4.
Activation of the C1 factor in sensory neurons. (Left) The two primary models for the activation of the C1 factor are illustrated. For each, alterations in processing, modification, or protein–protein interactions may play a role in modulating the activity of the protein in response to specific signals that initiate HSV reactivation from the latent state. (Right) Localization of the C1 factor in sensory neurons allows interaction with other transcription factors such as GABP. Coupled with the modification of GABP by stress-activated kinases (GABP+), the interaction results in C1-mediated GABP transcriptional stimulation of the basal level expression of the HSV IE genes.
Regardless of the biochemical mechanism, the localization of the C1 factor correlates directly with the reactivation of HSV from the latent state. Unlike other cellular factors that have been analyzed in this context, the induction of C1 is not transcriptional. In addition, the time course of this activation strongly suggests that it is an initial event that may provide the link between generic activation signals (i.e., stress) and HSV reactivation. Presently, two models have been presented for reactivation of HSV from the latent state. The first model suggests that reactivation proceeds through a gene expression pattern that is distinct from the lytic phase. This hypothesis is based on the observation that elevated levels of HSV IE mRNA have not been detected in reactivated ganglia before the detection of late gene messages (26–28). However, it is important to consider that the stimulation of IE gene basal transcription may not result in detectably elevated levels of IE mRNA in a ganglion. Rather, the coordinated stimulation of the basal level expression of the five IE genes within a given neuron may be the most significant requirement for the initiation of reactivation.
The second model suggests that reactivation is caused by the coordinated activation of transcription of the five IE genes by cellular transcription factors in an αTIF-independent manner. Previous studies have clearly demonstrated that the transcriptional activation potential of αTIF is not required for the reactivation of HSV from latency (29–30). However, transcription of the IE genes is controlled by the reiterated enhancer domains that provide binding sites for numerous cellular transcription factors, including Oct-1 and GABP. Several studies have illustrated the significance of the GABP binding sites present in the IE enhancer domains for the basal transcription of these genes (13–14, 31). Strikingly, recent studies demonstrate that the C1 factor interacts with and mediates the apparent transcriptional stimulation by GABP (J.L.V. and T.M.K., unpublished work). The altered localization of the C1 factor in stimulated ganglia may therefore result in interaction with GABP and coordinated regulation of the basal level expression of the IE genes in an αTIF-independent manner (Fig. 4 Right). This model is further strengthened by studies that demonstrate that GABP is present in sensory neurons and is regulated by stress-activated protein kinase pathways (15, 17–18). While the relocation of the C1 factor may represent a critical step in the reactivation process, additional factors may also be important in determining a productive viral reactivation.
Control of the HSV latency cycle by C1 is an interesting model, because the factor is an unusual transcription coactivator that has a critical role in the regulation of HSV IE genes. In addition, its role in the regulation of cell-cycle progression may be related to mechanisms evolved by the virus to determine the cell state. Finally, the protein is involved in the regulation of the IE genes of other neurotrophic herpes viruses (32) and may, therefore, also represent the critical switch component of these viral lytic–latent cycles. As such, the regulated localization of the C1 factor may represent an important target for novel antiviral therapeutics.
Acknowledgments
We thank J. Yewdell and J. Bennink for expert microscopy assistance; P. Sharp, B. Moss, and members of the Laboratory of Viral Diseases for helpful discussions; Yelena Besnovataya for technical assistance, and B. Moss, A. McBride, L. D’Adamio, J. Yewdell, and J. L. C. McKnight for critical reading of this manuscript. These studies were supported by the Laboratory of Viral Diseases, National Institute of Allergy and Infectious Diseases, National Institutes of Health (T.M.K.) and National Institutes of Health Grant A124009 (A.S.).
ABBREVIATIONS
- HCF
host cell factor
- HSV
herpes simplex virus
- IE
immediate early gene
- NGF
nerve growth factor
- αTIF
α-trans-induction-factor
- GABP
GA binding protein
References
- 1.Roizman B, Sears A E. In: Fundamental Virology. Fields B, Knipe D M, Howley P M, editors. Philadelphia: Lippincott; 1996. pp. 1043–1107. [Google Scholar]
- 2.Whitley R J. In: Fields Virology. Fields B, Knipe D M, Howley P M, editors. Philadelphia: Lippincott; 1996. pp. 2297–2342. [Google Scholar]
- 3.Gerster T, Roeder R G. Proc Natl Acad Sci USA. 1988;85:6347–6351. doi: 10.1073/pnas.85.17.6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kristie T M, LeBowitz J H, Sharp P M. EMBO J. 1989;8:4229–4238. doi: 10.1002/j.1460-2075.1989.tb08608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kristie T M, Sharp P A. Genes Dev. 1990;4:2383–2396. doi: 10.1101/gad.4.12b.2383. [DOI] [PubMed] [Google Scholar]
- 6.O’Hare P, Goding C R. Cell. 1988;52:435–445. doi: 10.1016/s0092-8674(88)80036-9. [DOI] [PubMed] [Google Scholar]
- 7.Preston C M, Frame M C, Campbell M E M. Cell. 1988;52:425–434. doi: 10.1016/s0092-8674(88)80035-7. [DOI] [PubMed] [Google Scholar]
- 8.Kristie T M, Sharp P A. J Biol Chem. 1993;268:6525–6534. [PubMed] [Google Scholar]
- 9.Wilson A C, LaMarco K, Peterson M G, Herr W. Cell. 1993;74:115–125. doi: 10.1016/0092-8674(93)90299-6. [DOI] [PubMed] [Google Scholar]
- 10.Kristie T M, Pomerantz J L, Twomey T C, Parent S A, Sharp P A. J Biol Chem. 1995;270:4387–4394. doi: 10.1074/jbc.270.9.4387. [DOI] [PubMed] [Google Scholar]
- 11.Wilson A C, Peterson M G, Herr W. Genes Dev. 1995;9:2445–2458. doi: 10.1101/gad.9.20.2445. [DOI] [PubMed] [Google Scholar]
- 12.McKnight J L C, Kristie T M, Roizman B. Proc Natl Acad Sci USA. 1987;84:7061–7065. doi: 10.1073/pnas.84.20.7061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Triezenberg S J, LaMarco K L, McKnight S L. Genes Dev. 1988;2:730–742. doi: 10.1101/gad.2.6.730. [DOI] [PubMed] [Google Scholar]
- 14.LaMarco K L, McKnight S L. Genes Dev. 1989;3:1372–1383. doi: 10.1101/gad.3.9.1372. [DOI] [PubMed] [Google Scholar]
- 15.Hagmann M, Georgiev O, Schaffner W, Douville P. Nucleic Acids Res. 1995;23:4978–4985. doi: 10.1093/nar/23.24.4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valyi-Nagy T, Deshmane S, Dillner A, Fraser N W. J Virol. 1991;65:4142–4152. doi: 10.1128/jvi.65.8.4142-4152.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffmeyer A, Avots A, Flory E, Weber C K, Serfling E, Rapp U R. J Biol Chem. 1988;273:10112–10119. doi: 10.1074/jbc.273.17.10112. [DOI] [PubMed] [Google Scholar]
- 18.Flory E, Hoffmeyer A, Smola U, Rapp R, Bruder J T. J Virol. 1996;70:2260–2268. doi: 10.1128/jvi.70.4.2260-2268.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goto H, Motomura S, Wilson A C, Freiman R N, Nakabeppu Y, Fukushima K, Fujishima M, Herr W, Nishimoto T H. Genes Dev. 1997;11:726–737. doi: 10.1101/gad.11.6.726. [DOI] [PubMed] [Google Scholar]
- 20.Kristie T M. J Biol Chem. 1997;272:26749–26755. doi: 10.1074/jbc.272.42.26749. [DOI] [PubMed] [Google Scholar]
- 21.Wilson A C, Parrish J E, Massa H F, Nelson D L, Trask B J, Herr W. Genomics. 1995;25:462–468. doi: 10.1016/0888-7543(95)80046-o. [DOI] [PubMed] [Google Scholar]
- 22.Fawl R L, Roizman B. J Virol. 1993;67:7025–7031. doi: 10.1128/jvi.67.12.7025-7031.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilcox C L, Johnson E M. J Virol. 1988;62:393–399. doi: 10.1128/jvi.62.2.393-399.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilcox C L, Johnson E M. J Virol. 1987;61:2311–2315. doi: 10.1128/jvi.61.7.2311-2315.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilcox C L, Smith R L, Freed C R, Johnson E M. J Neurosci. 1990;4:1268–1275. doi: 10.1523/JNEUROSCI.10-04-01268.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tal-Singer R, Lasner T M, Podrzucki W, Skokotas A, Leary J J, Berger S L, Fraser N W. J Virol. 1997;71:5268–5276. doi: 10.1128/jvi.71.7.5268-5276.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kosz-Vnenchak M, Jacobson J, Coen D M, Knipe D M. J Virol. 1993;67:5383–5393. doi: 10.1128/jvi.67.9.5383-5393.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kramer M F, Coen D M. J Virol. 1995;69:1389–1399. doi: 10.1128/jvi.69.3.1389-1399.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sears A E, Hukkanen V, Labow M A, Levine A J, Roizman B. J Virol. 1991;65:2929–2935. doi: 10.1128/jvi.65.6.2929-2935.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steiner I, Spivack J G, Deshmane S L, Ace C I, Preston C M, Fraser N W. J Virol. 1990;64:1630–1638. doi: 10.1128/jvi.64.4.1630-1638.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kristie T M, Roizman B. Proc Natl Acad Sci USA. 1984;81:4065–4069. doi: 10.1073/pnas.81.13.4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moriuchi H, Moriuchi M, Cohen J I. J Virol. 1995;69:4693–4701. doi: 10.1128/jvi.69.8.4693-4701.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]