In the early 1970s, physicians were finally forced to abandon their belief that, given the vast array of effective antimicrobial agents, virtually all bacterial infections were treatable. Their optimism was shaken by the emergence of resistance to multiple antibiotics among such pathogens as Staphylococcus aureus, Streptococcus pneumoniae, Pseudomonas aeruginosa, and Mycobacterium tuberculosis. The evolution of increasingly antimicrobial-resistant bacterial species stems from a multitude of factors that includes the widespread and sometimes inappropriate use of antimicrobials, the extensive use of these agents as growth enhancers in animal feed, and, with the increase in regional and international travel, the relative ease with which antimicrobial-resistant bacteria cross geographic barriers (1–3).
The irony of this trend toward progressively more resistant bacteria is that it coincides with a period of dramatically increased understanding of the molecular mechanisms of antimicrobial resistance. Unfortunately, while this insight has resulted in the identification of novel drug targets, it has not yet resulted in effective new chemotherapeutic agents. This paradox stands in sharp contrast to the dramatic progress made in antiviral (notably antiretroviral) therapy in the past ten years, where a number of newly discovered molecular targets have resulted in clinically effective therapeutic agents.
Nowhere has this issue been of greater concern than with the Gram-positive bacteria pneumococci, enterococci, and staphylococci. Multidrug resistance is now the norm among these pathogens. S. aureus is perhaps the pathogen of greatest concern because of its intrinsic virulence, its ability to cause a diverse array of life-threatening infections, and its capacity to adapt to different environmental conditions (4, 5). The mortality of S. aureus bacteremia remains approximately 20–40% despite the availability of effective antimicrobials (6). S. aureus is now the leading overall cause of nosocomial infections and, as more patients are treated outside the hospital setting, is an increasing concern in the community (7, 8).
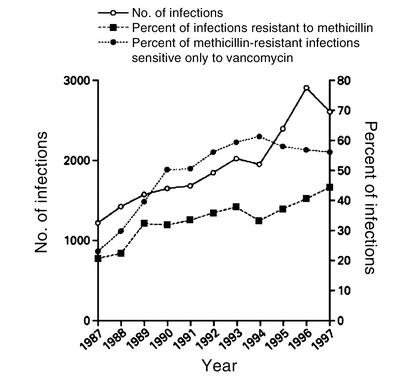
S. aureus isolates from intensive care units across the country and from blood culture isolates worldwide are increasingly resistant to a greater number of antimicrobial agents (4, 8). Inevitably this has left fewer effective bactericidal antibiotics to treat these often life-threatening infections (Figure 1). As rapidly as new antibiotics are introduced, staphylococci have developed efficient mechanisms to neutralize them (Table 1).
Figure 1.
S. aureus infections in intensive care units in the National Nosocomial Infections Surveillance System. Data include the total number of infections from 1987 to 1997. Isolates were tested for sensitivity to the following antimicrobial agents: gentamicin, tobramycin, amikacin, ciprofloxacin, clindamycin, erythromycin, chloramphenicol, trimethoprim-sulfamethoxazole, and vancomycin. Some hospitals did not test for all of the antibiotics. Reproduced with permission from New England Journal of Medicine (4).
Table 1.
Mechanisms of S. aureus resistance to antimicrobialsA
Recent reports of S. aureus isolates with intermediate or complete resistance to vancomycin portend a chemotherapeutic era in which effective bactericidal antibiotics against this organism may no longer be readily available (9, 10). This review will focus on the emergence of antimicrobial resistance in S. aureus. It will review the historical evolution of resistant strains, their spread, the molecular mechanisms of resistance for selected antibiotics, and progress toward the development of alternative drug targets or novel approaches for therapeutic or prophylactic intervention.
Penicillin resistance
History and epidemiology.
The mortality of patients with S. aureus bacteremia in the pre-antibiotic era exceeded 80%, and over 70% developed metastatic infections (11). The introduction of penicillin in the early 1940s dramatically improved the prognosis of patients with staphylococcal infection. However, as early as 1942, penicillin-resistant staphylococci were recognized, first in hospitals and subsequently in the community (12). By the late 1960s, more than 80% of both community- and hospital-acquired staphylococcal isolates were resistant to penicillin. This pattern of resistance, first emerging in hospitals and then spreading to the community, is now a well-established pattern that recurs with each new wave of antimicrobial resistance (13).
Kirby first demonstrated that penicillin was inactivated by penicillin-resistant strains of S. aureus (14). Bondi and Dietz (15) subsequently identified the specific role of penicillinase. More than 90% of staphylococcal isolates now produce penicillinase, regardless of the clinical setting. The gene for β-lactamase is part of a transposable element located on a large plasmid, often with additional antimicrobial resistance genes (e.g., gentamicin and erythromycin). Spread of penicillin resistance primarily occurs by spread of resistant strains.
Mechanisms of resistance.
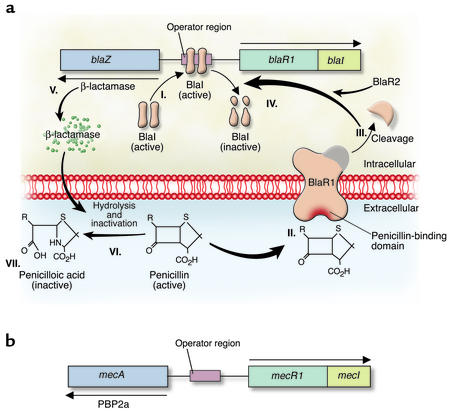
Staphylococcal resistance to penicillin is mediated by blaZ, the gene that encodes β-lactamase (Figure 2a). This predominantly extracellular enzyme, synthesized when staphylococci are exposed to β-lactam antibiotics, hydrolyzes the β-lactam ring, rendering the β-lactam inactive. blaZ is under the control of two adjacent regulatory genes, the antirepressor blaR1 and the repressor blaI (16). Recent studies have demonstrated that the signaling pathway responsible for β-lactamase synthesis requires sequential cleavage of the regulatory proteins BlaR1 and BlaI. Following exposure to β-lactams, BlaR1, a transmembrane sensor-transducer, cleaves itself (17, 18). Zhang et al. (18) hypothesize that the cleaved protein functions as a protease that cleaves the repressor BlaI, directly or indirectly (an additional protein, BlaR2, may be involved in this pathway) and allows blaZ to synthesize enzyme.
Figure 2.
(a) Induction of staphylococcal β-lactamase synthesis in the presence of the β-lactam antibiotic penicillin. I. The DNA-binding protein BlaI binds to the operator region, thus repressing RNA transcription from both blaZ and blaR1-blaI. In the absence of penicillin, β-lactamase is expressed at low levels. II. Binding of penicillin to the transmembrane sensor-transducer BlaR1 stimulates BlaR1 autocatalytic activation. III–IV. Active BlaR1 either directly or indirectly (via a second protein, BlaR2) cleaves BlaI into inactive fragments, allowing transcription of both blaZ and blaR1-blaI to commence. V–VII. β-Lactamase, the extracellular enzyme encoded by blaZ (V), hydrolyzes the β-lactam ring of penicillin (VI), thereby rendering it inactive (VII). (b) Mechanism of S. aureus resistance to methicillin. Synthesis of PBP2a proceeds in a fashion similar to that described for β-lactamase. Exposure of MecR1 to a β-lactam antibiotic induces MecR1 synthesis. MecR1 inactivates MecI, allowing synthesis of PBP2a. MecI and BlaI have coregulatory effects on the expression of PBP2a and β-lactamase.
Methicillin resistance
History and epidemiology.
Methicillin, introduced in 1961, was the first of the semisynthetic penicillinase-resistant penicillins. Its introduction was rapidly followed by reports of methicillin-resistant isolates (19). For clinicians, the spread of these methicillin-resistant strains has been a critical one. The therapeutic outcome of infections that result from methicillin-resistant S. aureus (MRSA) is worse than the outcome of those that result from methicillin-sensitive strains (20). The difference has been ascribed to the underlying medical problems of the often sicker and older MRSA-infected patients as well as the less effective bactericidal drugs available to treat these infections, rather than to enhanced virulence of the MRSA strains.
First reported in a British hospital, MRSA clones rapidly spread across international borders. Waves of clonal dissemination with different dominant phage types (e.g., 83 complex) were reported in the 1960s and were responsible for a large proportion of cases (21, 22). Once identified in a new setting, these unique MRSA clones rapidly spread, often becoming the resident clones and accounting for an increasing percentage of nosocomial infections (23, 24). Like the penicillin-resistant strains, the MRSA isolates also frequently carried resistance genes to other antimicrobial agents (25).
The spread of methicillin-resistant clones is reminiscent of the emergence of penicillin resistance in the 1940s. First detected in hospitals in the 1960s, methicillin resistance is now increasingly recognized in the community (13). While many of these infections occurred in patients with some antecedent hospital experience, recently there has been an increasing number of subjects with no prior hospital exposure. These community-based infections have been reported in patients from both rural and urban settings (26–29). Of concern is the high mortality associated with some of these community-acquired MRSA infections. In one instance, clonally related MRSA strains caused the deaths of four otherwise healthy children (28). The empiric selection of β-lactams as initial therapy may have contributed to the increased morbidity in these infections. In addition, the presence of virulence genes such as the enterotoxins or the Panton-Valentine leukocidin may also have contributed to patient morbidity in some of the community-acquired infections (30, 31).
The mecA gene (the gene responsible for methicillin resistance) is part of a mobile genetic element found in all MRSA strains. Katayama et al. (32) demonstrated that mecA is part of a genomic island designated staphylococcal cassette chromosome mec (SCCmec). To date, four different SCCmec elements varying in size from 21 to 67 kb have been characterized (33). In contrast to the numerous different strains of methicillin-susceptible S. aureus (MSSA) that cause infections, only a limited number of clones are responsible for the epidemic spread of MRSA. This distinction reflects the genetic constraints of horizontal transfer of the mec element from related staphylococcal species into S. aureus. The frequency of this transfer event is subject to some debate, but it is clearly uncommon (34–37). The mec element SCCmec has been identified in several different MSSA genetic backgrounds. Using the DNA fingerprint of the recipient MSSA in combination with the genetic organization of the particular SCCmec, an MRSA biotype as well as an evolutionary profile of a particular clone can be established (33, 35–38). Oliveira et al. identified a putative pathway for the evolution of several pandemic clones in Europe (38). By comparing the genetic backgrounds of epidemic MSSA clones, these investigators also identified the likely ancestral MSSA recipients of the mec element. These studies suggest that the emergence of “epidemic” MRSA clones was in part the result of the successful horizontal transfer of the mec gene into an ecologically fit and transmission-efficient MSSA clone (37, 39).
The recent upsurge of community-acquired MRSA infections reported in patients from different countries was associated with the detection of a unique SCCmec, type IV (40). This element, smaller than the other elements, appears more genetically mobile and does not, at present, carry additional antimicrobial resistance genes. It also appears to occur in a more diverse range of MSSA genetic backgrounds, suggesting that it has been heterologously transferred more readily from other staphylococcal species (41).
Mechanisms of resistance.
Methicillin resistance requires the presence of the chromosomally localized mecA gene (16, 42) (Figure 2b). mecA is responsible for synthesis of penicillin-binding protein 2a (PBP2a; also called PBP2′) a 78-kDa protein (43–45). PBPs are membrane-bound enzymes that catalyze the transpeptidation reaction that is necessary for cross-linkage of peptidoglycan chains (46). Their activity is similar to that of serine proteases, from which they appear to have evolved. PBP2a substitutes for the other PBPs and, because of its low affinity for all β-lactam antibiotics, enables staphylococci to survive exposure to high concentrations of these agents. Thus, resistance to methicillin confers resistance to all β-lactam agents, including cephalosporins. Recent studies determined the crystal structure of a soluble derivative of PBP2a. PBP2a differs from other PBPs in that its active site blocks binding of all β-lactams but allows the transpeptidation reaction to proceed (47).
Phenotypic expression of methicillin resistance is variable, and each MRSA strain has a characteristic profile of the proportion of bacterial cells that grow at specific concentrations of methicillin (48). Expression of resistance in some MRSA strains is regulated by homologues of the regulatory genes for blaZ. These genes, mecI and mecR1, regulate the mecA response to β-lactam antibiotics in a fashion similar to that of the regulation of blaZ by the genes blaR1 and blaI upon exposure to penicillin (Figure 2). In fact, the DNA sequences bound by the repressor genes to achieve inhibition of gene activation are identical (49). The sequence homology of mecI-mecR1 with the blaR1-blaI regulatory genes results in the induction of mecA expression from this leaky alternative system. Deletions or mutations in mecI or the promoter region of mecA result in constitutive expression rather than variable expression of mec (50). Rosato et al. (51) have recently found that either mecI or blaI must be functional in all MRSA, and they suggest that this may be a protective mechanism preventing overproduction of a toxic protein. An additional series of genes, the fem genes (factor essential for resistance to methicillin resistance, play a role in cross-linking peptidoglycan strands and also contribute to the heterogeneity of expression of methicillin resistance (52).
As noted earlier, the mecA gene is invariably part of a larger unique, mobile genetic element, SCCmec. These islands may also contain additional genes for antimicrobial resistance and insertion sequences, as well as genes of uncertain function. The four SCCmec’s contain two recombinases, ccrA and ccrB from the invertase/resolvase family, that are responsible for site-specific integration and excision from the chromosome at attBscc, a part of an open reading frame of unknown function near the origin of replication (32, 53). The genetic mechanisms responsible for the transfer of these large mobile elements are uncertain.
Since no homologue of mecA exists in methicillin-susceptible staphylococci, it has been assumed that mecA was acquired from one of several coagulase-negative staphylococcal species (54). Couto et al. (55) identified a mecA gene in a methicillin-sensitive Staphylococcus sciuri with 88% homology on the amino acid level to MRSA. Transduction of the S. sciuri mecA into an MSSA resulted in increased resistance to methicillin coupled with the detection of PBP2a (56). These studies therefore suggest one possible source of the mecA element in S. aureus. Hiramatsu and associates (41, 57) have speculated that the simultaneous detection of the new type IV SCCmec in different geographic regions of the world potentially reflects its enhanced mobility and multiple simultaneous transmissions from another coagulase-negative staphylococcus.
Quinolone resistance
History and epidemiology.
Fluoroquinolones were initially introduced for the treatment of Gram-negative bacterial infections in the 1980s. However, because of their Gram-positive bacterial spectrum, they have also been used to treat bacterial infections caused by pneumococci and staphylococci. Quinolone resistance among S. aureus emerged quickly, more prominently among the methicillin-resistant strains. As a result, the ability to use fluoroquinolones as antistaphylococcal agents was dramatically reduced. The reasons for the disparity in rates of quinolone resistance between MSSA and MRSA strains are uncertain. One contributing factor is likely antibiotic selective pressure, especially in the hospital setting, resulting in the selection and spread of the more antibiotic-resistant MRSA strains.
Fluoroquinolone resistance develops as a result of spontaneous chromosomal mutations in the target of the antibiotic, topoisomerase IV or DNA gyrase, or by the induction of a multidrug efflux pump. When quinolones are used to treat infections caused by other bacterial pathogens, subjects colonized with S. aureus (e.g., on their skin or mucosal surfaces) are likely exposed to subtherapeutic antibiotic concentrations and are therefore at risk of becoming colonized with resistant mutants (58). These resident, resistant strains then become the reservoir for future infections. Høiby et al. (59) demonstrated that ciprofloxacin therapy rapidly increased the proportion of coagulase-negative staphylococcal strains colonizing the nares and skin that were resistant to both ciprofloxacin and methicillin. Since S. aureus is also a part of our commensal flora, a similar selection process is likely to occur.
Mechanisms of resistance.
Resistance to quinolones results from the stepwise acquisition of chromosomal mutations. The confluence of high bacterial density, the likely preexistence of resistant subpopulations, and the sometimes limited quinolone concentrations achieved at sites of staphylococcal infections creates an environment that fosters selection of resistant mutants (58). The quinolones act on DNA gyrase, which relieves DNA supercoiling, and topoisomerase IV, which separates concatenated DNA strands. Amino acid changes in critical regions of the enzyme-DNA complex (quinolone resistance–determining region [QRDR]) reduce quinolone affinity for both of its targets. The ParC subunit (GrlA in S. aureus) of topoisomerase IV and the GyrA subunit in gyrase are the most common sites of resistance mutations; topoisomerase IV mutations are the most critical, since they are the primary drug targets in staphylococci (58, 60). Single amino acid mutations are sometimes sufficient to confer clinical resistance, but for the more active fluoroquinolones additional mutations appear necessary. Resistance mutations can accumulate in the QRDR sites, increasing the levels of resistance. It is fairly common for both targets to have resistance mutations. An additional mechanism of resistance in S. aureus is induction of the NorA multidrug resistance efflux pump. Increased expression of this pump in S. aureus can result in low-level quinolone resistance (61). In an interesting linkage of virulence to antimicrobial resistance, a recent study showed that exposure of a quinolone-resistant isolate to a quinolone increased the organism’s expression of fibronectin-binding protein, a surface protein that mediates adherence to tissue surfaces (62).
While newer fluoroquinolones (8-methoxyquinolones such as moxifloxacin) retain in vitro activity against ciprofloxacin-resistant staphylococci and appear less likely to select for resistant mutants (63), it is not clear that this translates into therapeutic efficacy. Entenza et al. (64) reported that while moxifloxacin retained a low minimal inhibitory concentration (MIC) against ciprofloxacin-resistant strains of S. aureus, treatment with this newer quinolone failed in an experimental model of endocarditis. This suggests a potential disparity between in vitro susceptibility testing and in vivo therapeutic efficacy.
Vancomycin resistance
History and epidemiology.
The dramatic increase in use of vancomycin to treat infections caused by methicillin-resistant staphylococci (both coagulase-positive and -negative), Clostridium difficile, and enterococcal infections preceded the emergence of vancomycin-resistant staphylococci (65). Staphylococcal resistance to vancomycin in a clinical isolate was first reported in a strain of Staphylococcus haemolyticus (66). In 1997, the first report of vancomycin intermediate-resistant S. aureus (VISA) came from Japan, and additional cases were subsequently reported from other countries (9, 67). The VISA isolates were all MRSA and were not clonal. Many of the patients had received vancomycin therapy and had MRSA infections (68).
Two recent reports of infections caused by vancomycin-resistant S. aureus (VRSA) are of great concern because they reflect both complete resistance and a different mechanism for dissemination. In contrast to the chromosomally mediated resistance for VISA strains, the VRSA strains acquire resistance by conjugal transfer of the vanA operon from an Enterococcus faecalis, raising the specter of a far more efficient means for dissemination of the resistance gene among strains of staphylococci (10, 69).
Mechanisms of resistance.
Two forms of S. aureus resistance to vancomycin have now been identified (70). One form has been identified in the VISA strains, which have MICs to vancomycin of 8–16 μg/ml (9). A pre-VISA stage of resistance, heterogeneously resistant, has also been identified. The heteroresistant strains remain susceptible to vancomycin but contain resistant subpopulations. It is hypothesized that, on exposure to vancomycin, the VISA isolates are selected from the vancomycin-resistant subpopulations (71). The reduced susceptibility to vancomycin appears to result from changes in peptidoglycan biosynthesis (Figure 3). The VISA strains are notable for the additional quantities of synthesized peptidoglycan that result in irregularly shaped, thickened cell walls. There is also decreased cross-linking of peptidoglycan strands, which leads to the exposure of more D-Ala-D-Ala residues (72, 73). The altered cross-linking results from reduced amounts of L-glutamine that are available for amidation of D-glutamate in the pentapeptide bridge (70). As a result there are more D-Ala-D-Ala residues available to bind and trap vancomycin (Figure 3). The bound vancomycin then acts as a further impediment to drug molecules reaching their target on the cytoplasmic membrane. HPLC provided further proof of this novel resistance mechanism by showing that large quantities of vancomycin become trapped in the abnormal peptidoglycan (74). The molecular mechanisms for these alterations in peptidoglycan biosynthesis are unexplained.
Figure 3.
Mechanisms of S. aureus resistance to vancomycin: VISA strains. VISA strains appear to be selected from isolates that are heterogeneously resistant to vancomycin. These VISA strains synthesize additional quantities of peptidoglycan with an increased number of D-Ala-D-Ala residues that bind vancomycin, preventing the molecule from getting to its bacterial target. Adapted from ref. 98.
The second form of vancomycin resistance has resulted from the probable conjugal transfer of the vanA operon from a vancomycin-resistant E. faecalis. Showsh et al. (75) reported that the enterococcal plasmid containing vanA also encodes a sex pheromone that is synthesized by S. aureus, suggesting a potential facilitator of conjugal transfer. These VRSA isolates demonstrate complete vancomycin resistance, with MICs of ≥128 μg/ml. Resistance in these isolates is caused by alteration of the terminal peptide to D-Ala-D-Lac instead of D-Ala-D-Ala (Figure 4). Synthesis of D-Ala-D-Lac occurs only with exposure to low concentrations of vancomycin. As a result, the additional biosynthetic demands are limited and the VRSA strain is ecologically fit (76). This ecological fitness, the possibility that this plasmid exchange will occur more frequently (due to the ever increasing likelihood of patients being colonized with both MRSA and vancomycin-resistant enterococci), and the resistance of these strains to both β-lactams and glycopeptides all increase the likelihood that VRSA strains will rapidly become more prevalent.
Figure 4.
Mechanisms of S. aureus resistance to vancomycin: VRSA strains. VRSA strains are resistant to vancomycin because of the acquisition of the vanA operon from an enterococcus that allows synthesis of a cell wall precursor that ends in D-Ala-D-Lac dipeptide rather than D-Ala-D-Ala. The new dipeptide has dramatically reduced affinity for vancomycin. In the presence of vancomycin, the novel cell wall precursor is synthesized, allowing continued peptidoglycan assembly. Adapted from ref. 99.
Prospects for the future: new antimicrobials, new approaches
Currently available agents.
Quinupristin-dalfopristin and linezolid are two of the newer antimicrobial agents currently available with activity against drug-resistant staphylococci (including most VISA and VRSA strains in vitro). Both agents are protein synthesis inhibitors with a Gram-positive spectrum. Quinupristin-dalfopristin retains bactericidal antistaphylococcal activity if the strain is susceptible to erythromycin and lincosamide. Linezolid is bacteriostatic. Cross-resistance has not been noted for linezolid, but at least one clinical isolate has developed resistance during therapy (77). Daptomycin, a novel bactericidal agent that damages the cytoplasmic membrane, is currently undergoing clinical trials (78). Other agents with antistaphylococcal activity in varying stages of development include modified glycopeptides, carbapenems, oxazolidinones, quinolones, and tetracyclines.
Novel approaches and targets.
The supply of new agents with novel mechanisms of action is limited, however, and emphasizes the need for the development of new drug targets (79, 80). Unfortunately, an increasing number of pharmaceutical companies have either eliminated or dramatically reduced their anti-infectives units. This results partly from financial considerations but also from frustration that target-based biochemical screening has failed to develop any clinically useful products. The failure has been attributed, in part, to the realization that target-based strategies do not take into account the “intrinsic” mechanisms of bacterial resistance (e.g., biofilms, multidrug efflux pumps) that contribute to in vivo bacterial resistance (79).
Despite these developments, a number of interesting models for identification of new drug targets have emerged. One approach has been to integrate genomic information on potential drug targets with high-throughput screening followed by chemical modification and efficacy animal testing. There has been a renewed interest in characterization of essential components of critical biosynthetic pathways (e.g., peptidoglycan assembly or fatty acid biosynthesis) as potential targets. Several different techniques, including in vivo expression technology, signature-tagged mutagenesis, and recognition of expressed S. aureus antigens, have been used to identify potential targets that are expressed during infection (80–82). Analysis of the crystal structure of drug targets (e.g., modifications of β-lactams that attack the active site of PBP2a) and synthesis of carbohydrate-modified compounds (glycopeptide analogues with altered carbohydrates) are increasingly being used to develop alternative agents (47, 83). Modification of S. aureus genes associated with virulence reduces infectivity (84, 85). Whether these genes can be successfully used as potential targets is uncertain.
Possible preventive measures.
Prevention of S. aureus infections has to date been limited to the application of infection control measures. In some countries, such as The Netherlands and Denmark, where strict isolation policies have prevailed, these precautions have been effective in preventing dissemination of MRSA, while in the USA and England, the success of infection control procedures has been limited. The potential of strict infection control programs to curb MRSA transmission suggests that, given recent trends, stricter infection control guidelines are warranted (86). Newer, more rapid diagnostic methods that can detect the presence of S. aureus or other pathogens and allow for rapid identification and isolation of colonized patients should enhance the efficacy of these programs.
Because nasal carriage of S. aureus raises the risk of subsequent infection, efforts have been directed to the elimination of carriage using topical antimicrobials (87–89). These approaches have been variably successful. More recently, the potential use of novel agents for this purpose, such as endopeptidase, lysostaphin, or phage lytic enzymes has also been considered (90–92).
Finally, several S. aureus vaccine candidates are under investigation. A capsular polysaccharide protein conjugate vaccine underwent a clinical trial with hemodialysis patients, with encouraging but inconclusive results (93). Other candidate vaccines directed at S. aureus virulence determinants such as the surface adhesins or enterotoxins are in varying stages of development (94).
The difficult therapeutic problem of multidrug-resistant S. aureus is just one example of the diminishing efficacy of antimicrobial agents for the treatment of bacterial infections. This trend is particularly alarming for S. aureus because of the severity and diversity of disease caused by this uniquely versatile pathogen. While effective antistaphylococcal agents still exist, their shelf-life is likely to be increasingly limited. Novel approaches to therapy and prevention will become more and more important, especially with the diminishing availability of new “wonder drugs.”
Acknowledgments
This work was supported by NIH grants DA-09656, DA-11868, and DA-15018. Abigail Zuger’s critical review of this manuscript is gratefully acknowledged.
Footnotes
Conflict of interest: The author has received research support from GlaxoSmithKline and is involved in ongoing clinical trials with Cubist Pharmaceuticals Inc.
Nonstandard abbreviations used: methicillin-resistant Staphylococcus aureus (MRSA); staphylococcal cassette chromosome mec (SCCmec); methicillin-susceptible S. aureus (MSSA); penicillin-binding protein (PBP); quinolone resistance–determining region (QRDR); minimal inhibitory concentration (MIC); vancomycin intermediate-resistant S. aureus (VISA); vancomycin-resistant S. aureus (VRSA).
The Science in Medicine series is supported in part by a generous grant from the Doris Duke Charitable Foundation.
References
- 1.Cohen ML. Epidemiology of drug resistance: implications for a post-antimicrobial era. Science. 1992;257:1050–1055. doi: 10.1126/science.257.5073.1050. [DOI] [PubMed] [Google Scholar]
- 2.Tomasz A. Multiple-antibiotic-resistant pathogenic bacteria. A report on the Rockefeller University Workshop. N. Engl. J. Med. 1994;330:1247–1251. doi: 10.1056/NEJM199404283301725. [DOI] [PubMed] [Google Scholar]
- 3.Swartz MN. Use of antimicrobial agents and drug resistance. N. Engl. J. Med. 1997;337:491–492. doi: 10.1056/NEJM199708143370709. [DOI] [PubMed] [Google Scholar]
- 4.Lowy FD. Staphylococcus aureus infections. N. Engl. J. Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 5.Waldvogel, F.A. 2000. Staphylococcus aureus (including staphylococcal toxic shock). In Principles and practice of infectious diseases. G.L. Mandell, J.E. Bennett, and R. Dolin, editors. Churchill Livingstone. Philadelphia, Pennsylvania, USA. 2069–2092.
- 6.Mylotte JM, McDermott C, Spooner JA. Prospective study of 114 consecutive episodes of Staphylococcus aureus bacteremia. Rev. Infect. Dis. 1987;9:891–907. doi: 10.1093/clinids/9.5.891. [DOI] [PubMed] [Google Scholar]
- 7.CDC NNIS System: National Nosocomial Infections Surveillance (NNIS) system report, data summary from January 1992–April 2001, issued August 2001. Am. J. Infect. Control. 2001;29:400–421. doi: 10.1067/mic.2001.119952. [DOI] [PubMed] [Google Scholar]
- 8.Diekema DJ, et al. Survey of infections due to Staphylococcus species: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, Latin America, Europe, and the Western Pacific region for the SENTRY Antimicrobial Surveillance Program, 1997–1999. Clin. Infect. Dis. 2001;32(Suppl. 2):S114–S132. doi: 10.1086/320184. [DOI] [PubMed] [Google Scholar]
- 9.Hiramatsu K, et al. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J. Antimicrob. Chemother. 1997;40:135–136. doi: 10.1093/jac/40.1.135. [DOI] [PubMed] [Google Scholar]
- 10.Staphylococcus aureus resistant to vancomycin. United States, 2002. MMWR. 2002;51:565–567. [PubMed] [Google Scholar]
- 11.Skinner D, Keefer CS. Significance of bacteremia caused by Staphylococcus aureus. Arch. Intern. Med. 1941;68:851–875. [Google Scholar]
- 12.Rammelkamp CH, Maxon T. Resistance of Staphylococcus aureus to the action of penicillin. Proc. Royal Soc. Exper. Biol. Med. 1942;51:386–389. [Google Scholar]
- 13.Chambers HF. The changing epidemiology of Staphylococcus aureus? Emerg. Infect. Dis. 2001;7:178–182. doi: 10.3201/eid0702.010204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kirby WMM. Extraction of a higly potent penicillin inactivator from penicillin resistant staphylococci. Science. 1944;99:452–453. doi: 10.1126/science.99.2579.452. [DOI] [PubMed] [Google Scholar]
- 15.Bondi JA, Dietz CC. Penicillin resistant staphylococci. Proc. Royal Soc. Exper. Biol. Med. 1945;60:55–58. doi: 10.3181/00379727-60-15089. [DOI] [PubMed] [Google Scholar]
- 16.Kernodle, D.S. 2000. Mechanisms of resistance to β-lactam antibiotics. In Gram-positive pathogens. V.A. Fischetti, R.P. Novick, J.J. Ferretti, D.A. Portnoy, and J.I. Rood, editors. American Society for Microbiology. Washington, DC, USA. 609–620.
- 17.Gregory PD, Lewis RA, Curnock SP, Dyke KG. Studies of the repressor (BlaI) of beta-lactamase synthesis in Staphylococcus aureus. Mol. Microbiol. 1997;24:1025–1037. doi: 10.1046/j.1365-2958.1997.4051770.x. [DOI] [PubMed] [Google Scholar]
- 18.Zhang HZ, Hackbarth CJ, Chansky KM, Chambers HF. A proteolytic transmembrane signaling pathway and resistance to beta-lactams in staphylococci. Science. 2001;291:1962–1965. doi: 10.1126/science.1055144. [DOI] [PubMed] [Google Scholar]
- 19.Jevons MP. “Celbenin”-resistant staphylococci. Br. Med. J. 1961;1:124–125. [Google Scholar]
- 20.Cosgrove SE, et al. Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus bacteremia: a meta-analysis. Clin. Infect. Dis. 2003;36:53–59. doi: 10.1086/345476. [DOI] [PubMed] [Google Scholar]
- 21.Jessen O, Rosendal K, Bulow P, Faber V, Eriksen KR. Changing staphylococci and staphylococcal infections. A ten-year study of bacteria and cases of bacteremia. N. Engl. J. Med. 1969;281:627–635. doi: 10.1056/NEJM196909182811201. [DOI] [PubMed] [Google Scholar]
- 22.Parker MT, Hewitt JH. Methicillin resistance in Staphylococcus aureus. Lancet. 1970;1:800–804. doi: 10.1016/s0140-6736(70)92408-6. [DOI] [PubMed] [Google Scholar]
- 23.Panlilio AL, et al. Methicillin-resistant Staphylococcus aureus in U.S. hospitals, 1975–1991. Infect. Control Hosp. Epidemiol. 1992;13:582–586. doi: 10.1086/646432. [DOI] [PubMed] [Google Scholar]
- 24.Couto I, et al. Unusually large number of methicillin-resistant Staphylococcus aureus clones in a Portuguese hospital. J. Clin. Microbiol. 1995;33:2032–2035. doi: 10.1128/jcm.33.8.2032-2035.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lyon BR, Iuorio JL, May JW, Skurray RA. Molecular epidemiology of multiresistant Staphylococcus aureus in Australian hospitals. J. Med. Microbiol. 1984;17:79–89. doi: 10.1099/00222615-17-1-79. [DOI] [PubMed] [Google Scholar]
- 26.Moreno F, Crisp C, Jorgensen JH, Patterson JE. Methicillin-resistant Staphylococcus aureus as a community organism. Clin. Infect. Dis. 1995;21:1308–1312. doi: 10.1093/clinids/21.5.1308. [DOI] [PubMed] [Google Scholar]
- 27.Herold BC, et al. Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. J. Am. Med. Assoc. 1998;279:593–598. doi: 10.1001/jama.279.8.593. [DOI] [PubMed] [Google Scholar]
- 28.Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus: Minnesota and North Dakota, 1997–1999. MMWR. 1999;48:707–710. [PubMed] [Google Scholar]
- 29.Groom AV, et al. Community-acquired methicillin-resistant Staphylococcus aureus in a rural American Indian community. J. Am. Med. Assoc. 2001;286:1201–1205. doi: 10.1001/jama.286.10.1201. [DOI] [PubMed] [Google Scholar]
- 30.Baba T, et al. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet. 2002;359:1819–1827. doi: 10.1016/s0140-6736(02)08713-5. [DOI] [PubMed] [Google Scholar]
- 31.Fey PD, et al. Comparative molecular analysis of community- or hospital-acquired methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2003;47:196–203. doi: 10.1128/AAC.47.1.196-203.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katayama Y, Ito T, Hiramatsu K. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 2000;44:1549–1555. doi: 10.1128/aac.44.6.1549-1555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hiramatsu K, Cui L, Kuroda M, Ito T. The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends Microbiol. 2001;9:486–493. doi: 10.1016/s0966-842x(01)02175-8. [DOI] [PubMed] [Google Scholar]
- 34.Kreiswirth B, et al. Evidence for a clonal origin of methicillin resistance in Staphylococcus aureus. Science. 1993;259:227–230. doi: 10.1126/science.8093647. [DOI] [PubMed] [Google Scholar]
- 35.Hiramatsu K. Molecular evolution of MRSA. Microbiol. Immunol. 1995;39:531–543. doi: 10.1111/j.1348-0421.1995.tb02239.x. [DOI] [PubMed] [Google Scholar]
- 36.Fitzgerald JR, Sturdevant DE, Mackie SM, Gill SR, Musser JM. Evolutionary genomics of Staphylococcus aureus: insights into the origin of methicillin-resistant strains and the toxic shock syndrome epidemic. Proc. Natl. Acad. Sci. U. S. A. 2001;98:8821–8826. doi: 10.1073/pnas.161098098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Enright MC, et al. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA) Proc. Natl. Acad. Sci. U. S. A. 2002;99:7687–7692. doi: 10.1073/pnas.122108599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oliveira DC, Tomasz A, de Lencastre H. The evolution of pandemic clones of methicillin-resistant Staphylococcus aureus: identification of two ancestral genetic backgrounds and the associated mec elements. Microb. Drug Resist. 2001;7:349–361. doi: 10.1089/10766290152773365. [DOI] [PubMed] [Google Scholar]
- 39.Crisostomo MI, et al. The evolution of methicillin resistance in Staphylococcus aureus: similarity of genetic backgrounds in historically early methicillin-susceptible and -resistant isolates and contemporary epidemic clones. Proc. Natl. Acad. Sci. U. S. A. 2001;98:9865–9870. doi: 10.1073/pnas.161272898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma XX, et al. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 2002;46:1147–1152. doi: 10.1128/AAC.46.4.1147-1152.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okuma K, et al. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J. Clin. Microbiol. 2002;40:4289–4294. doi: 10.1128/JCM.40.11.4289-4294.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chambers HF. Methicillin resistance in staphylococci: molecular and biochemical basis and clinical implications. Clin. Microbiol. Rev. 1997;10:781–791. doi: 10.1128/cmr.10.4.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hartman BJ, Tomasz A. Low-affinity penicillin-binding protein associated with beta-lactam resistance in Staphylococcus aureus. J. Bacteriol. 1984;158:513–516. doi: 10.1128/jb.158.2.513-516.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Utsui Y, Yokota T. Role of an altered penicillin-binding protein in methicillin- and cephem-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 1985;28:397–403. doi: 10.1128/aac.28.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song MD, Wachi M, Doi M, Ishino F, Matsuhashi M. Evolution of an inducible penicillin-target protein in methicillin-resistant Staphylococcus aureus by gene fusion. FEBS Lett. 1987;221:167–171. doi: 10.1016/0014-5793(87)80373-3. [DOI] [PubMed] [Google Scholar]
- 46.Ghuysen JM. Molecular structures of penicillin-binding proteins and beta-lactamases. Trends Microbiol. 1994;2:372–380. doi: 10.1016/0966-842x(94)90614-9. [DOI] [PubMed] [Google Scholar]
- 47.Lim D, Strynadka NC. Structural basis for the beta lactam resistance of PBP2a from methicillin-resistant Staphylococcus aureus. Nat. Struct. Biol. 2002;9:870–876. doi: 10.1038/nsb858. [DOI] [PubMed] [Google Scholar]
- 48.Tomasz A, Nachman S, Leaf H. Stable classes of phenotypic expression in methicillin-resistant clinical isolates of staphylococci. Antimicrob. Agents Chemother. 1991;35:124–129. doi: 10.1128/aac.35.1.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Archer GL, Bosilevac JM. Microbiology. Signaling antibiotic resistance in staphylococci. Science. 2001;291:1915–1916. doi: 10.1126/science.1059671. [DOI] [PubMed] [Google Scholar]
- 50.Niemeyer DM, Pucci MJ, Thanassi JA, Sharma VK, Archer GL. Role of mecA transcriptional regulation in the phenotypic expression of methicillin resistance in Staphylococcus aureus. J. Bacteriol. 1996;178:5464–5471. doi: 10.1128/jb.178.18.5464-5471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rosato AE, et al. mecA-blaZ corepressors in clinical Staphylococcus aureus isolates. Antimicrob. Agents Chemother. 2003;47:1460–1463. doi: 10.1128/AAC.47.4.1460-1463.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berger-Bachi B. Expression of resistance to methicillin. Trends Microbiol. 1994;2:389–393. doi: 10.1016/0966-842x(94)90617-3. [DOI] [PubMed] [Google Scholar]
- 53.Ito T, Katayama Y, Hiramatsu K. Cloning and nucleotide sequence determination of the entire mec DNA of pre-methicillin-resistant Staphylococcus aureus N315. Antimicrob. Agents Chemother. 1999;43:1449–1458. doi: 10.1128/aac.43.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Archer GL, Niemeyer DM. Origin and evolution of DNA associated with resistance to methicillin in staphylococci. Trends Microbiol. 1994;2:343–347. doi: 10.1016/0966-842x(94)90608-4. [DOI] [PubMed] [Google Scholar]
- 55.Couto I, et al. Ubiquitous presence of a mecA homologue in natural isolates of Staphylococcus sciuri. Microb. Drug Resist. 1996;2:377–391. doi: 10.1089/mdr.1996.2.377. [DOI] [PubMed] [Google Scholar]
- 56.Couto I, Wu SW, Tomasz A, de Lencastre H. Development of methicillin resistance in clinical isolates of Staphylococcus sciuri by transcriptional activation of the mecA homologue native to the species. J. Bacteriol. 2003;185:645–653. doi: 10.1128/JB.185.2.645-653.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hiramatsu K, et al. New trends in Staphylococcus aureus infections: glycopeptide resistance in hospital and methicillin resistance in the community. Curr. Opin. Infect. Dis. 2002;15:407–413. doi: 10.1097/00001432-200208000-00009. [DOI] [PubMed] [Google Scholar]
- 58.Hooper DC. Fluoroquinolone resistance among Gram-positive cocci. Lancet Infect. Dis. 2002;2:530–538. doi: 10.1016/s1473-3099(02)00369-9. [DOI] [PubMed] [Google Scholar]
- 59.Høiby N, et al. Excretion of ciprofloxacin in sweat and multiresistant Staphylococcus epidermidis. Lancet. 1997;349:167–169. doi: 10.1016/S0140-6736(96)09229-X. [DOI] [PubMed] [Google Scholar]
- 60.Ng EY, Trucksis M, Hooper DC. Quinolone resistance mutations in topoisomerase IV: relationship to the flqA locus and genetic evidence that topoisomerase IV is the primary target and DNA gyrase is the secondary target of fluoroquinolones in Staphylococcus aureus. Antimicrob. Agents Chemother. 1996;40:1881–1888. doi: 10.1128/aac.40.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ng EY, Trucksis M, Hooper DC. Quinolone resistance mediated by norA: physiologic characterization and relationship to flqB, a quinolone resistance locus on the Staphylococcus aureus chromosome. Antimicrob. Agents Chemother. 1994;38:1345–1355. doi: 10.1128/aac.38.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bisognano C, Vaudaux P, Rohner P, Lew DP, Hooper DC. Induction of fibronectin-binding proteins and increased adhesion of quinolone-resistant Staphylococcus aureus by subinhibitory levels of ciprofloxacin. Antimicrob. Agents Chemother. 2000;44:1428–1437. doi: 10.1128/aac.44.6.1428-1437.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ince D, Zhang X, Hooper DC. Activity of and resistance to moxifloxacin in Staphylococcus aureus. Antimicrob. Agents Chemother. 2003;47:1410–1415. doi: 10.1128/AAC.47.4.1410-1415.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Entenza JM, Que YA, Vouillamoz J, Glauser MP, Moreillon P. Efficacies of moxifloxacin, ciprofloxacin, and vancomycin against experimental endocarditis due to methicillin-resistant Staphylococcus aureus expressing various degrees of ciprofloxacin resistance. Antimicrob. Agents Chemother. 2001;45:3076–3083. doi: 10.1128/AAC.45.11.3076-3083.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kirst HA, Thompson DG, Nicas TI. Historical yearly usage of vancomycin. Antimicrob. Agents Chemother. 1998;42:1303–1304. doi: 10.1128/aac.42.5.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schwalbe RS, Stapleton JT, Gilligan PH. Emergence of vancomycin resistance in coagulase-negative staphylococci. N. Engl. J. Med. 1987;316:927–931. doi: 10.1056/NEJM198704093161507. [DOI] [PubMed] [Google Scholar]
- 67.Smith TL, et al. Emergence of vancomycin resistance in Staphylococcus aureus. Glycopeptide-Intermediate Staphylococcus aureus Working Group. N. Engl. J. Med. 1999;340:493–501. doi: 10.1056/NEJM199902183400701. [DOI] [PubMed] [Google Scholar]
- 68.Fridkin SK, et al. Epidemiological and microbiological characterization of infections caused by Staphylococcus aureus with reduced susceptibility to vancomycin, United States, 1997–2001. Clin. Infect. Dis. 2003;36:429–439. doi: 10.1086/346207. [DOI] [PubMed] [Google Scholar]
- 69.Vancomycin-resistant Staphylococcus aureus: Pennsylvania, 2002. MMWR. 2002;51:902. [PubMed] [Google Scholar]
- 70.Walsh TR, Howe RA. The prevalence and mechanisms of vancomycin resistance in Staphylococcus aureus. Annu. Rev. Microbiol. 2002;56:657–675. doi: 10.1146/annurev.micro.56.012302.160806. [DOI] [PubMed] [Google Scholar]
- 71.Hiramatsu K, et al. Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet. 1997;350:1670–1673. doi: 10.1016/S0140-6736(97)07324-8. [DOI] [PubMed] [Google Scholar]
- 72.Hanaki H, et al. Increase in glutamine-non-amidated muropeptides in the peptidoglycan of vancomycin-resistant Staphylococcus aureus strain Mu50. J. Antimicrob. Chemother. 1998;42:315–320. doi: 10.1093/jac/42.3.315. [DOI] [PubMed] [Google Scholar]
- 73.Hanaki H, et al. Activated cell-wall synthesis is associated with vancomycin resistance in methicillin-resistant Staphylococcus aureus clinical strains Mu3 and Mu50. J. Antimicrob. Chemother. 1998;42:199–209. doi: 10.1093/jac/42.2.199. [DOI] [PubMed] [Google Scholar]
- 74.Sieradzki K, Roberts RB, Haber SW, Tomasz A. The development of vancomycin resistance in a patient with methicillin- resistant Staphylococcus aureus infection. N. Engl. J. Med. 1999;340:517–523. doi: 10.1056/NEJM199902183400704. [DOI] [PubMed] [Google Scholar]
- 75.Showsh SA, De Boever EH, Clewell DB. Vancomycin resistance plasmid in Enterococcus faecalis that encodes sensitivity to a sex pheromone also produced by Staphylococcus aureus. Antimicrob. Agents Chemother. 2001;45:2177–2178. doi: 10.1128/AAC.45.7.2177-2178.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gonzalez-Zorn B, Courvalin P. vanA-mediated high level glycopeptide resistance in MRSA. Lancet Infect. Dis. 2003;3:67–68. doi: 10.1016/s1473-3099(03)00510-3. [DOI] [PubMed] [Google Scholar]
- 77.Tsiodras S, et al. Linezolid resistance in a clinical isolate of Staphylococcus aureus. Lancet. 2001;358:207–208. doi: 10.1016/S0140-6736(01)05410-1. [DOI] [PubMed] [Google Scholar]
- 78.Fuchs PC, Barry AL, Brown SD. In vitro bactericidal activity of daptomycin against staphylococci. J. Antimicrob. Chemother. 2002;49:467–470. doi: 10.1093/jac/49.3.467. [DOI] [PubMed] [Google Scholar]
- 79.Projan SJ, Youngman PJ. Antimicrobials: new solutions badly needed. Curr. Opin. Microbiol. 2002;5:463–465. doi: 10.1016/s1369-5274(02)00364-8. [DOI] [PubMed] [Google Scholar]
- 80.Projan SJ. New (and not so new) antibacterial targets: from where and when will the novel drugs come? Curr. Opin. Pharmacol. 2002;2:513–522. doi: 10.1016/s1471-4892(02)00197-2. [DOI] [PubMed] [Google Scholar]
- 81.McDevitt D, Rosenberg M. Exploiting genomics to discover new antibiotics. Trends Microbiol. 2001;9:611–617. doi: 10.1016/s0966-842x(01)02235-1. [DOI] [PubMed] [Google Scholar]
- 82.Etz H, et al. Identification of in vivo expressed vaccine candidate antigens from Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 2002;99:6573–6578. doi: 10.1073/pnas.092569199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ge M, et al. Vancomycin derivatives that inhibit peptidoglycan biosynthesis without binding D-Ala-D-Ala. Science. 1999;284:507–511. doi: 10.1126/science.284.5413.507. [DOI] [PubMed] [Google Scholar]
- 84.Cheung AL, et al. Diminished virulence of a sar-/agr- mutant of Staphylococcus aureus in the rabbit model of endocarditis. J. Clin. Invest. 1994;94:1815–1822. doi: 10.1172/JCI117530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mazmanian SK, Liu G, Jensen ER, Lenoy E, Schneewind O. Staphylococcus aureus sortase mutants defective in the display of surface proteins and in the pathogenesis of animal infections. Proc. Natl. Acad. Sci. U. S. A. 2000;97:5510–5515. doi: 10.1073/pnas.080520697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Verhoef J, et al. A Dutch approach to methicillin-resistant Staphylococcus aureus. Eur. J. Clin. Microbiol. Infect. Dis. 1999;18:461–466. doi: 10.1007/s100960050324. [DOI] [PubMed] [Google Scholar]
- 87.Yu VL, et al. Staphylococcus aureus nasal carriage and infection in patients on hemodialysis. Efficacy of antibiotic prophylaxis. N. Engl. J. Med. 1986;315:91–96. doi: 10.1056/NEJM198607103150204. [DOI] [PubMed] [Google Scholar]
- 88.Kluytmans J, van Belkum A, Verbrugh H. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin. Microbiol. Rev. 1997;10:505–520. doi: 10.1128/cmr.10.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Perl TM, et al. Intranasal mupirocin to prevent postoperative Staphylococcus aureus infections. N. Engl. J. Med. 2002;346:1871–1877. doi: 10.1056/NEJMoa003069. [DOI] [PubMed] [Google Scholar]
- 90.Peacock SJ, de Silva I, Lowy FD. What determines nasal carriage of Staphylococcus aureus? Trends Microbiol. 2001;9:605–610. doi: 10.1016/s0966-842x(01)02254-5. [DOI] [PubMed] [Google Scholar]
- 91.Fischetti VA. Phage antibacterials make a comeback. Nat. Biotechnol. 2001;19:734–735. doi: 10.1038/90777. [DOI] [PubMed] [Google Scholar]
- 92.Climo MW, Patron RL, Goldstein BP, Archer GL. Lysostaphin treatment of experimental methicillin-resistant Staphylococcus aureus aortic valve endocarditis. Antimicrob. Agents Chemother. 1998;42:1355–1360. doi: 10.1128/aac.42.6.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shinefield H, et al. Use of a Staphylococcus aureus conjugate vaccine in patients receiving hemodialysis. N. Engl. J. Med. 2002;346:491–496. doi: 10.1056/NEJMoa011297. [DOI] [PubMed] [Google Scholar]
- 94.Michie CA. Staphylococcal vaccines. Trends Immunol. 2002;23:461–463. doi: 10.1016/s1471-4906(02)02270-6. [DOI] [PubMed] [Google Scholar]
- 95.Lyon BR, Skurray R. Antimicrobial resistance of Staphylococcus aureus: genetic basis. Microbiol. Rev. 1987;51:88–134. doi: 10.1128/mr.51.1.88-134.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lina G, et al. Distribution of genes encoding resistance to macrolides, lincosamides, and streptogramins among staphylococci. Antimicrob. Agents Chemother. 1999;43:1062–1066. doi: 10.1128/aac.43.5.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Allignet J, Aubert S, Morvan A, el Solh N. Distribution of genes encoding resistance to streptogramin A and related compounds among staphylococci resistant to these antibiotics. Antimicrob. Agents Chemother. 1996;40:2523–2528. doi: 10.1128/aac.40.11.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sieradzki K, Pinho MG, Tomasz A. Inactivated pbp4 in highly glycopeptide-resistant laboratory mutants of Staphylococcus aureus. J. Biol. Chem. 1999;274:18942–18946. doi: 10.1074/jbc.274.27.18942. [DOI] [PubMed] [Google Scholar]
- 99.Murray BE. Vancomycin-resistant enterococcal infections. N. Engl. J. Med. 2000;342:710–721. doi: 10.1056/NEJM200003093421007. [DOI] [PubMed] [Google Scholar]