Abstract
CD40 signaling modulates the immune response at least in part by activation of nuclear factor κB (NFκB). It has been shown that two distinct domains in the CD40 cytoplasmic tail (cyt), namely cyt-N and cyt-C, independently activate NFκB. Although four members of the tumor necrosis factor receptor-associated factor (TRAF) family, including TRAF2, TRAF3, TRAF5, and TRAF6, bind to the CD40 cyt, how each TRAF protein contributes to the NFκB activation by CD40 is not clear. Here we report that TRAF2, TRAF3, and TRAF5 bind cyt-C, whereas TRAF6 binds cyt-N. cyt-N is conserved poorly between human and mouse CD40, while cyt-C is highly conserved. However, single aa substitution of Glu-235 in cyt-N of human CD40 with Ala abolishes the binding of TRAF6 to cyt-N and NFκB activation by cyt-N. Conservation of this Glu between mouse and human CD40 strongly suggests that TRAF6 could link cyt-N to signals essential for CD40-mediated immune response. Furthermore, NFκB activation by cyt-C is inhibited by a kinase-negative form of NFκB-inducing kinase more efficiently than that by cyt-N, consistent with the result that NFκB activation by TRAF2 and TRAF5 is inhibited by a kinase-negative form of NFκB-inducing kinase more efficiently than that by TRAF6. These results indicate that NFκB activating signals emanating from cyt-N and cyt-C are mediated by the different members of the TRAF family and could be regulated in a distinct manner.
Keywords: tumor necrosis factor receptor-associated factor, nuclear factor κB-inducing kinase, protein–protein interaction
CD40 is a member of the tumor necrosis factor receptor (TNFR) superfamily, which includes TNFR-1, TNFR-2, Fas, lymphotoxin-β receptor, CD27, CD30, OX40, and the low-affinity nerve growth factor receptor (1). CD40 is expressed in late B cells in bone marrow, mature B cells, and certain accessory cells, including bone marrow-derived dendritic cells and follicular dendritic cells (2, 3), and is a receptor for CD40 ligand (CD40L), which is present on activated CD4+ T cells (4). Signaling through CD40 rescues B cells from apoptosis induced by crosslinking of the surface IgM complex (5), induces B cells to undergo Ig isotype switching (6, 7), and activates antigen-presenting cells to prime cytotoxic T lymphocytes (8–10).
CD40 signaling events are reported to include modulation of the activity of nonreceptor-type tyrosine kinases such as Lyn, Fyn, and Syk, activation of phosphatidylinositol-3-kinase, phosphorylation of phospholipase Cγ2 (11–13), and activation of the Rel/nuclear factor κB (NFκB) transcription factors (14). CD40 signaling is also linked to the induction of the B7 (15), intercellular adhesion molecule-1 (16, 17), Fas (18), CD23 (19), lymphocyte function-associated antigen-1 (16), Bcl-XL, Cdk4 and Cdk6 proteins (20). However, the molecular mechanism of signal transduction from CD40 is not clear. The cytoplasmic tail (cyt) of TNFR superfamily members lacks sequences indicative of catalytic activity, but is associated with a family of signal transducer proteins, the TNFR-associated factor (TRAF) family. Among the six members of the TRAF family, TRAF2 (21), TRAF3 (also known as CD40bp, LAP-1, or CRAF1) (22–25), TRAF5 (26), and TRAF6 (27) are reported to associate with the CD40cyt. Overexpression of TRAF2, TRAF5, or TRAF6 leads to the activation of NFκB. It has been demonstrated that both the amino-terminal (aa 220–245, cyt-N; our aa numbering starts from the first methionine) and the carboxyl-terminal halves (aa 246–277, cyt-C) of the CD40cyt can independently activate NFκB (20, 28). cyt-C, which is required for the binding of TRAF2, TRAF3, and TRAF5 to CD40, is identical between hCD40 and mCD40. Conversion of Thr-254 to Ala abolishes the binding of TRAFs to cyt-C (22, 26, 27) and disables CD40 signaling linked to growth inhibition (29). cyt-C is required to inhibit apoptosis of B cells induced by activation of antigen receptor signaling (20). Although the physiological significance of cyt-C has been demonstrated, whether cyt-C is sufficient for both the association of TRAF2 and TRAF5 with CD40 and the activation of downstream events was not addressed. cyt-N, which binds TRAF6, is poorly conserved between hCD40 and mCD40, and the biological role of interaction of TRAF6 with cyt-N has not been well characterized. Therefore, how each TRAF contributes to the biological consequences of CD40 signaling remains to be elucidated.
In this paper, we focus on NFκB activation as an output of CD40 signaling and demonstrate that NFκB activation by cyt-C and by cyt-N are mediated by the different members of the TRAF family. Our results also suggest that these two NFκB activation domains could be linked to NFκB activation by means of distinct signaling pathways.
MATERIALS AND METHODS
Cell Culture, Antibodies, and Plasmids.
Jurkat cells were cultured in RPMI medium 1640 supplemented with 10% fetal bovine serum, and 293T cells were cultured in DMEM supplemented with 10% fetal bovine serum. The anti-Flag epitope monoclonal antibody M2 was purchased from Eastman Kodak. The cDNAs encoding human CD40 (hCD40) (2), human CD40L (4), mouse CD40 (mCD40) (30), human NFκB-inducing kinase (NIK) (31), and mouse TRAF2 (21) were either amplified by reverse transcriptase–PCR or cloned from cDNA libraries. Human TRAF3 cDNA and κB site-driven luciferase reporter (3xκB-Luc) (unpublished data) were gifts of George Mosialos and Shigeki Miyamoto, respectively. Isolation of TRAF5 (26) and TRAF6 (27) was reported previously. aa substitution mutations were generated by the method of Kunkel by using appropriate oligonucleotides (32). For the construction of expression vectors, each cDNA was subcloned into SRα promoter-driven pME18S plasmid (33).
In Vivo Binding Assay.
For in vivo binding assay, 293T cells were cotransfected with expression vectors for Flag-tagged TRAF and glutathione S-transferase (GST)-tagged cyt of CD40 or its mutants. Thirty-six hours after transfection, transfected cells were harvested and lysed with TNE buffer (26) followed by centrifugation. The supernatant was incubated with glutathione-Sepharose beads for 1 hr at 4°C. After the beads were washed, the GST fusion protein complexes were separated on a 8.5% polyacrylamide/SDS gel. A part of the lysate before the GST pulldown assay was separated on 8.5% polyacrylamide/SDS gel to analyze the expression level of each TRAF protein. The Flag-tagged TRAF protein was detected by Western blotting by using anti-Flag antibody M2 and horseradish peroxidase (HRP)-conjugated anti-mouse IgG. The one-tenth of GST fusion proteins attached to beads was separated on 12.5% polyacrylamide/SDS gel and visualized by Coomassie brilliant blue R-250 staining.
Transient Transfections and Reporter Gene Assays.
Jurkat cells (2 × 106) were transfected with 0.5 μg of the reporter plasmid, 3xκB-Luc, 0.5 μg of β-galactosidase (β-gal) expression vector driven by β-actin promoter (β-actin-β-gal) and the indicated amounts of various expression plasmids by the DEAE-dextran method. The total amount of DNA transfected was always adjusted to 5 μg with a control expression vector. 293T cells (4 × 105) were transfected with 1 ng of 3xκB-Luc, 10 ng of β-actin-β-gal, and the indicated amounts of various expression plasmids by the calcium phosphate method. The total amount of DNA transfected was always adjusted to 10 μg with a control expression vector. Thirty-six hours after transfection, cell extracts were prepared by PicaGene Reporter Lysis Buffer (TOYO INK, Tokyo) followed by centrifugation. Luciferase activity was measured by PicaGene luciferase assay system (TOYO INK) and β-gal activity was used to standardize transfection efficiency (34).
RESULTS AND DISCUSSION
Two Independent TRAF-Binding Domains in CD40cyt.
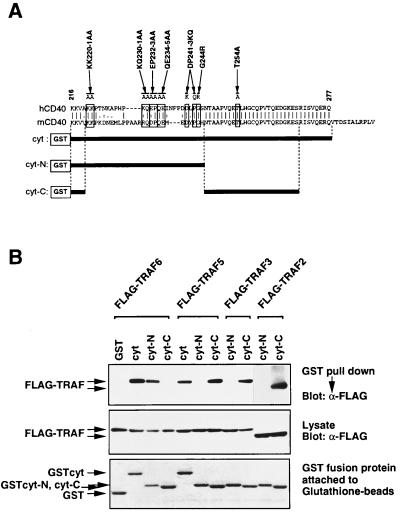
To understand the role of each TRAF protein in CD40 signaling, we have mapped the binding domain for each TRAF protein in CD40cyt. Fig. 1A shows alignment of the cyt of hCD40 (2) and mCD40 (30). The carboxyl-terminal half of cyt (aa 246–277 in hCD40) is completely conserved between hCD40 and mCD40, whereas the amino-terminal half (aa 216–245) is poorly conserved. It has been demonstrated that TRAF6 binds aa 216–245 of hCD40 (cyt-N) (20), whereas TRAF3 binds aa 246–269 (cyt-C) (28). However, whether cyt-C is sufficient for association with TRAF2 and TRAF5 was not addressed. Furthermore, the possibility that each TRAF protein might recognize the other parts of cyt was not systematically tested. To clarify this point, we carried out in vivo GST pulldown experiments using cell extracts prepared from 293T cells cotransfected with expression vectors for Flag-tagged TRAF proteins and GST-tagged cyt, cyt-N, or cyt-C (Fig. 1B). TRAF2, TRAF3, and TRAF5 bind cyt-C, but they do not bind cyt-N. In contrast, TRAF6 binds cyt-N, but not cyt-C. These data indicate that CD40cyt has two independent TRAF-binding domains: one, which binds TRAF6, is located between aa 216 and 245 (cyt-N), and the other, which binds TRAF2, TRAF3, and TRAF5, is located between aa 246 and 269 (cyt-C).
Figure 1.
TRAF2, TRAF3, and TRAF5 bind to the carboxyl-terminal conserved domain, but TRAF6 binds to the N-terminal domain in CD40cyt. (A) Alignment of human and mouse CD40cyt. The carboxyl-terminal part of CD40cyt is identical between hCD40 and mCD40, but the N-terminal part is poorly conserved. Conserved residues substituted in mutagenesis described in the text are indicated by boxes, and the resulting aas by the mutagenesis are shown above these boxes. GST-tagged hCD40cyt constructs (cyt, cyt-N and cyt-C) are shown below the sequences. (B) In vivo association of TRAFs with GST-hCD40 in 293T cells. Cell extracts from 293T cells cotransfected with expression plasmids for Flag-tagged TRAFs and GST-tagged hCD40cyt or its deletion mutants were prepared. The GST pulldown assays were performed and TRAF proteins bound to beads were then analyzed by Western blotting by using anti-Flag antibody M2 (Top). One-tenth of GST fusion proteins attached to beads was separated on 12.5% polyacrylamide/SDS gel and visualized by Coomassie brilliant blue R-250 staining (Bottom). A portion of the lysate used for the GST pulldown assay was separated on 8.5% polyacrylamide/SDS gel and expression level of Flag-tagged TRAFs was analyzed by Western blotting by using anti-Flag antibody M2 (Middle).
Conversion of Glu-235 in cyt-N to Ala (E235A) Abolishes the Binding of TRAF6 to CD40.
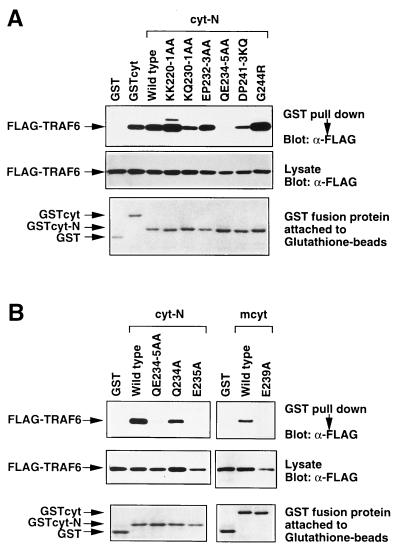
It has been demonstrated that conversion of Thr-254 to Ala (T254A) in cyt-C abolishes the binding of TRAF2, TRAF3, and TRAF5 to cyt-C (22, 26), and also disables CD40-induced growth inhibition (29). Although cyt-N is poorly conserved between hCD40 and mCD40, TRAF6 binds both species of CD40 (27). This binding specificity of TRAF6 led us to search for aa residues critical for both TRAF6 binding and biological output of CD40 signaling. To identify residues critical for TRAF6 binding, we introduced aa substitution mutations into residues conserved between hCD40 and mCD40. The following mutations were introduced into cyt-N: KK220–1AA (both Lys-220 and Lys-221 were changed to Ala), KQ230–1AA, EP232–3AA, QE234–5AA, DP241–3KQ, and G244R (Fig. 1A). These cyt-N mutants were expressed as GST fusion proteins in 293T cells and were used in GST pulldown assays (Fig. 2A). Among the mutations introduced, only the QE234–5AA mutation completely abolished TRAF6 binding. To further identify the residue responsible for TRAF6 binding, we generated the mutants Q234A and E235A. Q234A bound TRAF6, whereas E235A did not bind at all (Fig. 2B Left). Substitution of the corresponding residue in mCD40, Glu-239, with Ala abolished binding of TRAF6 to mouse cyt (Right), suggesting that this Glu is critical for coupling to TRAF6-dependent signals. During the preparation of the manuscript, Pullen et al. (35) reported that an 8-mer peptide, QEPQEINF, derived from aa 231–238 of hCD40, could bind to TRAF6, which is consistent with our results shown here. The aa sequence around Glu-235 does not fit the PXQXT (36), the EXGKE (37), or the VXXSXEE (38) motif, which has been demonstrated to be a consensus sequence required for binding to TRAF1, TRAF2, TRAF3, and TRAF5. However, this is not surprising, since the TRAF-C domain of TRAF6 is the most divergent among the members of the TRAF family. Although aas surrounding Glu-235 could be the direct binding site for TRAF6, no such sequence was found in IL-1 receptor-associated kinase, which has been demonstrated to be associated with TRAF6 on IL-1 stimulation (39).
Figure 2.
Glu-235 in hCD40 is critical for TRAF6 binding. (A) TRAF6 binding with cyt-N was abolished by the QE234–5AA mutation. Conserved residues in the N-terminal poorly conserved region in hCD40cyt were mutated in the cyt-N deletion mutant, and each construct was tagged with GST. GST-tagged CD40 cytoplasmic domain or its mutants and Flag-tagged TRAF6 were transiently expressed in 293T cells and in vivo binding assays were performed. Association between Flag-TRAF6 and GST-tagged CD40 mutants (Top) and expression level of Flag-TRAF6 (Middle) and that of GST-tagged CD40 mutants (Bottom) are shown. (B) Single point mutation E235A abolished TRAF6 association with GST-CD40. (Left) Either Gln-234 or Glu-235 alone was mutated to Ala in the GST-tagged cyt-N deletion mutant. GST-tagged CD40cyt or its mutants and Flag-tagged TRAF6 were transiently coexpressed in 293T cells and in vivo binding assays were performed. Association between Flag-TRAF6 and GST-tagged CD40 mutants (Top) and expression level of Flag-tagged TRAF6 (Middle) and that of GST-tagged CD40 mutants (Bottom) are shown. (Right) Glu-239 in mCD40, corresponding to Glu-235 in hCD40, was mutated to Ala in GST-tagged full-length cyt of mCD40. In vivo binding assay between mCD40 and TRAF6 in 293T cells was performed as described above.
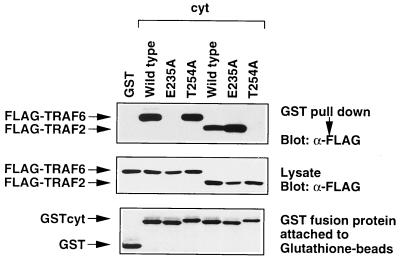
To further confirm the independence of two TRAF-binding domains in CD40, we addressed whether the E235A mutation affects the binding of TRAF2 to CD40 and also whether the T254A mutation affects the binding of TRAF6 to CD40 in the context of the full-length CD40cyt. Fig. 3 shows that E235A and T254A mutations have no effect on the binding of CD40 to TRAF2 and TRAF6, respectively. These data further support the idea that the two domains, cyt-N and cyt-C, are independent in terms of TRAF association.
Figure 3.
Effects of E235A and T254A mutations on TRAF6 or TRAF2 association with GST-cyt. GST-cyt or its mutants (E235A or T254A) and Flag-tagged TRAF6 or TRAF2 were transiently expressed in 293T cells and binding assays were performed. Association of Flag-tagged TRAF6 and Flag-tagged TRAF2 with GST-tagged CD40 mutants (Top) and expression level of Flag-tagged TRAF6 and Flag-tagged TRAF2 (Middle) and that of GST-tagged CD40 mutants (Bottom) are shown.
E235A Mutation Abolishes NFκB Activation Triggered by an Expression Vector for CD40 Lacking cyt-C (CD40cyt-N).
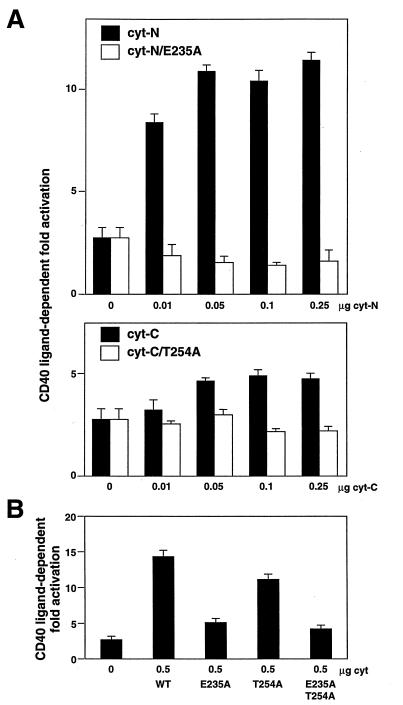
We have previously shown that cyt-N, which binds TRAF6, activates NFκB (20). To further define the biological role of Glu-235 and TRAF6 in CD40 signaling, we analyzed the effect of the E235A mutation on the ability of cyt-N to activate NFκB. CD40L-dependent NFκB activation by cyt-N was measured by cotransfection of the 3xκB-Luc with CD40cyt-N, with or without an expression vector for CD40L (Fig. 4A Upper). NFκB activation was induced by CD40cyt-N in a dose-dependent manner. However, CD40cyt-N carrying the E235A mutation (CD40cyt-N/E235A) did not activate NFκB, even if the amount of CD40cyt-N/E235A expression vector was increased up to 1.0 μg. These results indicate that Glu-235 plays a critical role in NFκB activation as well as TRAF6 binding. CD40L-dependent NFκB activation was also induced by CD40cyt-C expression in a dose-dependent manner, but not by CD40cyt-C carrying the T254A mutation (Fig. 4A Lower). These results strongly suggest that the cyt-N and cyt-C domains of CD40 mediate NFκB activation independently by using different members of the TRAF family: cyt-N uses TRAF6 and cyt-C uses TRAF2 or TRAF5. NFκB activation by full-length CD40 carrying the E235A mutation (CD40cyt/E235A) was about 30% of that by wild-type CD40, and NFκB activation by CD40cyt/T254A was about 70% of that by wild-type CD40 in Jurkat human T cell line (Fig. 4B), suggesting that both domains are used in a redundant manner to mediate NFκB activation in Jurkat cells. The full-length CD40 carrying both E235A and T254A mutations (CD40cyt/E235A, T254A) still weakly activates NFκB. Because introducing these two mutations into the subdomain of CD40 almost completely abolished NFκB activation by each domain (Fig. 4A), it is possible that another NFκB activation domain, which could be destroyed in cyt-N and cyt-C constructs, could be present.
Figure 4.
Effects of E235A and T254A mutations on the CD40-mediated NFκB activation. (A) NFκB activation by CD40cyt-N was completely abolished by the E235A mutation. (Upper) CD40L-dependent fold activation of NFκB by CD40cyt-N (closed bar) and that by CD40cyt-N E235A (open bar) are shown. (Lower) CD40L-dependent fold activation of NFκB by CD40cyt-C (closed bar) and that by CD40cyt-C T254A (open bar) are shown. (B) Effects of E235A and T254A mutations on NFκB activation by the full-length CD40. CD40L-dependent fold activation of NFκB by the full-length CD40 with various mutatons is shown. Jurkat cells (2 × 106) were transfected with 0.5 μg of 3xκB-Luc, 0.5 μg of β-actin-β-gal, the indicated amounts of pME-hCD40 construct, 0 μg or 0.5 μg of pME-hCD40L, and enough pME18S control plasmid to give 5 μg of total DNA by the DEAE-dextran method. Cell extracts prepared at 36 hr after transfection were used for the luciferase assay. Fold activation was expressed as the ratio of luciferase activity with CD40L expression to that without CD40L expression. Values correspond to means ± SEMs of at least three independent experiments.
A Kinase-Negative Form of NIK Inhibits NFκB Activation by CD40cyt-C More Efficiently than That by CD40cyt-N.
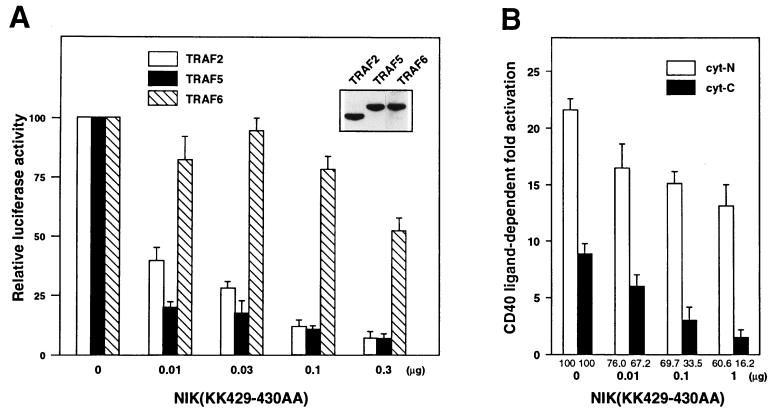
NIK is a mitogen-activated protein kinase kinase kinase-related kinase that associates with various members of the TRAF family and activates NFκB when overexpressed (31, 40). A kinase negative form of NIK lacking the two lysine residues in its catalytic domain, NIK(KK429–430AA), behaves as a dominant negative inhibitor that suppresses TNF- and IL-1-induced NFκB activation. To investigate the role of NIK in CD40-mediated NFκB activation, we examined the effect of NIK(KK429–430AA) expression on NFκB activation by TRAFs, which can bind the cyt of CD40 (Fig. 5A). The indicated amount of NIK(KK429–430AA) expression vector was transiently cotransfected into 293T cells with Flag-tagged TRAF2, TRAF5, or TRAF6 expression vectors in the presence of the κB site-driven luciferase reporter plasmid. TRAF2- and TRAF5-mediated NFκB activation was dramatically inhibited by NIK(KK429–430AA) expression, whereas TRAF6-mediated NFκB activation was minimally inhibited. Expression levels of each TRAF protein were almost equal (Fig. 5A Inset). The amount of NIK(KK429–430AA) expression plasmid required for 50% inhibition was less than 0.01 μg for TRAF2 and TRAF5, while that for TRAF6 is more than 0.3 μg. The results suggest that NFκB activation by TRAF2 and TRAF5 is mediated principally by NIK, while a different NFκB-activating kinase in addition to NIK is involved in TRAF6-mediated NFκB activation. This difference has not been described in previous papers, probably because of the use of an excess amount of NIK(KK429–430AA) expression plasmid in previous studies (40, 41). In fact, when we use 1 μg of NIK(KK429–430AA) expression plasmid, TRAF6-mediated NFκB activation is also significantly suppressed (data not shown). It is possible that a large excess of NIK(KK429–430AA) might sequester a factor that is essential for the activation of both NIK and a putative NFκB-activating kinase. The putative kinase is unlikely to be mitogen-activated protein kinase/extracellular signal-regulated kinase-1 (MEKK-1), since a kinase-negative form of MEKK-1 did not inhibit TRAF6-mediated NFκB activation efficiently (data not shown).
Figure 5.
Effect of kinase-negative NIK on the NFκB activation by TRAF or CD40. 293T cells (4 × 105) were transfected with 1 ng of 3xκB-Luc, 10 ng of β-actin-β-gal, and various expression vectors by the calcium phosphate method. The total amount of DNA transfected was adjusted to 10 μg with a control expression vector. Thirty-six hours after transfection, cell extracts were prepared and used for luciferase assay. (A) Kinase-negative NIK inhibits NFκB activation by TRAF2 or TRAF5 more efficiently than that by TRAF6. pME-Flag TRAF2 (2 μg), TRAF5 (2 μg), or TRAF6 (1 μg) and indicated amounts of pME-NIK(KK429–430AA) were transfected. Luciferase activity in the absence of kinase-negative NIK was set to 100. Actual luciferase activities induced by each TRAF protein in the absence of kinase-negative NIK were comparable: TRAF2 (5210 ± 150), TRAF5 (4930 ± 110), TRAF6 (6650 ± 180). Expression levels of each TRAF protein in transfected cells were measured by Western blotting by using anti-Flag antibody (Inset). (B) Kinase-negative NIK inhibits NFκB activation by CD40cyt-C more efficiently than that by CD40cyt-N. 0.03 μg of pME-hCD40cyt-N or pME-hCD40cyt-C and 0 μg or 0.03 μg of pME-hCD40L were transfected. CD40L-dependent fold activation of NFκB is shown. Relative values in which fold activation in the absence of kinase-negative NIK was set to 100 are shown below each column. Values correspond to means ± SEMs of at least three independent experiments.
Because TRAF2 and TRAF5 bind cyt-C, whereas TRAF6 binds cyt-N, NFκB activation by cyt-N and that by cyt-C may be regulated differently by NIK. To address this issue, we measured the effect of NIK(KK429–430AA) expression on NFκB activation by CD40cyt-N or CD40cyt-C (Fig. 5B). NFκB activation by CD40cyt-C was more efficiently inhibited by NIK(KK429–430AA) expression than NFκB activation by CD40cyt-N. Similar results were obtained by using the Jurkat T cell line (data not shown). These data also suggest that NFκB activation by cyt-C is predominantly mediated by NIK, and that activation of the putative NFκB-activating kinase is triggered by the interaction of cyt-N with TRAF6 (Fig. 6). Therefore, full-length CD40 could activate NFκB either by means of an NIK-dependent or -independent pathway. It is possible that NIK and the putative NFκB-activating kinase are regulated by different cellular factors in addition to TRAFs. In such a case, NFκB activation derived from cyt-N and cyt-C might be regulated in a distinct manner and could be cell type- or differentiation stage-dependent. Distinct regulation of cyt-N and cyt-C was also demonstrated in the case of CD40-mediated extracellular signal-regulated kinase activation. It has been demonstrated that the extracellular signal-regulated kinase activation by cyt-C is Ras-dependent, whereas that by cyt-N is Ras-independent (42). Taken together, the results strongly suggest that signals emanating from cyt-N and those from cyt-C are differentially regulated in a cell type or differentiation stage-dependent manner, although both were mediated by the TRAF family of proteins. This dual signal control might explain the pleiotropic roles of CD40 in the immune system (43). Analysis of CD40 mutants in vivo in transgenic mice may clarify the different roles of cyt-N and cyt-C in CD40 signaling.
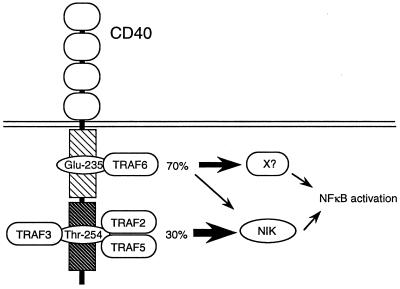
Figure 6.
A model for NFκB activation by the CD40cyt. There are two domains (cyt-N and cyt-C) in the CD40cyt for the NFκB activation. The cyt-N domain activates the NFκB through TRAF6 and the contribution to the activation by this domain is approximately 70%, whereas the cyt-C domain activates the NFκB through TRAF2 and/or TRAF5 and the contribution to the activation is approximately 30%. Although the NFκB activation by the cyt-C domain is thought to be mediated mainly by NIK, it is possible that the NFκB activation by cyt-N is mediated by an unknown molecule (kinase) in addition to NIK.
Acknowledgments
We are grateful to Dr. George Mosialos (Infectious Disease Division, Brigham and Women’s Hospital) for TRAF3 cDNA and to Dr. Shigeki Miyamoto for 3xκB-Luc. We also thank Drs. Ronald Wisdom and Taishin Akiyama for critical reading of the manuscript. This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports and Culture of Japan and by a Grant-in-Aid for AIDS Research from the Japan Health Science Foundation.
ABBREVIATIONS
- NFκB
nuclear factor κ B
- NIK
NFκB-inducing kinase
- cyt
cytoplasmic tail
- TNFR
tumor necrosis factor receptor
- TRAF
TNFR-associated factor
- β-gal
β-galactosidase
- β-actin-β-gal
β-gal expression vector driven by β-actin promoter
- CD40L
CD40 ligand
- hCD40
human CD40
- mCD40
mouse CD40
- GST
glutathione S-transferase
- HRP
horseradish peroxidase
References
- 1.Beutler B, van Huffel C. Science. 1994;264:667–668. doi: 10.1126/science.8171316. [DOI] [PubMed] [Google Scholar]
- 2.Stamenkovic I, Clark E A, Seed B. EMBO J. 1989;8:1403–1410. doi: 10.1002/j.1460-2075.1989.tb03521.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schriever F, Freedman A S, Freeman G, Messner E, Lee G, Daley J, Nadler L M. J Exp Med. 1989;169:2043–2058. doi: 10.1084/jem.169.6.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armitage R J, Fanslow W C, Strockbine L, Sato T A, Clifford K N, Macduff B M, Anderson D M, Gimpel S D, Davis-Smith T, Maliszewski C R, et al. Nature (London) 1992;357:80–82. doi: 10.1038/357080a0. [DOI] [PubMed] [Google Scholar]
- 5.Tsubata T, Wu J, Honjo T. Nature (London) 1993;364:645–648. doi: 10.1038/364645a0. [DOI] [PubMed] [Google Scholar]
- 6.Jabara H H, Fu S M, Geha R S, Vercelli D. J Exp Med. 1990;172:1861–1864. doi: 10.1084/jem.172.6.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Defrance T, Vanbervliet B, Durand I, Briolay J, Banchereau J. Eur J Immunol. 1992;22:2831–2839. doi: 10.1002/eji.1830221112. [DOI] [PubMed] [Google Scholar]
- 8.Ridge J P, Di Rosa F, Matzinger P. Nature (London) 1998;393:474–478. doi: 10.1038/30989. [DOI] [PubMed] [Google Scholar]
- 9.Bennett S R, Carbone F R, Karamalis F, Flavell R A, Miller J F, Heath W R. Nature (London) 1998;393:478–480. doi: 10.1038/30996. [DOI] [PubMed] [Google Scholar]
- 10.Schoenberger S P, Toes R E, van der Voort E I, Offringa R, Melief C J. Nature (London) 1998;393:480–483. doi: 10.1038/31002. [DOI] [PubMed] [Google Scholar]
- 11.Uckun F M, Schieven G L, Dibirdik I, Chandan-Langlie M, Tuel-Ahlgren L, Ledbetter J A. J Biol Chem. 1991;266:17478–17485. [PubMed] [Google Scholar]
- 12.Faris M, Gaskin F, Parsons J T, Fu S M. J Exp Med. 1994;179:1923–1931. doi: 10.1084/jem.179.6.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ren C L, Morio T, Fu S M, Geha R S. J Exp Med. 1994;179:673–680. doi: 10.1084/jem.179.2.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berberich I, Shu G L, Clark E A. J Immunol. 1994;153:4357–4366. [PubMed] [Google Scholar]
- 15.Ranheim E A, Kipps T J. J Exp Med. 1993;177:925–935. doi: 10.1084/jem.177.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bjorck P, Paulie S. Immunology. 1993;78:218–225. [PMC free article] [PubMed] [Google Scholar]
- 17.Barrett T B, Shu G, Clark E A. J Immunol. 1991;146:1722–1729. [PubMed] [Google Scholar]
- 18.Schattner E J, Elkon K B, Yoo D H, Tumang J, Krammer P H, Crow M K, Friedman S M. J Exp Med. 1995;182:1557–1565. doi: 10.1084/jem.182.5.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burlinson E L, Graber P, Bonnefoy J Y, Ozanne B W, Cushley W. Eur J Immunol. 1996;26:1069–1073. doi: 10.1002/eji.1830260517. [DOI] [PubMed] [Google Scholar]
- 20.Ishida T, Kobayashi N, Tojo T, Ishida S, Yamamoto T, Inoue J. J Immunol. 1995;155:5527–5535. [PubMed] [Google Scholar]
- 21.Rothe M, Wong S C, Henzel W J, Goeddel D V. Cell. 1994;78:681–692. doi: 10.1016/0092-8674(94)90532-0. [DOI] [PubMed] [Google Scholar]
- 22.Hu H M, O’Rourke K, Boguski M S, Dixit V M. J Biol Chem. 1994;269:30069–30072. [PubMed] [Google Scholar]
- 23.Cheng G, Cleary A M, Ye Z S, Hong D I, Lederman S, Baltimore D. Science. 1995;267:1494–1498. doi: 10.1126/science.7533327. [DOI] [PubMed] [Google Scholar]
- 24.Mosialos G, Birkenbach M, Yalamanchili R, VanArsdale T, Ware C, Kieff E. Cell. 1995;80:389–399. doi: 10.1016/0092-8674(95)90489-1. [DOI] [PubMed] [Google Scholar]
- 25.Sato T, Irie S, Reed J C. FEBS Lett. 1995;358:113–118. doi: 10.1016/0014-5793(94)01406-q. [DOI] [PubMed] [Google Scholar]
- 26.Ishida T K, Tojo T, Aoki T, Kobayashi N, Ohishi T, Watanabe T, Yamamoto T, Inoue J. Proc Natl Acad Sci USA. 1996;93:9437–9442. doi: 10.1073/pnas.93.18.9437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishida T, Mizushima S, Azuma S, Kobayashi N, Tojo T, Suzuki K, Aizawa S, Watanabe T, Mosialos G, Kieff E, et al. J Biol Chem. 1996;271:28745–28748. doi: 10.1074/jbc.271.46.28745. [DOI] [PubMed] [Google Scholar]
- 28.Cheng G, Baltimore D. Genes Dev. 1996;10:963–973. doi: 10.1101/gad.10.8.963. [DOI] [PubMed] [Google Scholar]
- 29.Inui S, Kaisho T, Kikutani H, Stamenkovic I, Seed B, Clark E A, Kishimoto T. Eur J Immunol. 1990;20:1747–1753. doi: 10.1002/eji.1830200819. [DOI] [PubMed] [Google Scholar]
- 30.Torres R M, Clark E A. J Immunol. 1992;148:620–626. [PubMed] [Google Scholar]
- 31.Malinin N L, Boldin M P, Kovalenko A V, Wallach D. Nature (London) 1997;385:540–544. doi: 10.1038/385540a0. [DOI] [PubMed] [Google Scholar]
- 32.Kunkel T A. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shiio Y, Yamamoto T, Yamaguchi N. Proc Natl Acad Sci USA. 1992;89:5206–5210. doi: 10.1073/pnas.89.12.5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herbomel P, Bourachot B, Yaniv M. Cell. 1984;39:653–662. doi: 10.1016/0092-8674(84)90472-0. [DOI] [PubMed] [Google Scholar]
- 35.Pullen S S, Miller H G, Everdeen D S, Dang T T A, Crute J J, Kehry M R. Biochemistry. 1998;37:11836–11845. doi: 10.1021/bi981067q. [DOI] [PubMed] [Google Scholar]
- 36.Devergne O, Hatzivassiliou E, Izumi K M, Kaye K M, Kleijnen M F, Kieff E, Mosialos G. Mol Cell Biol. 1996;16:7098–7108. doi: 10.1128/mcb.16.12.7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gedrich R W, Gilfillan M C, Duckett C S, Van Dongen J L, Thompson C B. J Biol Chem. 1996;271:12852–12858. doi: 10.1074/jbc.271.22.12852. [DOI] [PubMed] [Google Scholar]
- 38.Lee S Y, Lee S Y, Kandala G, Liou M L, Liou H C, Choi Y. Proc Natl Acad Sci USA. 1996;93:9699–9703. doi: 10.1073/pnas.93.18.9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cao Z, Xiong J, Takeuchi M, Kurama T, Goeddel D V. Nature (London) 1996;383:443–446. doi: 10.1038/383443a0. [DOI] [PubMed] [Google Scholar]
- 40.Song H Y, Regnier C H, Kirschning C J, Goeddel D V, Rothe M. Proc Natl Acad Sci USA. 1997;94:9792–9796. doi: 10.1073/pnas.94.18.9792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakano H, Shindo M, Sakon S, Nishinaka S, Mihara M, Yagita H, Okumura K. Proc Natl Acad Sci USA. 1998;95:3537–3542. doi: 10.1073/pnas.95.7.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kashiwada M, Shirakata Y, Inoue J I, Nakano H, Okazaki K, Okumura K, Yamamoto T, Nagaoka H, Takemori T. J Exp Med. 1998;187:237–244. doi: 10.1084/jem.187.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hollenbaugh D, Ochs H D, Noelle R J, Ledbetter J A, Aruffo A. Immunol Rev. 1994;138:23–37. doi: 10.1111/j.1600-065x.1994.tb00845.x. [DOI] [PubMed] [Google Scholar]