Abstract
The 16S rRNA-binding ribosomal protein S15 is a key component in the assembly of the small ribosomal subunit in bacteria. We have shown that S15 from the extreme thermophile Thermus thermophilus represses the translation of its own mRNA in vitro, by interacting with the leader segment of its mRNA. The S15 mRNA-binding site was characterized by footprinting experiments, deletion analysis and site-directed mutagenesis. S15 binding triggers a conformational rearrangement of its mRNA into a fold that mimics the conserved three-way junction of the S15 rRNA-binding site. This conformational change masks the ribosome entry site, as demonstrated by direct competition between the ribosomal subunit and S15 for mRNA binding. A comparison of the T.thermophilus and Escherichia coli regulation systems reveals that the two regulatory mRNA targets do not share any similarity and that the mechanisms of translational inhibition are different. Our results highlight an astonishing plasticity of mRNA in its ability to adapt to evolutionary constraints, that contrasts with the extreme conservation of the rRNA-binding site.
Keywords: induced-fit mechanism/molecular mimicry/ribosomal protein S15/RNA–protein interaction/translational regulation
Introduction
Feedback regulation at the translational level is commonly used in bacteria and bacteriophages for fast adaptation of highly expressed proteins to environmental variations and cellular requirements. A typical example is the coordinated synthesis of ribosomal components during ribosome biogenesis in Escherichia coli. When synthesized in excess over rRNAs, primary rRNA-binding proteins interact with their mRNA and inhibit translation of their own genes and the other genes in their operon. The similarities observed between mRNA and rRNA targets for a number of E.coli ribosomal proteins, such as S7, S8, L1 (reviewed in Zengel and Lindahl, 1994) and L20 (Guillier et al., 2002), sustain the mechanism based on mimicry and competition previously proposed by Nomura et al. (1980). However, in other cases, i.e. E.coli proteins S4 (Deckman and Draper, 1987; Vartikar and Draper, 1989) and L4 (Freedman et al., 1987; Stelzl et al., 2000), analogies between both target RNAs could not be obviously detected, at least at sequence and secondary structure levels. Although interactions of ribosomal proteins with their mRNA targets have been studied intensively for many years, molecular details of translational repression mechanisms in the expression of ribosomal protein gene operons remain scarce.
Ribosomal protein S15 is highly conserved among prokaryotes. It plays a pivotal role in the assembly of the central domain of the small ribosomal subunit (Held et al., 1974) and forms one of the bridges between the two subunits in 70S ribosomes (Culver et al., 1999). The interaction of S15 with 16S rRNA has been characterized at high resolution, based on extensive biochemical investigations (Powers and Noller, 1995; Batey and Williamson, 1996a,b; Serganov et al., 1996, 2001) and crystal structures of small ribosomal subunits (Schluenzen et al., 2000; Wimberly et al., 2000) and isolated complexes containing Thermus thermophilus S15 (TtS15) (Agalarov et al., 2000; Nikulin et al., 2000). Briefly, S15 binds to 16S rRNA at two highly conserved recognition sites, namely a three-way junction and a G–U/G–C motif separated from the junction by one helical turn. This triggers a conformational realignment that is required for subsequent binding of other proteins, resulting in the formation of the subunit platform. The binding of E.coli S15 (EcS15) to its own mRNA in a region that forms a pseudoknot causes translation inhibition, as a result of trapping the ribosome on its loading site (Philippe et al., 1993). Structural information on the S15–16S rRNA complex, coupled with the biochemical data on the mRNA site (Philippe et al., 1990, 1995; Bénard et al., 1994, 1998; Serganov et al., 2002) have allowed the characterization of the extent of mimicry between the two RNA targets. It turns out that EcS15 recognizes two sites on the pseudoknot that, unlike in rRNA, are equally important for binding. The first is a G–U/G–C motif analogous to its rRNA counterpart, and the second differs completely from the rRNA junction (Serganov et al., 2002). Recently, the mRNA target for Bacillus stearothermophilus S15 (BsS15) was identified (Scott and Williamson, 2001) and found to contain a bipartite site for S15 binding. This RNA contains a three-way junction, which shares no significant similarity with either the E.coli mRNA or 16S rRNA target. Thus, regulatory systems identified in E.coli are probably not universal for all bacteria.
The S15 gene promoter is one of the strongest in T.thermophilus (Maseda and Hoshino, 1995; Serganov et al., 1997) and its expression requires negative regulation. We established that TtS15 inhibits its own translation and determined the thermodynamic and kinetic parameters of the interaction. The TtS15 mRNA-binding site was localized and characterized using footprinting, deletion analysis and site-directed mutagenesis. We found that ThS15 triggers formation of the RNA junction that partially mimics 16S rRNA and, in so doing, masks the ribosome-binding site. Such interplay of the protein with the junctional conformation within its mRNA adds a new variety to existing mechanisms of translational inhibition. Furthermore, our results highlight an astonishing versatility and adaptability of regulatory mechanisms that contrast with the strong constraints associated with ribosome assembly.
Results
TtS15 represses its synthesis on the level of translation
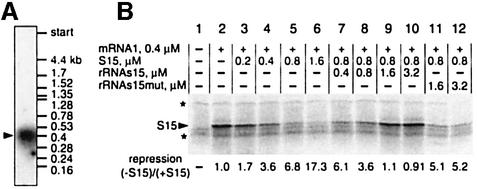
The TtS15 gene transcript(s) were analyzed by northern blot hybridization using RNA isolated from T.thermophilus cells and 32P-labeled TtS15 DNA as a probe. A single RNA transcript of ∼400 nucleotides in size was detected (Figure 1A). Since the transcription start point was mapped at positions –67/68 (Serganov et al., 1997), the TtS15 mRNA should end ∼50 nucleotides downstream of the coding region, in a potential ρ-independent terminator located between genes coding for S15 and polynucleotide phosphorylase. Thus, in contrast to the E.coli system (Portier and Regnier, 1984), TtS15 is most probably translated from a monocistronic mRNA, and regulation of its translation cannot be coupled with the control of upstream or downstream genes.
Fig. 1. Autorepression of TtS15 translation in vitro. (A) Northern blot analysis of T.thermophilus mRNA revealed a single RNA species encoding S15. (B) Autorepression analysis. mRNA1 was incubated in the E.coli extract in the presence of [35S]methionine and non-labeled TtS15 (lanes 3–6). Rescue of TtS15 synthesis was done by either rRNAs15 (lanes 7–10) or rRNAs15mut (lanes 11 and 12) in the presence of 0.8 µM TtS15. The repression ratio was calculated after background subtraction (lane 1) and normalization of the S15 amount using extract-specific protein expression (indicated by asterisks) as an internal reference.
By analogy to what happens in E.coli, we assumed that TtS15 is able to repress its own translation through a feedback regulatory mechanism. Due to a lack of reliable vector for gene expression in T.thermophilus and toxicity of TtS15 for E.coli cells (Serganov et al., 1997), we used an in vitro E.coli cell-free translation system. The natural mRNA transcript (mRNA1) was translated in the cell extract in the presence of [35S]methionine, resulting in a major protein corresponding to labeled TtS15. Addition of TtS15 to the translation mixture decreased TtS15 synthesis in a dose-dependent manner (Figure 1B), while background synthesis of cell-specific proteins was unaffected. Inhibition of TtS15 translation was rescued efficiently by addition of a 16S rRNA fragment from T.thermophilus containing the TtS15-binding site (rRNAs15), while an rRNA fragment carrying a mutation that abolishes S15 binding (rRNAs15mut) (Serganov et al., 2001) failed to derepress translation (Figure 1B). We verified that the stability of the mRNA was unaffected during the experiment time, and that a spliceosomal protein U1A was unable to repress translation (data not shown).
TtS15 specifically interacts with its mRNA
We tested the ability of TtS15 to bind its own mRNA. The apparent dissociation constant (Kd) for this interaction was determined at various temperatures by titrating 32P-labeled mRNA1 with TtS15, using the nitrocellulose filter-binding assay. We found that TtS15 binds mRNA with high affinity (Kd ∼5 nM) over a wide range of temperatures (10–55°C) (Figure 2A and B, Table I). Experimental data for moderate and high temperatures were easily fit to a binding isotherm that assumes one-site binding, in agreement with the measured 1:1 stoichiometry (data not shown). However, at temperatures <30°C, we obtained biphasic curves, with the high affinity component of the mRNA binding curve decreasing progressively. This reduction affected the fitting of experimental data to the theoretical isotherm, and we were unable to calculate high affinity dissociation constants at low temperatures (0–10°C). The more marked temperature dependence was observed for the binding of TtS15 to rRNAs15 (Figure 2A and B). Remarkably, the affinity of TtS15 for mRNA1 is 10-fold lower than for rRNA at intermediate temperatures (20–45°C), whereas these differences tend to decrease at higher temperatures (60–70°C) (Figure 2B), probably due to partial melting of RNA secondary structures and RNA degradation.
Fig. 2. Interaction between TtS15 and its RNA targets. (A) Saturation binding curves with 32P-labeled mRNA1 and rRNAs15 at the indicated temperature. (B) Temperature dependence of equilibrium binding constants for complexes of TtS15 with mRNA1 and rRNAs15. (C) Dissociation kinetics of the S15–RNA complexes. Complexes were formed with 32P-labeled RNAs, and dissociation was measured by dilution with excess unlabeled RNA. (D) Initial rate of association of S15–RNA complexes. The bimolecular rate parameter defined in equation 7 of Riggs et al. (1970) is  , where R, P and PR are RNA, protein and complex concentrations, respectively.
, where R, P and PR are RNA, protein and complex concentrations, respectively.
Table I. Binding constants for TtS15 interaction with mRNA and rRNA of T.thermophilus.
| RNA | t (°C) | Kd direct (×10–9 M) | Kd compet. (×10–9 M) | koff (×10–4/s) | kon pred. (×105/M/s) | kon exper. (×105/M/s) |
|---|---|---|---|---|---|---|
| rRNAs15 | 0 | 2.20 ± 1.30 | – | – | – | – |
| rRNAs15 | 37 | 0.23 ± 0.00 | 0.28 ± 0.09 | – | – | – |
| rRNAs15 | 45 | 0.23 ± 0.00 | – | 2.13 ± 0.25 | 9.26 | 25.0 ± 8.0 |
| mRNA1 | 35 | 4.48 ± 0.16 | – | – | – | – |
| mRNA1 | 45 | 4.90 ± 0.18 | – | 9.57 ± 2.50 | – | – |
| mRNA2 | 37 | 3.50 ± 0.00 | 3.30 ± 1.32 | – | – | – |
| mRNA2 | 45 | 2.86 ± 0.47 | – | 9.10 ± 1.23 | 3.18 | 1.47 ± 0.5 |
Kd direct and Kd compet., apparent dissociation constants from direct RNA–protein binding and competition experiments, respectively; kon pred. and kon exper., association rate constants calculated from the koff/Kd ratio and determined in the experiments, respectively.
Removal of the 3′ half of mRNA (mRNA2) did not alter its affinity for TtS15 (Table I). Both mRNA1 and mRNA2 were able to compete with the 16S rRNA fragment (Table I). The deduced apparent Kds were consistent with those measured from direct binding assays (Table I). Control 5S rRNA from T.thermophilus was unable to compete with mRNA and rRNAs15.
Kinetics of S15–mRNA binding
Rate constants for TtS15–mRNA and TtS15–rRNA complexes (Figure 2C and D; Table I) were determined by filter-binding assay. Since high temperatures resulted in RNA degradation, the following experiments were conducted at 45°C, a temperature still permitting growth of T.thermophilus, although lower than the optimal temperature of 70°C. Dissociation was monitored by displacement of the bound labeled RNA by unlabeled RNA, and the data were fit to a single exponential. The dissociation rate curves showed monophasic behavior, indicative of unimolecular dissociation reactions for all complexes (Figure 2C). The two mRNA complexes dissociate at the same rate (koff ∼9 × 10–4/s) with a half-life of 15 min, while the rRNA complex dissociates 4.5-fold slower (koff ∼2 × 10–4/s) with a half-life of 65 min (Table I).
Association rate constants (Table I) were estimated from the initial rate data by applying the integrated rate equation for a bimolecular reaction (Figure 2D). TtS15 binds mRNA1 with an apparent rate constant (kon) of 25 × 105/M/s, 16-fold slower than for rRNAs15. Due to limitations of the filter-binding technique for direct measurements of the fast association reaction, this difference might represent a small overestimation, since the predicted kons calculated from the ratio between koffs and Kds are close, but not identical, to the experimental kon values.
Footprinting reveals TtS15-induced changes in the 5′ region of mRNA
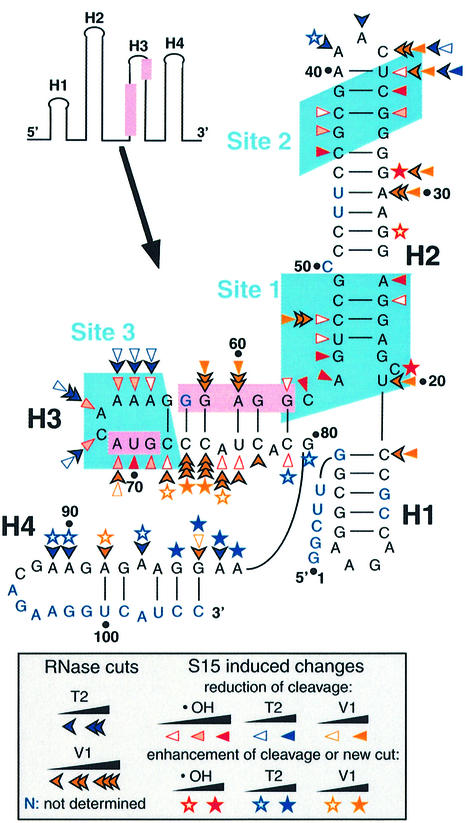
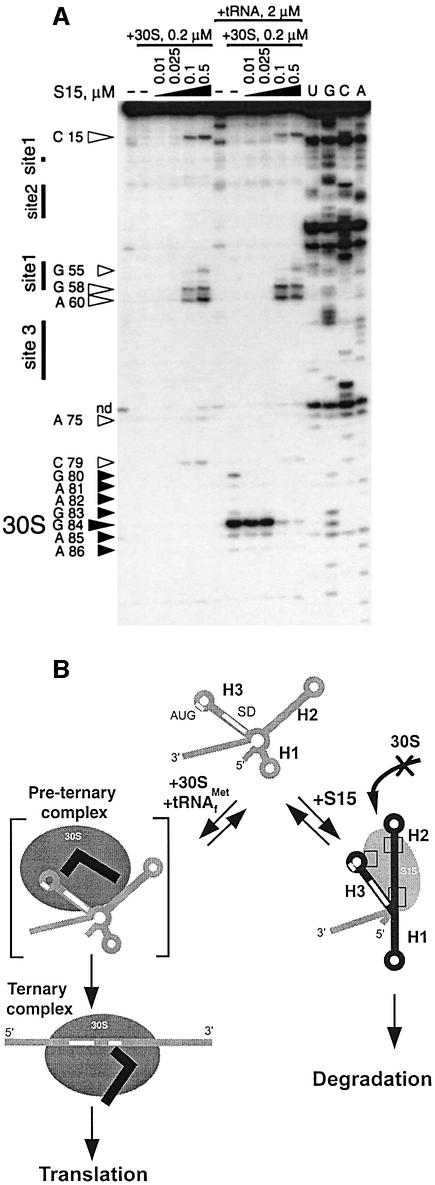
The binding site on mRNA2 of TtS15 was mapped by hydroxyl radical and RNase footprinting. Hydroxyl radical-induced cleavage is insensitive to the secondary structure of RNA and reveals a precise footprint of bound proteins, whereas RNases V1 and T2 that cleave double- and single-stranded RNA regions, respectively, are sensitive to steric accessibility. Representative gels are shown in Figure 3, and the results are summarized in Figure 4. The 5′-terminal region can be folded into three stem–loop folds (1–3) involving helices H1–H3, consistent with RNase V1 and T2 cleavages and secondary structure prediction (Jaeger et al., 1990) (Figure 4). In the free RNA, the Shine–Dalgarno (SD) sequence appears to be sequestered within helix H3, as confirmed by strong RNase V1 cuts. However, the presence of two bulged residues (A75 and A78) could weaken the stability of this helix, thus facilitating interaction with the anti-SD sequence, an event required for 30S subunit binding.
Fig. 3. Footprinting of TtS15 on mRNA2. Hydroxyl radical footprint, short migration (A) and long migration (B). Footprinting with RNase V1 (C) and RNase T2 (D). TtS15-induced protections and enhancements are shown by arrows and asterisks, respectively. Black, gray and white symbols reflect the intensity of changes (strong, medium and low). Assignment of G83–84, migrating as three bands, is tentative.
Fig. 4. Summary of hydroxyl radical and nuclease footprinting experiments on mRNA2. The SD sequence and initiation codon are highlighted in pink, and the three determined binding sites (sites 1–3) in light blue. The code is indicated in the insert.
TtS15 induced reactivity changes to hydroxyl radical treatment between positions 21 and 79 of mRNA at 45°C, but not at 0°C, as expected from binding experiments. The TtS15-induced protections from hydroxyl radical cleavage are clustered into three distinct regions, which coincide with more extended nuclease footprints. The first site (nucleotides 25–26/53–58) covers the bottom of helix H2 and the junction with helix H3 (Figure 3A and B). Protections can be correlated with the reduction of RNase V1 cleavage at C53. The second site (nucleotides 34–36/42–44) is localized in the upper part of helix H2 (Figure 3A). The simultaneous occurrence of RNases V1 and T2 cuts at U36 and C37 in the free RNA most probably reflects a local unstable conformation. The cleavage reduction observed toward the 5′ side of the loop might result from either protection or conformational rearrangement of the loop induced by TtS15. Moreover, the 3′ part of the loop remained fully accessible to RNase T2, with an even enhanced cleavage at A39. The third site (nucleotides 64–72) is located in the apical part of helix H3 and overlaps the initiation codon of the TtS15 gene (Figure 3B–D). Protections correlate with the decrease of RNase T2 cleavage.
Notably, the SD sequence is surrounded directly by sites 1 and 3 and protected from RNase V1 cleavage upon ThS15 binding. Thus, these results suggest that TtS15 binding most probably prevents access to the ribosome loading site. Unexpectedly, TtS15 binding also enhanced RNase V1 cleavages within the 3′ strand of helix H3. This could reflect stabilization of helix H3 or/and increased accessibility of this side of the stem following reorientation of helices. Changes in the RNase V1 cleavage pattern were also accompanied by the appearance of new RNase T2 cuts downstream of helix H3 (Figure 3D). Such pronounced accessibility changes are generally reflective of conformational rearrangements. These data are consistent with TthS15 binding inducing a reorganization of the spatial arrangement of helices in the mRNA target.
The minimal mRNA contains two S15-binding regions
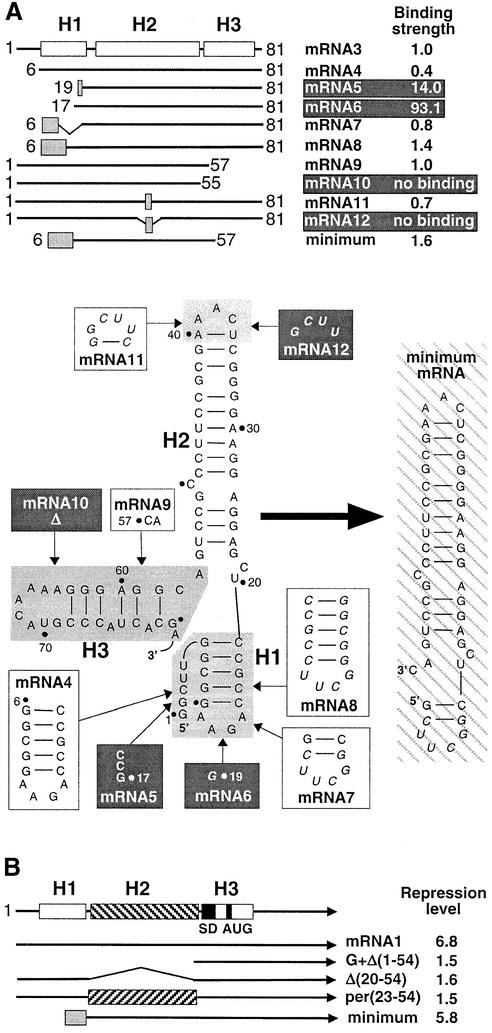
A deletion analysis was used to identify further the structural elements essential for TtS15 binding and to determine the minimal mRNA fragment still retaining wild-type-like affinity for TtS15. The results are summarized in Figure 5A. The full-length mRNA could be reduced to the first 81 nucleotides (mRNA3) without loss of TtS15 binding, as expected from footprinting data. On the other hand, deletion of the 5′-terminal 80 nucleotides of mRNA abolished TtS15 binding.
Fig. 5. Deletion analysis of mRNA. (A) Minimization of the TtS15-binding site by filter-binding assay. The upper part represents a schematic of mRNA3, mutant mRNAs and their effects on binding TtS15. The relative binding strength (Krel) is expressed as the ratio of the Kds of mRNA2 and mutant RNA. mRNAs, with diminishing binding, are shown on a dark background. Shaded boxes represent RNA regions with nucleotide substitutions. The bottom part of the panel shows details of mutagenesis. The RNA stem–loops, which were subjected to truncation and mutagenesis, are shaded in gray. Nucleotide sequences, replacing the shaded regions, are shown in boxes (white and black for mutations not diminishing and diminishing TtS15 binding, respectively). Within the RNA sequences, substitutions are shown in italics. The minimum RNA fragment is shown on the right and hatched. (B) Schematic representation of the leader regions of selected mRNA1 mutants and their effects on autorepression determined as in Figure 1B.
Progressive deletions of the 5′ part of mRNA3 (mRNAs 4–6) clearly indicate that the short non-paired 5′ region can be omitted without reduction of S15 affinity, whereas the integrity of helix H1 is required for complex formation (Figure 5A). However, stem–loop 1 could be shortened or replaced by variant stem–loops (mRNAs 7 and 8). Thus, in agreement with the footprinting results, stem–loop 1 is not directly involved in S15 recognition.
Unexpectedly, stem–loop 3 is not essential for TtS15 binding although protected by the protein. However, nucleotides A56 and C57 are crucial for TtS15 binding (mRNAs 9 and 10). This, in turn, suggests that stem–loop 2 is indispensable for TtS15 interaction. Indeed, truncation of the upper part of stem–loop 2 (mRNA12) severely affected TtS15 binding. Nevertheless, substitution of the apical loop and the adjacent U36–A40 pair by a UUCG tetraloop closed by a C–G pair was tolerated (mRNA11). Therefore, all elements essential for S15 recognition are provided by nucleotides in helix H2 and downstream of A56–C57. As anticipated from these results, a 46 nucleotide fragment (minimum mRNA), restricted to these elements and the short version of stem–loop 1, bound TtS15 with wild-type affinity (Kd 4.51 ± 1.10 nM) and competed with rRNAs15 for S15 binding as well as mRNA2.
In addition, deletions found to inhibit TtS15 binding also decreased the repression level in in vitro translation assays, when introduced in mRNA1 (Figure 5B). This is the case for deletion of stem–loops 1 and 2 (G + Δ1–54), and deletion or permutation of stem–loop 2 [Δ21–54, per(23–54)]. On the other hand, replacing stem–loop 1 by the short hairpin as in the minimum fragment only slightly affected translation inhibition. Thus, our results clearly indicate that translational autorepression of TtS15 synthesis is mediated by binding of TtS15 to its mRNA 5′-untranslated region (UTR).
mRNA site 1 mimics the rRNA three-way junction
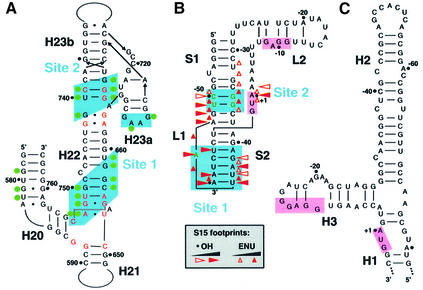
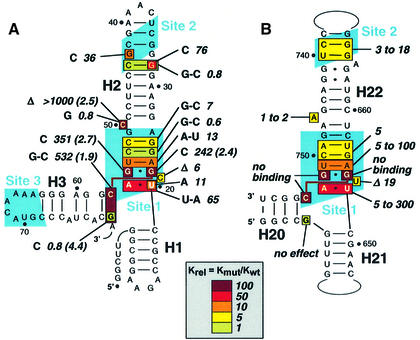
Comparison of the 16S rRNA- (Nikulin et al., 2000; Serganov et al., 2001) and mRNA-binding regions reveals a striking resemblance in the folding topology. Both RNAs contain a long helix (H22 and H2 in rRNA and mRNA, respectively) with the two binding sites separated by approximately the same distance. Notably, the invariant nucleotides constraining the three-way junction between helices H20, H21 and H22 in 16S rRNA (Serganov et al., 1996, 2001) are found in the bottom part of helix H2 and surrounding nucleotides that delineate site 1 (Figure 8A). Thus, it is tempting to speculate that helices H1, H2 and H3 of the mRNA might adopt, at least in the presence of S15, a conformation resembling the rRNA three-way junction that constitutes the major RNA-binding site of S15. By analogy to the rRNA site, helices H1 and H2 would correspond to the co-axially stacked helices H21 and H22 in 16S rRNA, and helix H3 to helix H20 that makes an acute angle with helix H22 (Figure 6A and B). However, helices H20–22 form a closed three-way junction in 16S rRNA, while helices H1 and H3 are not directly connected in the mRNA.
Fig. 8. Recognition of rRNA and mRNA targets by S15. (A) The T.thermophilus 16S rRNA-binding site. Nucleotides contacting TtS15 (shown by green circles) are from the crystallographic structures of S15–rRNA (Nikulin et al., 2000) and S15–S6–S18–rRNA (Agalarov et al., 2000) complexes, and from the 30S ribosomal subunit (Brodersen et al., 2002). Nucleotides shown in red are conserved in 16S rRNAs. (B) Proposed pseudoknot structure from E.coli S15 mRNA specifically binding EcS15. Hydroxyl radical and ethylnitrosourea (ENU) footprints (Philippe et al., 1995) are indicated. Nucleotides essential for EcS15 binding are shown in green (Bénard et al., 1994, 1998; Serganov et al., 2002). (C) Predicted secondary structure for the 5′-UTR of the B.stearothermophilus S15 mRNA interacting with the BsS15 protein (Scott and Williamson, 2001). The SD sequence and initiation codon are highlighted in pink in (B) and (C).
Fig. 6. Effects of RNA mutations on TtS15 binding. The color code (in the insert) reflects binding strength changes, from no effect (green) to a strong effect (dark red). (A) Site-directed mutagenesis on T.thermophilus mRNA. The mutations are shown and the corresponding Krels (calculated as in Figure 5A) are indicated. The number in parentheses indicates the repression ratio (calculated as in Figure 1B), which is, for reference, 6.8 for mRNA1. (B) Summary of site-directed mutagenesis on E.coli rRNA from Serganov et al. (2001) given for comparison.
To test this ‘rRNA mimicry’ hypothesis, we mutated those nucleotides in mRNA that are potentially involved in forming the junction, in order to check whether they affect S15 binding as in 16S rRNA (Figure 6A and B). Indeed, a previous study showed that the rRNA three-way junction is constrained by an invariant C754–G654·G752 base triple and that mutation of any of these nucleotides was deleterious for S15 binding (Serganov et al., 2001). Here we found that mutations G22C, G55C and substitution of C57–G80 by G–C dramatically reduced S15 binding, consistent with potential formation of the C57–G22·G55 base triple. Moreover, these mutations also abolished autorepression of S15 synthesis in our in vitro translation assay (Figure 6A). As a corollary to the formation of a C57–G22·G55 base triple, G80 should not pair with G57. Indeed, the G80C replacement is tolerated, as in rRNA (Serganov et al., 2001), and one RNase T2 cut is even induced at this position upon S15 binding (Figure 4). In 16S rRNA, U652 and A753 form a reverse Hoogsteen pair, and substitutions of these nucleotides affected S15 binding to various degrees. Here we found that replacing U20–A56 by A–A or U–A decreased binding by 11- to 65-fold, a more pronounced effect than that induced by the corresponding mutations in rRNA (∼5-fold) (Serganov et al., 2001). On the other hand, deletion of C21, the equivalent of the non-conserved ‘spacer’ U653, reduced binding by a factor of six, 3-fold less than in rRNA. The moderate effect of the inversion of the A23–U54 base pair (13-fold) fell in the same range as those observed for mutations of the corresponding A655–U751 base pair in rRNA. Mutations of the G24–C53 and G25–C52 base pairs located above the junction did not significantly affect S15 binding, as observed in rRNA.
The present data clearly show that mutations in site 1 of mRNA and in the three-way junction of 16S rRNA induce similar effects on S15 binding. In particular, they strongly support the formation of an mRNA C57–G22·G55 base triple analogous to its rRNA C754–G654·G752 counterpart. Subtle differences observed at adjacent positions most probaby reflect more or less stringent constraints associated with the different topologies of the junctions (open in mRNA, closed in rRNA). Taken together, our results suggest that S15 binding induces folding of a 16S rRNA-like three-way junction in TtS15 mRNA.
mRNA site 2 is not equivalent to rRNA site 2
Site 2 in mRNA does not show significant similarity to the corresponding site in 16S rRNA. In particular, the conserved G–U/G–C motif, which makes base-specific contacts with S15 in the rRNA crystallographic structures, is replaced in mRNA by a G33·G43 mismatch flanked by G–C base pairs (Figure 6A and B). Notably, the conserved rRNA purine-rich motif, which is not required for primary S15 recognition but is important for binding of S6 and S18 proteins in the subsequent steps of ribosomal subunit assembly (Agalarov et al., 2000), is also missing. The importance of mRNA site 2 was confirmed by the loss of S15 binding resulting from truncation of this site in mRNA12 (Figure 5A). Then, we showed that reversing the G32–C44 base pair has no effect on S15 recognition, while its destabilization by the G32C substitution reduced S15 binding by 76-fold (Figure 6A). Formation of a canonical G33–C43 base pair also significantly affected binding (by a factor of 36). This contrasts with the corresponding rRNA site that contributes only modestly to the stability of the interaction with S15 (Batey and Williamson, 1996a; Serganov et al., 1996, 2001), but is used to trigger a conformational rearrangement required for subunit assembly (Agalarov et al., 2000; Serganov et al., 2001). A bulged residue (C50) is present between sites 1 and 2 in TtS15 mRNA, and its deletion severely affected S15 binding and in vitro autorepression (Figure 6A). A bulged nucleotide also exists in 16S rRNAs in which it introduces some flexibility within the helical axis trajectory, thus allowing proper positioning of the two sites relative to each other. Indeed, deletion of A746 in E.coli 16S rRNA prevents binding at site 2, while the affinity for S15 was only moderately reduced, since binding at site 1 was unaffected (Serganov et al., 2001). This suggests that in mRNA, as in rRNA, the bulged nucleotide is required to allow binding at both sites. However, in contrast to rRNA, binding at both sites is essential to anchor TtS15 to its mRNA target.
TtS15 inhibits its translation through a ‘displacement’ mechanism
The question of whether TtS15 uses a mechanism similar to that used by EcS15 to inhibit its translation was addressed by toeprinting experiments. In such experiments, primer extension with MMLV reverse transcriptase is used to visualize the formation of binary 30S–mRNA and ternary 30S–mRNA–tRNAfMet complexes (Hartz et al., 1991). Under our experimental conditions, we failed to detect the transient binary 30S–mRNA complex (left part of the gel on Figure 7A). However, binding of TtS15 could be titrated by the appearance of specific pauses or toeprints. Notably, major stops were located on the 3′ side of site 3 (position 75) and site 1 (A60 and G58), indicating that reverse transcriptase progression is impeded by the bound protein and/or stabilization of the RNA structure. There is no obvious explanation for the pause at C15. Formation of the stable ternary 30S–mRNA–tRNAfMet complex yielded a canonical toeprint at G84, 16 nucleotides downstream of the initiation codon (Hartz et al., 1991), and much weaker pauses around it (right part of the gel in Figure 7A). Addition of increasing amounts of TtS15 during the formation of the ternary complex led to progressive disappearance of the +16 toeprint, while TtS15-specific pauses appeared on the gel. These results clearly indicate that TtS15 competes with the ribosome for binding, in contrast to EcS15 which does not prevent ribosome binding, but rather stabilizes an unproductive ternary complex (Philippe et al., 1993). These two mechanisms are referred to as ‘displacement’ and ‘entrapment’ models, respectively (Draper, 1993).
Fig. 7. Competition between TtS15 and 30S subunit for mRNA binding. (A) Effect of TtS15 on the formation of the ternary mRNA–30S-tRNAfMet complex revealed by toeprint experiments. Reverse transcription stops specific for S15 or the ternary complex are indicated by open and full arrows, respectively. The intensity of the toeprint is proportional to the size of the arrow. (B) A ‘displacement’ model for the S15 autorepression mechanism. The model assumes that TtS15 induces a conformational change of the mRNA that masks the ribosome entry site. The different S15–RNA contacts are shown by squares. The mRNA is either bound to TtS15 and degraded, or bound to the 30S subunit and committed to the formation of a productive ternary complex.
Discussion
S15 prevents ribosome binding through an induced-fit mechanism
Autoregulation of ribosomal protein synthesis through adjustment of their concentration to that of their rRNA targets was essentially defined through studies in E.coli. Here, we demonstrate that such a feedback control exists in extreme thermophilic bacteria. We showed that TtS15, encoded by a monocistronic mRNA, represses the translation of its own mRNA in vitro. TtS15 binds its mRNA with a high affinity (in the nanomolar range), 20-fold lower than to 16S rRNA at the same temperature. This is consistent with an understanding of autoregulation reflecting preferential binding of the repressor to rRNA over mRNA. Deletion analysis and hydroxyl radical footprinting localized the TtS15-binding site within the first 81 nucleotides of mRNA, mostly in the 5′-UTR. Thus, because transcription and translation are coupled in bacteria, translation of mRNA can be inhibited efficiently soon after transcriptional initiation. The mRNA regulatory site folds into a three stem–loop scaffold, which shares no resemblance with the pseudoknot fold formed by the E.coli mRNA site (Philippe et al., 1990). The protein was found to protect three distinct sites from hydroxyl radical cleavage and to induce a major conformational change of the mRNA, as revealed by dramatic accessibility changes to RNase hydrolysis. In this context, TtS15 binds mRNA at a slower rate than rRNA and fails to bind mRNA at low temperature. This suggests that the free mRNA probably adopts multiple or improper conformations and that one or both components have to undergo adaptive transitions in order to bind each other. Such an induced-fit mechanism is a common theme in RNA–protein recognition (Patel, 1999; Williamson, 2000).
A major finding is that mRNA site 1 folds into a 16S rRNA-like three-way junction upon TtS15 binding, and is recognized by the protein as the corresponding rRNA site. These results are supported by striking analogies in nucleotide composition and effect of mutations (Figure 6A and B) (Batey and Williamson, 1996b; Serganov et al., 1996, 2001). By analogy with the rRNA three-way junction, we can reasonably assume that helices H1 and H2 (corresponding to H21 and H22 in rRNA) are stacked co-axially, with helix H3 (the mRNA equivalent of H20) making an acute angle with helix H2. However, unlike the rRNA three-way junction, mRNA site 1 is not sufficient to ensure stable binding, and requires binding at site 2. This most probably reflects the fact that mRNA helices H1–H3 do not form a closed junction as in rRNA and could account for the lower stability of the mRNA complex. Site 2, located in the upper part of helix H2, contains a G·G/G–C motif instead of the G–U/G–C motif conserved in rRNAs. Since U740 makes base-specific contacts with S15 in rRNA, replacement by guanosine at this position in mRNA (found in <1% of prokaryotic 16S rRNAs) points to a different recognition pattern for mRNA compared with 16S rRNA. Strikingly, binding at site 3 that overlaps the ribosome-binding site does not contribute to the stability of the mRNA complex. Thus, contacts with this site are not required for primary interaction but should result from the S15-induced conformational change and a movement of helix H3 towards helix H2. Similarly, it was shown that the interaction of S15 with the tetraloop in helix H23a of 16S rRNA (Figure 8A) arises as a consequence of S15-induced conformation change and does not participate in the initial recognition (Batey and Williamson, 1996a; Serganov et al., 1996). The strong S15-induced protections in the initiation codon might correspond to the S15 contacts found around U580 (helix H20) in 16S rRNA within the mature 30S subunit (Brodersen et al., 2002).
Another major consequence of the S15-induced conformational rearrangement is the strong reduction in accessibility of the SD sequence to RNase V1. This is clearly due to a masking of the strand carrying this sequence, since the opposite strand becomes much more accessible to the same RNase, which in turn indicates that helix H3 is stabilized. By analogy with the three-dimensional structure of the rRNA complex, the SD sequence, directly adjacent to the sharp turn of the RNA strand caused by the C–G·G base triple, should become less accessible to RNase V1, due to partial shielding by the protein and the acute angle between helices H2 and H3. These results strongly suggest that binding of TtS15 to its regulatory site should mask the ribosome entry site and prevent ribosome binding. This was confirmed by toeprinting experiments, which clearly indicated a competition between S15 and the 30S subunit for mRNA binding. Thus, a model could be proposed (Figure 7B), in which stem–loops 1–3 are not structurally constrained in the absence of TtS15. Although the SD sequence is engaged in helix 3, the presence of the two bulged residues A75 and A78 probably lowers the stability of the helix, thus allowing formation of the initiation complex. When TtS15 is in excess over its rRNA target, it binds to its regulatory mRNA site and triggers formation of the rRNA-like three-way junction that renders the ribosome entry site inaccessible. Thus, a key element of the regulatory mechanism is that TtS15 is used to stabilize a conformation of mRNA that is non-competent for ribosome binding. In addition, the non-translated mRNA might be degraded immediately upon forming a complex with the repressor, as shown for the E.coli S15 mRNA (Braun et al., 1998).
The feedback mechanisms used by S15 in E.coli and T.thermophilus are notably different, since they involve ‘entrapment’ and ‘displacement’, respectively. This is also reflected by the 100-fold difference in affinity observed between EcS15 and TtS15 for their respective mRNA targets. Indeed, translation is initiated in prokaryotes by the binding of 30S subunits and initiator tRNAfMet in a random order to form a transient ‘ternary pre-initiation’ complex (Figure 7B), which subsequently is converted into an irreversible ternary complex (Gualerzi and Pon, 1990). To compete efficiently with the 30S subunit for mRNA binding, a repressor has to bind mRNA with a much higher affinity than the 30S subunit or be in large excess over the 30S subunit (Schlax et al., 2001). That the affinity of TtS15 for its mRNA target is ∼10-fold higher than the experimentally determined K30S (2 × 107/M) (Calogero et al., 1988; Schlax et al., 2001) is compatible with a displacement mechanism. At the other extreme, repressors acting by an entrapment mechanism, such as E.coli S4 (Deckman and Draper, 1987; Schlax et al., 2001) and EcS15 (Serganov et al., 2001), which only needs to stabilize an unproductive initiation complex without competing with 30S subunit, can inhibit translation efficiently with a modest affinity in the range of 107/M.
RNA mimicry and versatility of autoregulation mechanisms
The case of S15 offers an unprecedented opportunity to understand how a highly phylogenetic conserved protein recognizes different RNA ligands. Indeed, detailed pictures of interactions between S15s from mesophilic (E.coli) and thermophilic bacteria (T.thermophilus and B.stearothermophilus) with their rRNA and mRNA targets are available (Figure 8). An astonishing diversity is observed at the level of known mRNA regulatory sites. Remarkably, the three mRNAs all require a bipartite site, while utilization of mimicry with the rRNA site is limited and versatile. Indeed, mimicry was restricted to the G–U/G–C motif in the E.coli mRNA pseudoknot (Serganov et al., 2002) and to the three-way junction in the T.thermophilus mRNA (this work). These differences are probably related to the regulation mechanism used, since the three-way junction is expected to provide a higher affinity than the G–U/G–C motif and is therefore more adapted to the displacement mechanism (see above). The B.stearothermophilus mRNA contains a G–U/G–C motif and a purine-rich three-way junction (Scott and Williamson, 2001). However, the three-way junction does not display any sequence identity with the rRNA junction, while the localization of the translation initiation signals and the topology of the junction are different in both mRNAs. Although further investigation is required to draw definitive conclusions, it appears that substantial differences should exist in the recognition mode and related regulatory mechanisms.
Taken together, these results indicate that a general translational control has emerged through evolution, despite the observed differences. Although very diverse regulation of S15 is unique at the present time, it is not an exception. A general scheme of ribosomal protein L1-mediated control, based on mimicry with the L1-binding site on 23S rRNA, was shown in mesophilic and thermophilic archaea and bacteria (Kohrer et al., 1998; Kraft et al., 1999). A feedback mechanism is conserved in some proteobacteria in the case of ribosomal protein L4 (Allen et al., 1999), and in Gram-negative bacteria in the case of threonyl-tRNA synthetase expression (Torres-Larios et al., 2002). The latter is also based on a competition mechanism involving mimicry with tRNAThr, the usual target of the repressor (Caillet et al., 2003). Mimicry-based translational regulation in eukaryotes is rare, mainly because, in contrast to prokaryotes, transcription and translation are not coupled due to cellular compartmentalization. Nevertheless, the yeast ribosomal protein L30 has optimized its regulation by a feedback mechanism at the level of splicing and translation (Li et al., 1996). Furthermore, its mRNA-binding site mimics a conserved site in rRNA, and a similar mechanism appears to take place in archaea (Vilardell et al., 2000).
These examples highlight the role of RNA mimicry in a variety of regulatory mechanisms and stress the extraordinary plasticity of mRNAs to fulfill their regulatory functions. This diversity contrasts with the extreme conservation of the prokaryotic rRNA sites, which most probably reflects a strong evolutionary pressure dictated by strict structural and functional constraints linked to ribosome assembly. This also suggests that mechanisms responsible for optimizing expression of ribosomal components have emerged later in evolution and have had to adapt to environmental conditions.
Materials and methods
Preparation of biological material
Unlabeled and uniformly radioactively labeled RNAs were obtained by in vitro transcription with phage T7 RNA polymerase, and purified on denaturing gels (Serganov et al., 2001). The natural TtS15 transcript (mRNA1) (nucleotides –68 to +315) was transcribed from a DNA fragment obtained by PCR amplification from plasmid pUS1593-7 (Serganov et al., 1997). mRNA2, containing the first 147 nucleotides of mRNA1 and additional nucleotides AUC at the 3′ end, was transcribed from plasmid pUT7mRNA147 linearized by BamHI after cloning of the corresponding PCR fragment into pUT7 vector (Serganov et al., 1997). Short mRNAs were transcribed from DNA templates obtained by annealing of two complementary oligonucleotides, containing the T7 promoter and the sequence of interest. Fragment rRNAs15, corresponding to nucleotides 559–753 from T.thermophilus 16S rRNA, and a derived G654C mutant (rRNAs15mut) were prepared as described previously (Serganov et al., 1997, 2001). Protein TtS15 was expressed in E.coli cells and purified under non-denaturing conditions using consecutive chromatographies on CM–Sepharose, hydroxyapatite and Superdex75 (Serganov et al., 1997).
Northern blotting
Total RNA was isolated from T.thermophilus by the hot phenol method (Serganov et al., 1997), separated by electrophoresis on a 1.5% formaldehyde–agarose gel, and transferred onto Hybond N membrane (Amersham Pharmacia Biotech). Northern blot hybridization was performed with 32P-end labeled DNA 1–384 in 50 mM Na phosphate buffer pH 6.8 supplemented with 50% formamide, 1% SDS, 5× SSC, 5× Denhardt’s solution, 250 µg/ml salmon testes DNA and 100 µg/ml tRNA at 55°C overnight. The membrane was washed three times with 1% SDS and 5× SSC at the same temperature and exposed on Biomax MS film (Kodak).
In vitro autorepression assay
In vitro translation was conducted by incubating mRNAs (0.4 µM) in 25 µl of an E.coli S30 extract (Promega) complemented with [35S]methionine (New England Nuclear) for 1 h at 43°C. For translation repression assays, increasing amounts of TtS15 (0.2–1.6 µM) were added to the translation mixture. In competition experiments, rRNAs15 and G654C rRNA mutant were added at the specified concentrations. Translation products were separated by electrophoresis on a 10–25% SDS–polyacrylamide gel and quantified on a BAS 2000 BioImager (Fuji).
Protein-binding assays
Equilibrium and rate binding constants were determined by nitrocellulose filter-binding assays in 50 mM Tris–HCl pH 7.5, 20 mM MgCl2, 270 mM KCl, 5 mM dithiothreitol and 0.02% (w/v) bovine serum albumin (BSA). Direct binding measurements were conducted as described previously (Serganov et al., 1996), using 10 000 c.p.m. of labeled RNA (<0.1 pM) and increasing concentrations of TtS15 (0.01 nM to 0.1 mM). Incubation times were 90 min at 0–50°C, 60 min at 55°C and 15 min at 60–70°C. The apparent dissociation constant (Kd) was determined assuming that complex formation obeys a simple bimolecular equilibrium and the concentration of the labeled RNA is negligible. Results were fitted with the equation:
Θ = [S15]/(Kd + [S15])
where Θ represents the fraction of labeled RNA bound to the filter. Competition experiments were carried out as described above, using TtS15 at a concentration of 6 nM, a negligible amount of labeled RNA and a variable concentration (10 pM to 1 µM) of competitor unlabeled RNA. Data were fitted with equation 5 of Lin and Riggs (1972).
The association rates of TtS15–mRNA and TtS15–rRNAs15 complexes were measured under standard conditions with 30 and 3.9 nM TtS15, and 10 and 2 nM RNA, respectively. Aliquots (50 µl) were withdrawn at short time intervals and filtered. The association rate constant (kon) was estimated from the initial rate data using equation 7 of Riggs et al. (1970). To measure the dissociation rate, the complexes were formed by incubating negligible amounts (10 000 c.p.m./50 µl) of labeled mRNA or rRNAs15, with 5 or 0.5 nM of TtS15, respectively. After 60 min incubation, the corresponding unlabeled RNA was added at a 10-fold excess over protein, and 50 µl aliquots were filtered at various time intervals. The complexes without addition of unlabeled RNA remained stable during the experiment time. Alternatively, dissociation of the complex was achieved by diluting the reaction mixture 10-fold with the incubation buffer, yielding similar results. The dissociation rate constant (koff) was deduced from the initial rate data after fitting to equation 4 of Riggs et al. (1970). All constants are the average of at least two experiments with standard deviations <30%. Fitting was done using Microcal Origin software (Microcal Software, USA).
Footprinting
The S15–RNA complexes were formed by incubating mRNA2 (0.5 µM) and a 3- or 6-fold excess of TtS15 at 45°C for 10 min in 20 µl of buffer B (50 mM HEPES–Na pH 7.9, 20 mM MgCl2, 270 mM KCl) for nuclease footprinting and in buffer B supplemented with 0.025% (w/v) BSA and 0.05 µg/µl tRNA for hydroxyl radical footprinting. RNase V1 (0.0025 U) (Pierce) or T2 (0.17 U) (Sigma) was added to the complex and incubation was continued for 10 min. The Fe(II)-EDTA-generated hydroxyl radical cleavage reaction was performed as described previously (Serganov et al., 1996). The RNAs were reverse transcribed using 32P-labeled oligonucleotide complementary to positions 143–150 or 95–112 of mRNA2. The resulting cDNAs were analyzed by electrophoresis on 10% polyacrylamide–8 M urea gels together with sequencing reactions.
Extension inhibition (toeprint) assay
Toeprinting experiments were carried out essentially as described in Philippe et al. (1993) using 25 nM mRNA2. Primer extension was conducted with 20 U of MMLV reverse transcriptase (Promega) for 15 min at 37°C from the labeled primer complementary to positions 95–112.
Acknowledgments
Acknowledgements
We are indebted to P.Romby for discussion and for providing 30S subunits and tRNA. This work was supported by NIH grant CA46778 to D.P. and by the CNRS.
References
- Agalarov S.C., Prasad,G., Funke,P.M., Stout,C.D. and Williamson,J.R. (2000) Structure of the S15,S6,S18–rRNA complex: assembly of the 30S ribosome central domain. Science, 288, 107–113. [DOI] [PubMed] [Google Scholar]
- Allen T., Shen,P., Samsel,L., Liu,R., Lindahl,L. and Zengel,J.M. (1999) Phylogenetic analysis of L4-mediated autogenous control of the S10 ribosomal protein operon. J. Bacteriol., 181, 6124–6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batey R.T. and Williamson,J.R. (1996a) Interaction of the Bacillus stearothermophilus ribosomal protein S15 with 16S rRNA: I. Defining the minimal RNA site. J. Mol. Biol., 261, 536–549. [DOI] [PubMed] [Google Scholar]
- Batey R.T. and Williamson,J.R. (1996b) Interaction of the Bacillus stearothermophilus ribosomal protein S15 with 16S rRNA: II. Specificity determinants of RNA–protein recognition. J. Mol. Biol., 261, 550–567. [DOI] [PubMed] [Google Scholar]
- Bénard L., Philippe,C., Dondon,L., Grunberg-Manago,M., Ehresmann,B., Ehresmann,C. and Portier,C. (1994) Mutational analysis of the pseudoknot structure of the S15 translational operator from Escherichia coli. Mol. Microbiol., 14, 31–40. [DOI] [PubMed] [Google Scholar]
- Bénard L., Mathy,N., Grunberg-Manago,M., Ehresmann,B., Ehresmann,C. and Portier,C. (1998) Identification in a pseudoknot of a U·G motif essential for the regulation of the expression of ribosomal protein S15. Proc. Natl Acad. Sci. USA, 95, 2564–2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun F., Le Derout,J. and Regnier,P. (1998) Ribosomes inhibit an RNase E cleavage which induces the decay of the rpsO mRNA of Escherichia coli. EMBO J., 17, 4790–4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen D.E., Clemons,W.M.,Jr, Carter,A.P., Wimberly,B.T. and Ramakrishnan,V. (2002) Crystal structure of the 30S ribosomal subunit from Thermus thermophilus: structure of the proteins and their interactions with 16S RNA. J. Mol. Biol., 316, 725–768. [DOI] [PubMed] [Google Scholar]
- Caillet J. et al. (2003) The modular structure of Escherichia coli threonyl-tRNA synthetase as both an enzyme and a regulator of gene expression. Mol. Microbiol., 47, 961–974. [DOI] [PubMed] [Google Scholar]
- Calogero R.A., Pon,C.L., Canonaco,M.A. and Gualerzi,C.O. (1988) Selection of the mRNA translation initiation region by Escherichia coli ribosomes. Proc. Natl Acad. Sci. USA, 85, 6427–6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culver G.M., Cate,J.H., Yusupova,G.Z., Yusupov,M.M. and Noller,H.F. (1999) Identification of an RNA–protein bridge spanning the ribosomal subunit interface. Science, 285, 2133–2136. [DOI] [PubMed] [Google Scholar]
- Deckman I.C. and Draper,D.E. (1987) S4–α mRNA translation regulation complex. II. Secondary structures of the RNA regulatory site in the presence and absence of S4. J. Mol. Biol., 196, 323–332. [DOI] [PubMed] [Google Scholar]
- Draper D.E. (1993) Translational regulation of ribosomal proteins in Escherichia coli. Molecular mechanisms. In Ilan,J. (ed.), Translational Regulation of Gene Expression. Plenum Press, New York, NY, pp. 1–26.
- Freedman L.P., Zengel,J.M., Archer,R.H. and Lindahl,L. (1987) Autogenous control of the S10 ribosomal protein operon of Escherichia coli: genetic dissection of transcriptional and posttranscriptional regulation. Proc. Natl Acad. Sci. USA, 84, 6516–6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualerzi C.O. and Pon,C.L. (1990) Initiation of mRNA translation in prokaryotes. Biochemistry, 29, 5881–5889. [DOI] [PubMed] [Google Scholar]
- Guillier M., Allemand,F., Raibaud,S., Dardel,F., Springer,M. and Chiaruttini,C. (2002) Translational feedback regulation of the gene for L35 in Escherichia coli requires binding of ribosomal protein L20 to two sites in its leader mRNA: a possible case of ribosomal RNA–messenger RNA molecular mimicry. RNA, 8, 878–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartz D., McPheeters,D.S., Green,L. and Gold,L. (1991) Detection of Escherichia coli ribosome binding at translation initiation sites in the absence of tRNA. J. Mol. Biol., 218, 99–105. [DOI] [PubMed] [Google Scholar]
- Held W.A., Ballou,B., Mizushima,S. and Nomura,M. (1974) Assembly mapping of 30S ribosomal proteins from Escherichia coli. Further studies. J. Biol. Chem., 249, 3103–3111. [PubMed] [Google Scholar]
- Jaeger J.A., Turner,D.H. and Zuker,M. (1990) Predicting optimal and suboptimal secondary structure for RNA. Methods Enzymol., 183, 281–306. [DOI] [PubMed] [Google Scholar]
- Kohrer C., Mayer,C., Neumair,O., Grobner,P. and Piendl,W. (1998) Interaction of ribosomal L1 proteins from mesophilic and thermophilic Archaea and Bacteria with specific L1-binding sites on 23S rRNA and mRNA. Eur. J. Biochem., 256, 97–105. [DOI] [PubMed] [Google Scholar]
- Kraft A., Lutz,C., Lingenhel,A., Grobner,P. and Piendl,W. (1999) Control of ribosomal protein L1 synthesis in mesophilic and thermophilic archaea. Genetics, 152, 1363–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Vilardell,J. and Warner,J.R. (1996) An RNA structure involved in feedback regulation of splicing and of translation is critical for biological fitness. Proc. Natl Acad. Sci. USA, 93, 1596–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.Y. and Riggs,A.D. (1972) Lac repressor binding to non-operator DNA: detailed studies and a comparison of equilibrium and rate competition methods. J. Mol. Biol., 72, 671–690. [DOI] [PubMed] [Google Scholar]
- Maseda H. and Hoshino,T. (1995) Screening and analysis of DNA fragments that show promoter activities in Thermus thermophilus. FEMS Microbiol. Lett., 128, 127–134. [DOI] [PubMed] [Google Scholar]
- Nikulin A. et al. (2000) Crystal structure of the S15–rRNA complex. Nat. Struct. Biol., 7, 273–277. [DOI] [PubMed] [Google Scholar]
- Nomura M., Yates,J.L., Dean,D. and Post,L.E. (1980) Feedback regulation of ribosomal protein gene expression in Escherichia coli: structural homology of ribosomal RNA and ribosomal protein mRNA. Proc. Natl Acad. Sci. USA, 77, 7084–7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D.J. (1999) Adaptive recognition in RNA complexes with peptide and protein molecules. Curr. Opin. Struct. Biol., 9, 74–87. [DOI] [PubMed] [Google Scholar]
- Philippe C., Portier,C., Mougel,M., Grunberg-Manago,M., Ebel,J.P., Ehresmann,B. and Ehresmann,C. (1990) Target site of Escherichia coli ribosomal protein S15 on its messenger RNA. Conformation and interaction with the protein. J. Mol. Biol., 211, 415–426. [DOI] [PubMed] [Google Scholar]
- Philippe C., Eyermann,F., Bénard,L., Portier,C., Ehresmann,B. and Ehresmann,C. (1993) Ribosomal protein S15 from Escherichia coli modulates its own translation by trapping the ribosome on the mRNA initiation loading site. Proc. Natl Acad. Sci. USA, 90, 4394–4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe C., Bénard,L., Portier,C., Westhof,E., Ehresmann,B. and Ehresmann,C. (1995) Molecular dissection of the pseudoknot governing the translational regulation of Escherichia coli ribosomal protein S15. Nucleic Acids Res., 23, 18–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portier C. and Regnier,P. (1984) Expression of the rpsO and pnp genes: structural analysis of a DNA fragment carrying their control regions. Nucleic Acids Res., 12, 6091–6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers T. and Noller,H.F. (1995) Hydroxyl radical footprinting of ribosomal proteins on 16S rRNA. RNA, 1, 194–209. [PMC free article] [PubMed] [Google Scholar]
- Riggs A.D., Bourgeois,S. and Cohn,M. (1970) The lac repressor– operator interaction. 3. Kinetic studies. J. Mol. Biol., 53, 401–417. [DOI] [PubMed] [Google Scholar]
- Schlax P.J., Xavier,K.A., Gluick,T.C. and Draper,D.E. (2001) Translational repression of the Escherichia coli α operon mRNA: importance of an mRNA conformational switch and a ternary entrapment complex. J. Biol. Chem., 276, 38494–38501. [DOI] [PubMed] [Google Scholar]
- Schluenzen F. et al. (2000) Structure of functionally activated small ribosomal subunit at 3.3 Ångstroms resolution. Cell, 102, 615–623. [DOI] [PubMed] [Google Scholar]
- Scott L.G. and Williamson,J.R. (2001) Interaction of the Bacillus stearothermophilus ribosomal protein S15 with its 5′-translational operator mRNA. J. Mol. Biol., 314, 413–422. [DOI] [PubMed] [Google Scholar]
- Serganov A.A., Masquida,B., Westhof,E., Cachia,C., Portier,C., Garber,M., Ehresmann,B. and Ehresmann,C. (1996) The 16S rRNA binding site of Thermus thermophilus ribosomal protein S15: comparison with Escherichia coli S15, minimum site and structure. RNA, 2, 1124–1138. [PMC free article] [PubMed] [Google Scholar]
- Serganov A., Rak,A., Garber,M., Reinbolt,J., Ehresmann,B., Ehresmann,C., Grunberg-Manago,M. and Portier,C. (1997) Ribosomal protein S15 from Thermus thermophilus. Cloning, sequencing, overexpression of the gene and RNA-binding properties of the protein. Eur. J. Biochem., 246, 291–300. [DOI] [PubMed] [Google Scholar]
- Serganov A., Bénard,L., Portier,C., Ennifar,E., Garber,M., Ehresmann,B. and Ehresmann,C. (2001) Role of conserved nucleotides in building the 16S rRNA binding site for ribosomal protein S15. J. Mol. Biol., 305, 785–803. [DOI] [PubMed] [Google Scholar]
- Serganov A., Ennifar,E., Portier,C., Ehresmann,B. and Ehresmann,C. (2002) Do mRNA and rRNA binding sites of E.coli ribosomal protein S15 share common structural determinants? J. Mol. Biol., 320, 963–978. [DOI] [PubMed] [Google Scholar]
- Stelzl U., Spahn,C.M. and Nierhaus,K.H. (2000) Selecting rRNA binding sites for the ribosomal proteins L4 and L6 from randomly fragmented rRNA: application of a method called SERF. Proc. Natl Acad. Sci. USA, 97, 4597–4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Larios A. et al. (2002) Structural basis of translational control by Escherichia coli threonyl tRNA synthetase. Nat. Struct. Biol., 9, 343–347. [DOI] [PubMed] [Google Scholar]
- Vartikar J.V. and Draper,D.E. (1989) S4–16 S ribosomal RNA complex. Binding constant measurements and specific recognition of a 460-nucleotide region. J. Mol. Biol., 209, 221–234. [DOI] [PubMed] [Google Scholar]
- Vilardell J., Yu,S.J. and Warner,J.R. (2000) Multiple functions of an evolutionarily conserved RNA binding domain. Mol. Cell, 5, 761–766. [DOI] [PubMed] [Google Scholar]
- Williamson J.R. (2000) Induced fit in RNA–protein recognition. Nat. Struct. Biol., 7, 834–837. [DOI] [PubMed] [Google Scholar]
- Wimberly B.T., Brodersen,D.E., Clemons,W.M.,Jr, Morgan-Warren, R.J., Carter,A.P., Vonrhein,C., Hartsch,T. and Ramakrishnan,V. (2000) Structure of the 30S ribosomal subunit. Nature, 407, 327–339. [DOI] [PubMed] [Google Scholar]
- Zengel J.M. and Lindahl,L. (1994) Diverse mechanisms for regulating ribosomal protein synthesis in Escherichia coli. Prog. Nucleic Acid Res. Mol. Biol., 47, 331–370. [DOI] [PubMed] [Google Scholar]