Abstract
The eukaryotic translation initiation factor eIF4GI binds several proteins and acts as a scaffold to promote preinitiation complex formation on the mRNA molecule (48S). Following mRNA attachment this complex scans along the messenger in a 5′ to 3′ direction until it locates and recognizes the initiation start codon. By using a combination of retroviral and picornaviral proteases (HIV-2 and L respectively) in the reticulocyte lysate system, we have characterized a 40 amino acid (aa) region of eIF4GI (aa 642–681) that exhibits general RNA-binding properties. Removal of this domain by proteolytic processing followed by translational assays showed virtually no inhibition of internal ribosome entry on the encephalomyocarditis virus, but resulted in drastic impairment of ribosome scanning as demonstrated by studying poliovirus and foot-and-mouth disease virus translation. Based on these findings, we propose that this 40 aa motif of eIF4GI is critical for ribosome scanning.
Keywords: cap-independent/eIF4G/HIV protease/IRES/translation
Introduction
Translational control is a major contributor to the regulation of gene expression in eukaryotes, the initiation step being the primary determinant in controlling the rate of protein synthesis (reviewed in Preiss and Hentze, 1999). The initiation of translation in eukaryotes is a complex process involving many different eukaryotic initiation factors (eIFs) (Merrick, 1992; Gray and Wickens, 1998; Gingras et al., 1999b; Preiss and Hentze, 1999; Pestova et al., 2001). Among them, eIF4GI plays a central role in the assembly of the preinitiation complex, acting as a scaffold protein that interacts with several other initiation factors (Hentze, 1997; Morley et al., 1997; Dever, 1999). The N-terminal one-third of eIF4GI has the binding site for eIF4E, the cap-binding protein (Lamphear et al., 1995; Gingras et al., 1999a), and also interacts with the poly(A)-binding protein (PABP) (Imataka et al., 1998). The C-terminal region of eIF4GI binds the RNA helicase eIF4A (Lamphear et al., 1995; Morino et al., 2000; Korneeva et al., 2001) and also interacts with the MAPK-signal integrating kinase-1 (Mnk-1), a specific eIF4E kinase (Pyronnet et al., 1999). The central region of eIF4GI, described as the most important for translation initiation (Morino et al., 2000), contains another eIF4A-binding site (Imataka and Sonenberg, 1997), an eIF3-binding site (Lamphear et al., 1995; Korneeva et al., 2000) and one or several RNA-binding domains (Pestova et al., 1996; Lomakin et al., 2000). Thus, eIF4GI, eIF4E and eIF4A assemble the eIF4F complex (Gingras et al., 1999b), which interacts with the 40S ribosomal subunit via eIF3 (Trachsel et al., 1977) to form the 43S complex (also containing eIF5, eIF2, GTP and Met-tRNAi; Asano et al., 2000). The 43S complex then binds the mRNA molecule at the 5′ capped extremity to become the 48S complex and moves along in a 5′ to 3′ direction (scanning process) until it encounters the initiation codon.
Eukaryotic cells can use an alternative mechanism of translation initiation that was first reported for members of the picornavirus family such as poliovirus (PV), encephalomyocarditis virus (EMCV) and foot-and-mouth disease virus (FMDV). This mechanism, called internal initiation, is dependent upon an RNA structure named internal ribosome entry segment (IRES) that allows translation by direct ribosome binding within the 5′-untranslated region (UTR) (Jackson and Wickens, 1997; Sachs and Varani, 2000). Following internal entry at the AUG triplet at the 3′ end of the IRES, ribosomes initiate translation at this site for cardiovirus IRES (e.g. EMCV), while in the case of entero/rhinovirus IRES (e.g. PV) initiation takes place at the next AUG codon, which is reached by ribosome scanning from the original entry site. Initiation of translation on aphthovirus IRES occurs both at the landing site and the next downstream AUG (Jackson and Kaminski, 1995). During translation of IRES-containing mRNAs, eIF4GI directly interacts with mRNA (Pestova et al., 1996; Kolupaeva et al., 1998; Lopez de Quinto and Martinez-Salas, 2000; Saleh et al., 2001) and associates with eIF4A promoting 48S complex assembly (Lomakin et al., 2000).
Infection of cells with FMDV or PV results in a rapid inhibition of host cell protein synthesis. This inhibition has been attributed, at least in part, to the rapid cleavage of eIF4GI and the delayed cleavage of eIF4GII (Etchison et al., 1982; Gradi et al., 1998), by L (from FMDV) and 2A (from PV) picornaviral proteases. Proteolysis of eIF4GI occurs between amino acids (aa) 634–635 (L protease) and 641–642 (2A protease), which segregates the N-terminal one-third of eIF4GI, containing the eIF4E-binding site, from the C-terminal two-thirds of the molecule, thereafter named p100 (635 or 642 to 1560; Lamphear et al., 1995). Translation of capped mRNAs is strongly inhibited upon this cleavage (Etchison et al., 1982), whereas the C-terminal two-thirds of eIF4GI (p100) is able to support and even stimulate translation of IRES-driven mRNAs (Borman et al., 1997) by direct binding to the IRES (Pestova et al., 1996).
Besides IRES initiation, translation of uncapped mRNA has also been studied in vitro (Ohlmann et al., 1995; De Gregorio et al., 1998) and in vivo (Gunnery and Mathews, 1995; Thoma et al., 2001). Very little is known about this mechanism, except that it involves binding of the 43S complex at the 5′ end of the mRNA followed by linear scanning of the 5′-UTR (Gunnery et al., 1997; De Gregorio et al., 1998). The factors mediating ribosome attachment and progression of the 48S complex on the uncapped RNA have not been clearly defined. However, this process is strongly enhanced by cleavage of endogenous eIF4GI by picornaviral proteases or by addition of recombinant p100 fragment (Ohlmann et al., 1995; Borman et al., 1997; De Gregorio et al., 1998) suggesting that the C-terminal part of eIF4GI plays a role in mediating ribosome binding or ribosome scanning or both.
In this report, we use the protease (PR) from HIV-2 and show that, like HIV-1 PR, it can process eIF4GI into two C-termini fragments due to recognition of two cleavage sites. The cleavage site yielding the larger fragment (named Ch-1 thereafter) is located 47 aa downstream of the L proteolytic site; this fragment is further cleaved by addition of higher doses of protease. Thus, we have used the HIV-2 PR as a tool to investigate the role of these fragments in the translation of capped, uncapped and IRES-driven mRNAs. Our results show that, while capped and uncapped mRNAs translation was severely inhibited by HIV-2 PR-mediated cleavage of eIF4GI, translation driven by the EMCV IRES was marginally affected. By using UV-crosslinking assays we were able to show that a 40 aa region which is present on p100 but absent on the Ch-1 fragment binds RNA. Moreover, in vitro translation with RNA constructs driven by IRESes from picornaviral origin revealed that this eIF4GI RNA-binding domain is critical in the progression of the 48S complex to the AUG codon. Taken together, these results suggest that eIF4GI is not only involved in 43S complex formation on the mRNA but has a critical role in ribosome scanning.
Results
Recombinant HIV-2 PR cleaves eIF4GI from rabbit reticulocyte lysate
We have previously shown that the HIV-1 PR was able to cleave eIF4GI resulting in an inhibition of translation in the rabbit reticulocyte lysate (RRL; Ohlmann et al., 2002). Thus, we have investigated the cleavage of eIF4GI by HIV-2 PR. Incubation of RRL with recombinant HIV-1 PR or HIV-2 PR showed that eIF4GI was cleaved by the two enzymes, in a dose-dependent manner (Figure 1). In both cases, eIF4GI was fully processed with 2.5 ng/µl retroviral protease (lanes 3 and 8), leading to the appearance of two fragments recognized by C-terminus-directed antibodies (epitope E, see Figure 2B). These fragments migrate at ∼100 kDa (fragment named Ch-1 for C-terminal HIV PR-resulting fragment 1) and 55 kDa (Ch-2) on SDS–PAGE, suggesting that HIV-1 PR and HIV-2 PR share the same cleavage sites on the C-terminal region of eIF4GI. Interestingly, the amount of Ch-1 generated upon cleavage of eIF4GI was much higher with the HIV-2 PR (compare lanes 2–5 with lanes 7–9), with Ch-2 being detected only with a high amount of enzyme added (compare lanes 6 and 9). This could be due to a weaker activity of the HIV-2 recombinant protease, but complete processing of eIF4GI and the fact that the Ch-1 fragment is still present at high doses of HIV-2 PR seem to rule out this possibility.
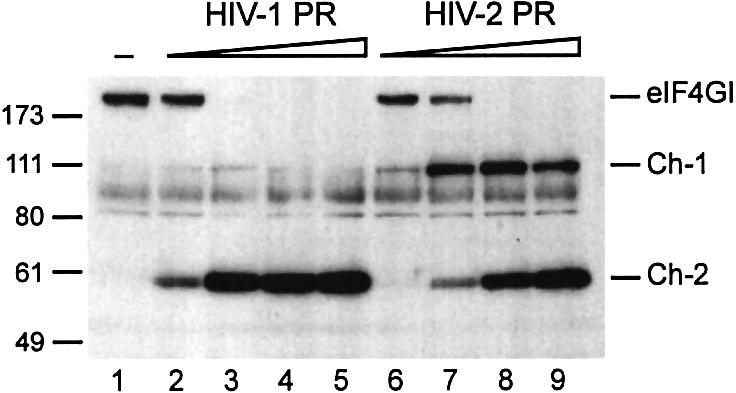
Fig. 1. eIF4GI is cleaved by recombinant HIV-1 and HIV-2 proteases. RRL (10 µl) was incubated with buffer (lane 1) or increasing amounts of recombinant HIV-1 PR (lanes 2–5: 0.25, 1.25, 5 and 12.5 ng/µl) or HIV-2 PR (lanes 6–9: 0.5, 2.5, 10 and 25 ng/µl) for 1 h at 30°C in a final volume of 20 µl. A 1 µl aliquot was resolved on 10% SDS–PAGE, proteins transferred to PVDF and the membrane was incubated with antibodies specific to the C-terminal part of eIF4GI (serum E, see Figure 2B). The resulting fragments and molecular weight markers (in kDa) are indicated on the figure.
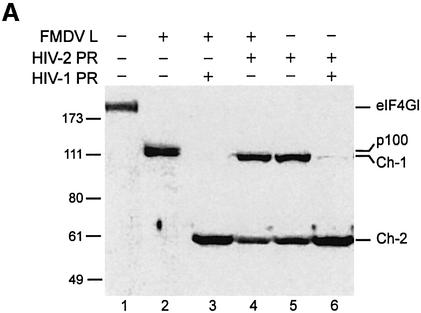
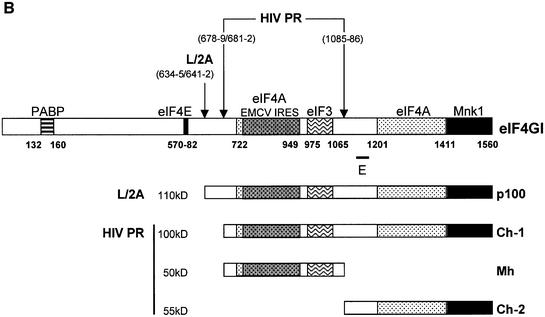
Fig. 2. Characterization of the C-terminal cleavage sites of eIF4GI. (A) RRL (10 µl) was incubated without (lane 1) or with L protease (1 µl, lanes 2–4) or HIV-2 PR (10 ng/µl, lanes 5 and 6), for 1 h at 30°C in a final volume of 20 µl. HIV-2 PR (20 ng/µl; lane 4) or 10 ng/µl HIV-1 PR (lanes 3 and 6) were then added and the mixture further incubated for 1 h. The samples were analysed by SDS–PAGE and western blotting as described in Figure 1. (B) The eIF4GI molecule is schematically represented with its different interaction domains for PABP and eIF4E (Gingras et al., 1999b), eIF4A (Lomakin et al., 2000; Morino et al., 2000), eIF3 (Korneeva et al., 2000), Mnk-1 (Morino et al., 2000) and the EMCV IRES (Lomakin et al., 2000). Cleavage sites for L/2A and HIV proteases, the obtained C-terminal fragments and the E epitope used for the western blot analysis are represented.
Mapping the HIV-2 PR cleavage sites on eIF4GI
These results suggest that HIV-2 PR could present a stronger affinity for the cleavage site yielding Ch-1 rather than Ch-2 (HIV-1 PR would have the opposite preferences). However, it is also possible that cleavage of eIF4GI by HIV-2 PR would lead to a Ch-1 fragment protected in some way from further digestion, this protection being inefficient when the cleavage is generated by HIV-1 PR. Therefore, in order to better characterize the fragments of eIF4GI generated upon cleavage by HIV-2 PR, we have used a combination of three different viral proteases in the RRL: FMDV L, HIV-1 PR and HIV-2 PR. As shown in Figure 2A, the p100 fragment obtained with L protease can be cleaved further in Ch-1 and Ch-2 by HIV proteases (compare lanes 3 and 4 with lane 2). This shows that the HIV-2 PR cleavage site is downstream of the L protease site and that the p100 fragment is not protected from further digestion by HIV proteases. It is important to note that, at this dose of HIV-1 PR, the Ch-1 species is barely detectable (lane 3). When RRL was first incubated with HIV-2 PR and then with HIV-1 PR, the Ch-1 fragment was further processed into Ch-2 (Figure 2A, compare lanes 5 and 6), indicating that the Ch-1 fragment generated by HIV-2 PR was not protected from further proteolysis by HIV-1 PR. This indicates that HIV-1 PR and HIV-2 PR present different affinities for the two cleavage sites of the eIF4GI substrate.
We and others have previously characterized the 55 kDa fragment (Ch-2) resulting from HIV-1 PR cleavage (Ventoso et al., 2001; Ohlmann et al., 2002), which corresponds to the very C-terminal domain of eIF4GI (aa 1087–1560). In addition, Ventoso and colleagues have also located two cleavage sites at positions 678–679 and 681–682 that yield the Ch-1 fragment (Ventoso et al., 2001). Thus, cleavage products generated by the HIV-2 PR were subjected to a mass spectrometry analysis. This analysis set out that the cleavage site leading to Ch-2 was located between aa 1086–1087, as already established for HIV-1 PR. Although we were unable to precisely identify the cleavage site leading to Ch-1, peptide analysis indicated that the latter was located between aa 670 and 707. Given that (i) this cleavage site is situated downstream of that of L protease, (ii) processing occurs between aa 670 and 707, (iii) cleavage of eIF4GI by HIV-1 PR and HIV-2 PR yields fragments migrating at the same size on SDS–PAGE, and (iv) HIV-1 PR and HIV-2 PR share the same cleavage site at position 1086; it is very likely that HIV-2 PR and HIV-1 PR both proteolyse eIF4GI at the same sites (aa 678–679 and 681–682) to yield Ch-1. At the highest amount of protease Ch-1 is further proteolysed into a middle fragment (Mh aa 679 or 682–1085) and Ch-2 (aa 1086–1560). It should be noted that this study was carried out with antibodies specific to the C-terminal region of eIF4GI (epitope 1139–1166) thus we could not detect the Mh fragment in western blot analysis, but we were able to visualize it by cleaving radiolabelled in vitro translated eIF4GI (data not shown).
Figure 2B shows a representation of the eIF4GI molecule with its functional domains and the positions of the different cleavage sites.
Impact of HIV-2 PR on translation in the RRL
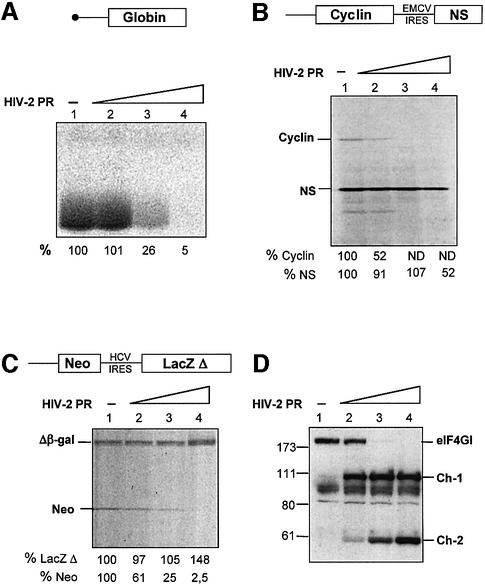
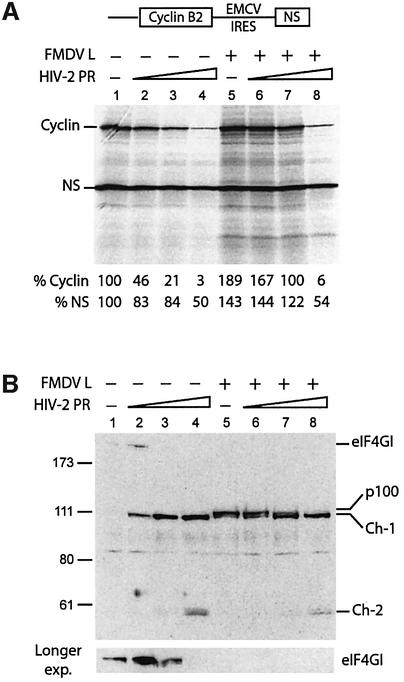
Our previous study in RRL showed that eIF4GI cleavage by the HIV-1 PR resulted in the inhibition of capped, uncapped and IRES-driven mRNA translation (Ohlmann et al., 2002). Although both HIV-1 PR and HIV-2 PR cleave eIF4GI at the same sites, the relative amount of cleavage products (Ch-1 and Ch-2) generated is rather different. Therefore, the next step was to investigate the impact of HIV-2 PR on in vitro translation. Translation was programmed with various mRNAs, including natural capped globin and an uncapped bicistronic construct coding for cyclin B2 and the NS protein of influenza driven by the EMCV IRES. As shown in Figure 3, addition of HIV-2 PR resulted in a dramatic inhibition of translation of capped (A) and uncapped (B) mRNAs, but only had a moderate effect on EMCV IRES-driven translation (B). Strikingly, translation of capped globin and uncapped cyclin B2 was almost abolished with the highest amount of protease (Figure 3A and B, lanes 4), while synthesis of NS was only partially impaired (Figure 3B, lane 4). At the lowest dose of protease used, the contrast between uncapped and IRES-driven translation was clear since uncapped cyclin translation was inhibited by 50%, whereas EMCV IRES translation was only slightly, if at all, impaired (Figure 3B, lane 2). To detect any non-specific effects due to general damage to the translation machinery, we have also employed a bicistronic mRNA construct containing the IRES of hepatitis C virus (HCV), which does not require eIF4GI for activity (Pestova et al., 1998b). Addition of HIV-2 PR strongly inhibited translation of the first cistron but did not affect HCV translation (Figure 3C), indicating no alteration of other key components of the translational apparatus at the amount of protease used. HCV-driven translation even shows a slight stimulation, which may reflect an increased availability of general translation factors after cleavage of eIF4GI by HIV-2 PR.
Fig. 3. HIV-2 PR abolishes capped and uncapped mRNAs translation, but moderately inhibits EMCV IRES-driven translation. (A, B and C) A RRL under full translation conditions (see Materials and methods) was pre-incubated without (lanes 1) or with different amounts of HIV-2 PR (lanes 2–4: 3.5, 7 and 14 ng/µl). After 1 h at 30°C, Palinavir (10 µM final concentration) was added to the reactions. Different transcripts (schematically represented on the upper part of each panel) including (A) natural capped globin mRNA (2.5 ng/µl), (B) XL–EMCV mRNA (10 ng/µl) and (C) DC–HCV mRNA (10 ng/µl) were translated under these conditions. Samples were processed on 15% SDS–PAGE, submitted to autoradiography, the relative intensities of the bands were quantified and the results are presented at the bottom of each panel. (D) Aliquots of the samples 1–4 from (A) were resolved on 10% SDS–PAGE, proteins transferred to PVDF and the membranes were incubated with antibodies specific to the C-terminal part of eIF4GI (serum E). The resulting fragments and molecular weight markers (in kDa) are indicated on the figure.
Western blot analysis was performed to visualize the impact of HIV-2 PR on eIF4GI in these translation reactions (Figure 3D). As expected, inhibition of capped globin mRNA correlated with the disappearance of intact eIF4GI (Figure 3A and D, compare lanes 2 and 3). However, uncapped cyclin translation was inhibited at low amounts of protease, when eIF4GI was only partially cleaved into the Ch-1 fragment (Figure 3B and D, compare lanes 1 and 2). This is rather surprising given the similarities between Ch-1 and the p100 fragment (see Figure 2B), which has been previously shown to stimulate translation of uncapped mRNAs (Ohlmann et al., 1995). On the contrary, inhibition of IRES-driven translation was only marginal upon cleavage of eIF4GI into Ch-1 (Figure 3B and D, compare lane 1 with 2 and 3), but was increased upon further cleavage of eIF4GI and detection of the Ch-2 species (lanes 3 and 4). These results suggest that cleavage of eIF4GI by the HIV-2 PR has different consequences on translation of capped, uncapped and IRES-driven mRNAs. While capped and uncapped translation is dramatically inhibited by processing of eIF4GI, it appears that IRES-driven initiation is not.
The inhibition of translation by HIV-2 PR is due to eIF4GI cleavage
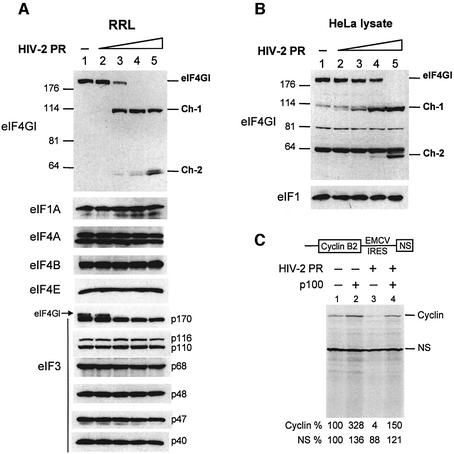
We wondered if HIV-2 PR could cleave other factors implicated in the general translation mechanism but unnecessary to HCV IRES initiation. To rule out this possibility, we incubated RRL with HIV-2 PR and then performed western blot analysis using antibodies against eIF1A, eIF3, eIF4A, eIF4B, eIF4E and the C-terminal region of eIF4GI. As shown in Figure 4A, no proteolysis could be detected with HIV-2 PR except the expected cleavage of eIF4GI at the doses of proteases we used. In this figure, it is important to note that we observed a doublet with the anti-eIF4A antibody, which may correspond to eIF4AI and eIF4II as suggested previously (Belsham et al., 2000). Moreover, as the p170 subunit of eIF3 was detected with an antibody directed against the whole eIF3 complex and eIF4GI, the cleavage of the latter can be observed on the blot. As our antibody against eIF1 recognized only the human protein, we performed a similar analysis with a lysate of HeLa cells. As shown in Figure 4B, no cleavage of eIF1 could be observed at a dose of HIV-2 PR where no intact eIF4GI can be detected (lane 5).
Fig. 4. Inhibition of translation observed with HIV-2 PR is due to cleavage of eIF4GI (A and B). (A) RRL (10 µl) or (B) HeLa lysate (127.5 µg) was incubated with buffer (lane 1) or increasing amounts of recombinant HIV-2 PR (lanes 2–5: 2.5, 5, 10 and 25 ng/µl) for 1 h at 30°C in a final volume of 20 µl. Aliquots were resolved on SDS–PAGE, proteins transferred to PVDF and the membranes were incubated with antibodies specific to eIF1, eIF1A, eIF3, eIF4A, eIF4B, eIF4E and the C-terminal part of eIF4GI, as indicated on the left side of each panel. For eIF4GI, resulting fragments and molecular weight markers (in kDa) are indicated on the figure. (C) RRL under full translation conditions was pre-incubated without (lanes 1 and 2) or with (lanes 3 and 4) 7 ng/µl HIV-2 PR. After 30 min at 30°C, Palinavir (10 µM final concentration) was added to the reactions with 1 ng/µl recombinant p100 fragment (lanes 2 and 4) or buffer (lanes 1 and 3). Uncapped XL–EMCV mRNA (10 ng/µl) was then translated under these conditions. Samples were processed on 15% SDS–PAGE, submitted to autoradiography, the relative intensities of the bands were quantified and the results are presented at the bottom of the figure.
In addition to checking the integrity of several initiation factors, we have also performed rescue experiments of uncapped and IRES-driven mRNA translation with a recombinant p100 C-terminal fragment. This fragment has been shown to support, and even stimulate, uncapped and IRES-driven translation (Ohlmann et al., 1995; Borman et al., 1997). For this, RRL was first submitted to HIV-2 PR, then the action of the enzyme was neutralized by the peptidomimetic Palinavir, a specific inhibitor of HIV proteases (Lamarre et al., 1997). Recombinant p100 fragment was added and translation of bicistronic mRNA encoding cyclin B2 (uncapped 5′-UTR-driven) and NS (EMCV IRES-driven) was monitored (Figure 4C). Whereas inhibition of translation was effective after pre-incubation with the HIV-2 PR (lane 3), addition of p100 restored translation of uncapped cyclin B2 as well as the EMCV IRES-driven cistron (lane 4). These results show that the translation inhibition of uncapped mRNA can be relieved by addition of recombinant p100 fragment, suggesting that this inhibition is due to eIF4GI processing.
‘Clipping’ of p100 to Ch-1 by HIV-2 PR leads to an inhibition of cap-independent translation
At this stage, it appears that Ch-1 and p100 exhibit very different properties on uncapped and IRES-driven mRNAs translation. This is particularly interesting as Ch-1 and p100 are very similar in sequence except for the very N-terminal end of the fragment (aa 635–681).
In an attempt to understand the role of this region, we have examined the effects of sequential cleavage of eIF4GI by both L and HIV-2 proteases. For this, endogenous eIF4GI from the RRL was processed by L protease to yield the p100 fragment, which was then further cleaved by HIV-2 PR. Translation of a bicistronic mRNA coding for cyclin B2 (uncapped 5′-UTR-driven) and NS (EMCV IRES-driven) was performed in RRL (Figure 5A), and western blot analysis was carried out in parallel to monitor conversion of p100 into the shorter Ch-1 fragment (Figure 5B). Whereas cleavage of eIF4GI into the p100 fragment by the FMDV L protease induced activation of uncapped mRNA translation and no change or slight stimulation of IRES-driven translation (Figure 5A, lane 5), the conversion of p100 into Ch-1 by HIV-2 PR led to a dramatic reduction of uncapped cyclin mRNA translation (20-fold inhibition), with only a moderate effect on IRES-dependent translation (2-fold inhibition; Figure 5A compare lanes 5 and 8). Interestingly, levels of translation observed with the highest dose of HIV-2 PR are the same whether RRL was first incubated with L protease or not (Figure 5A, compare lanes 4 and 8), corresponding to complete processing of either eIF4GI or p100 by HIV-2 PR (Figure 5B, compare lanes 4 and 8). An interesting feature is that uncapped translation only collapsed when processing of the p100 fragment was complete (Figure 5A and B, compare lanes 7 and 8), suggesting that small amounts of p100 fragment are sufficient to promote uncapped mRNA translation.
Fig. 5. ‘Clipping’ of p100 by HIV-2 PR inhibits uncapped translation. (A) RRL under full translation conditions was pre-incubated without (lanes 1–4) or with (lanes 5–8) 0.5 µl of L protease for 20 min at 30°C. Either buffer (lanes 1 and 5) or 1.75 ng/µl (lanes 2 and 6), 3.5 ng/µl (lanes 3 and 7) and 7 ng/µl (lanes 4 and 8) of HIV-2 PR were then added to the reactions. After 1 h at 30°C, Palinavir (10 µM final concentration) was added and translation of XL–EMCV mRNA (10 ng/µl) was performed under these conditions. Samples were analysed as described in Figure 5. (B) Western blot analysis of samples 1–8 of (A) was performed as described in Figure 1. A longer exposition of the upper part of the blot, corresponding to proteins of high molecular weight including intact eIF4GI is presented at the bottom of the panel.
Interestingly, moderate inhibition of IRES-driven translation occurs only at the highest dose of HIV-2 PR used (Figure 5A, lanes 4 and 8), and this also corresponds to partial conversion of Ch-1 into Ch-2 (Figure 5B, lanes 4 and 8), raising different possible explanations for this inhibition of IRES-driven translation: (i) an intrinsic reduced ability of Ch-1 to support this type of initiation; (ii) a reduction in the amount of Ch-1 fragment; or (iii) a dominant negative effect of the Ch-2 species. Once again, comparison of the two cistrons in Figure 5A shows that, at the highest dose of HIV-2 PR, inhibition of uncapped mRNA is 7–10 times stronger than IRES-driven translation (lanes 8), confirming that p100 and Ch-1 exhibit different properties, the latter being unable to support translation of uncapped mRNAs.
Ch-1 fragment can support translation of IRESs, but not that of uncapped mRNAs
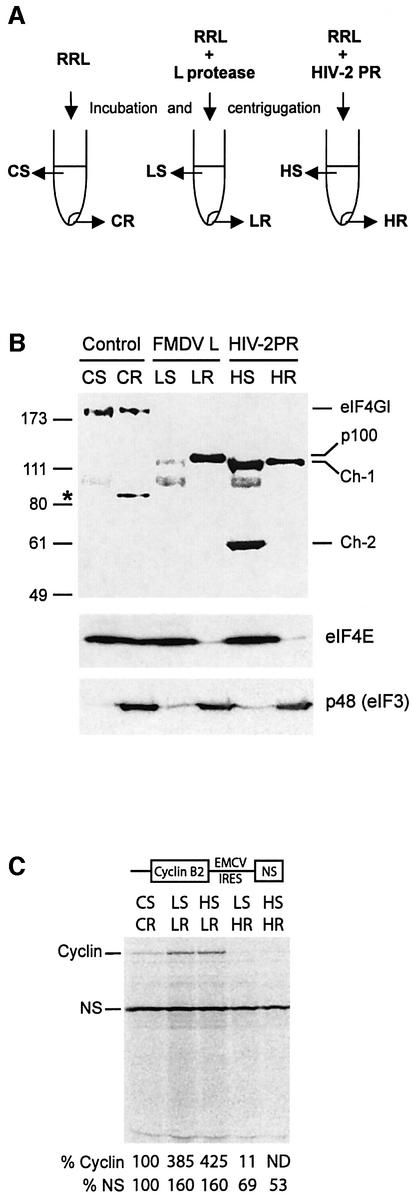
In order to investigate in more detail the biochemical properties of the eIF4GI cleavage products, we have analysed their distribution on ribosomes isolated from RRL. To this aim, lysate was incubated with FMDV L or HIV-2 proteases, and centrifuged after inhibition of the enzymatic activities. During this manipulation, the ribosomes and associated factors are pelleted, while other components remain in the supernatant. Figure 6A is a schematic representation of the supernatant (S) and ribosomal (R) fractions obtained from control RRL (CS and CR), RRL incubated with L protease (LS and LR), and RRL incubated with HIV-2 PR (HS and HR). Figure 6B shows a western blot analysis of these different fractions using antibodies against the C-terminal region of eIF4GI (E epitope, see Figure 2B), the eIF4E protein and the p48 subunit of eIF3. As shown previously (Rau et al., 1996), the p100 fragment resulting from cleavage of eIF4GI by FMDV L protease was almost entirely found associated with the ribosomes, with very little remaining in the supernatant (Figure 6B, top, LS and LR). In contrast, Ch-1 was distributed in both the ribosomal and supernatant fractions (lanes HS and HR), like intact eIF4GI (lanes CS and CR). It should be noted that we reproducibly observed that material from the ribosomal fraction appears to run slightly slower than samples from the S100 fraction (compare CS with CR, LS with LR, and HS with HR); the reason for this remains unknown. As expected, Ch-2 was exclusively found in the supernatant fraction (Figure 6B, lane HS), confirming the lack of an eIF3-binding site on this fragment (see Figure 2B). The eIF3 p48 subunit remained associated with the ribosomes (Figure 6B, bottom) indicating no change in the ribosome-binding ability of eIF3 following eIF4GI cleavage by L or HIV-2 proteases. As expected, eIF4E was released from the ribosomal fraction upon addition of L or HIV-2 proteases (Figure 6B, middle), due to the cleavage of the eIF4GI molecule between its eIF4E- and eIF3-binding sites.
Fig. 6. Ch-1 fragment can support IRES-mediated, but not uncapped mRNA translation. (A) Schematic representation of RRL incubation and fractionation. (B) RRL (200 µl) was incubated alone, with 15 µl of in vitro translated L protease or with 1.4 µg of HIV-2 PR. After 1 h at 30°C, L and HIV-2 proteases were inhibited by 0.5 mM E-64 or 10 µM Palinavir, respectively, and lysates were ultracentrifuged [see Materials and methods and (A)]. Aliquots of lysate corresponding to 2 µl of parent RRL were resolved on a 10% SDS–PAGE, and western blot analysis was performed with antibodies specific to the C-terminal part of eIF4GI (upper), eIF4E (middle) and p48 protein of eIF3 (bottom). The resulting proteins or fragments observed, and molecular weight markers (in kDa) are indicated on the figure. The asterisk denotes a non-specific reaction with the eIF4GI antibody. CS, supernatant control; CR, ribosomes control; LS, L-treated supernatant; LR, L-treated ribosomes; HS, HIV-2-treated supernatant; HR, HIV-2-treated ribosomes. (C) Different reconstituted lysates were obtained by combining 1 µl of ribosomes and 5 µl of supernatants described in (A). XL–EMCV mRNA (10 ng/µl) was then translated under these conditions. Samples were analysed as described in Figure 3.
The experiment described in Figure 6A and B provided us with an interesting tool to manipulate levels of Ch-1 and Ch-2 fragments and to investigate their respective role in translation. For this, ribosomal and supernatant fractions were utilized to make a reconstituted lysate as described previously (Rau et al., 1998). This allows four different combinations: L-treated supernatant (LS) with L or HIV-2-treated ribosomes (LR or HR), and HIV-2-treated supernatant (HS) with L or HIV-2-treated ribosomes.
The XL-EMCV uncapped bicistronic mRNA was translated in the reconstituted lysates (Figure 6C). As expected, reconstitution of the L protease-treated lysate (LSLR), resulted in translation enhancement of both cistrons, while reconstitution of HIV-2 PR-treated-lysate (HSHR) showed drastic inhibition of uncapped cyclin and slight impairment of EMCV-driven translation (Figure 6C, HSHR). Translation using HSLR lysate, in which p100 is mixed to Ch-1 and Ch-2, does not show any change compared with LSLR, indicating that the p100 present on the ribosomes is fully active on both uncapped and IRES-driven cistrons even in the presence of Ch-1 and Ch-2 in the post-ribosomal supernatant. This result allows us to rule out any dominant negative effect of Ch-2 as an explanation for the small inhibition of IRES translation observed with high amounts of HIV-2 PR. Interestingly, use of LSHR lysate, in which Ch-1 was the only C-terminal fragment of eIF4GI available, showed marginal reduction of NS translation (69%), confirming that the Ch-1 fragment is able to support IRES-driven translation, albeit to a lower extent than p100. In contrast, translation of uncapped cyclin B2 cistron was dramatically affected (to 11% of control conditions), confirming the inability of Ch-1 to maintain translation of uncapped mRNA. It should be noted that the slight difference observed for the two cistrons between the HSHR and the LSHR lysates may be attributed to the presence of a very low amount of p100 in the L supernatant (see Figure 6B, lane LS).
These results confirm that the Ch-1 fragment of eIF4GI is capable of maintaining IRES-driven translation under conditions where uncapped translation is almost abolished.
The 40 aa region of eIF4GI that is present on p100 but absent on Ch-1 exhibits general RNA-binding properties
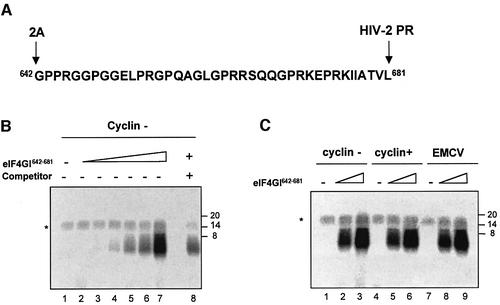
In a recent report, Lomakin and colleagues (Lomakin et al., 2000) have investigated the binding properties of a domain of eIF4GI spanning aa 643–696 and they found that the latter could bind β-globin RNA relatively strongly. Therefore, it is likely that the region of eIF4GI present on p100 and missing in Ch-1 may be involved in RNA binding. To test this, a 40 aa peptide corresponding to the region between the 2A and the HIV-2 PR cleavage site (aa 642–681) was chemically synthesized and purified (Figure 7A; thereafter named eIF4GI642–681) to perform UV-crosslinking assays. 32P-radiolabelled in vitro transcript corresponding to the first 172 nucleotides (nt) of cyclin B2 RNA was incubated with increasing amounts of eIF4GI642–681 and subjected to UV irradiation. Following RNase A treatment, samples were run on SDS–PAGE and subjected to autoradiography. As shown in Figure 7B, this resulted in efficient crosslinking of the peptide to the RNA (lanes 2–7) in a dose-dependent manner. Addition of unlabelled luciferase RNA (Promega) in a 35× molar excess competed for binding the peptide (compare lanes 7 and 8) suggesting that eIF4GI642–681 exhibits similar affinities for these two RNAs.
Fig. 7. eIF4GI642–681 can be crosslinked to various mRNAs. (A) Sequence of the 40 aa peptide (eIF4GI642–681), the 2A and HIV-2 cleavage sites are indicated on the figure. (B) Increasing amounts of chemically synthesized eIF4GI642–681 (lane 1, 0 µg; lane 2, 0.015 µg; lane 3, 0.030 µg; lane 4, 0.06 µg; lane 5, 0.12 µg; lane 6, 0.24 µg; lanes 7 and 8, 0.48 µg) was used in crosslinking assays with 32P-labelled uncapped cyclin probe (0.2 pmol; 75 000 c.p.m.) in the absence (lanes 1–7) or presence of unlabelled luciferase competitor (lane 8, 7 pmol). Following RNase A treatment, proteins were separated on 20% SDS–PAGE and the dried gel was exposed to autoradiography. The asterisk denotes a non-specific band which probably results from incomplete digestion of the radiolabelled transcript. (C) Crosslinking assays were performed in the absence (lanes 1, 4 and 7) or presence of chemically synthesized eIF4GI642–681 (lanes 2, 5 and 8, 0.25 µg; lanes 3, 6 and 9, 0.75 µg) with various 32P-labelled probes (75 000 c.p.m.): uncapped cyclin (lanes 1–3), capped cyclin (lanes 4–6) and uncapped EMCV (lanes 7–9). Following RNase A treatment, proteins were separated on 20% SDS–PAGE and the dried gel was subjected to autoradiography. Position of molecular weight markers are indicated on the right-hand side of the figure in kDa.
Other radiolabelled RNA probes including capped cyclin (172 nt) and the EMCV IRES (nt 270–836) were also used. As shown on Figure 7C, incubation of these RNAs with increasing concentrations of eIF4GI642–681 peptide also resulted in the formation of an RNA–peptide complex (uncapped cyclin, lanes 2 and 3; capped cyclin, lanes 5 and 6; EMCV, lanes 8 and 9).
These results extend previous findings (Lomakin et al., 2000), and show that eIF4GI642–681 is able to bind to different RNAs (capped, uncapped and IRES driven) with similar efficiency. As none of these constructs share a common RNA motif (data not shown), it is likely that eIF4GI642–681 exhibits general RNA-binding activity.
eIF4GI642–681 is critical for ribosomal scanning
EMCV IRES-driven initiation occurs by direct binding of eIF4GI (more specifically the region spanning from aa 746 to 969) and eIF4A upstream to the initiation codon. Following ribosomal attachment, initiation takes place to the AUG triplet and this does not involve ribosomal scanning (Pestova et al., 2001). Our results show that cleavage of eIF4GI does not inhibit significantly EMCV-driven translation, suggesting that Ch-1 can mediate ribosomal entry on this RNA, and imply that the RNA-binding activity of eIF4GI642–681 would not be involved in EMCV IRES translation. This sharply contrasts with data obtained with uncapped RNA transcripts whereby removal of this region abrogates translation initiation suggesting its involvement in ribosomal entry or scanning or both. Based on these observations, we then went on to look for a function of eIF4GI642–681 in ribosomal scanning.
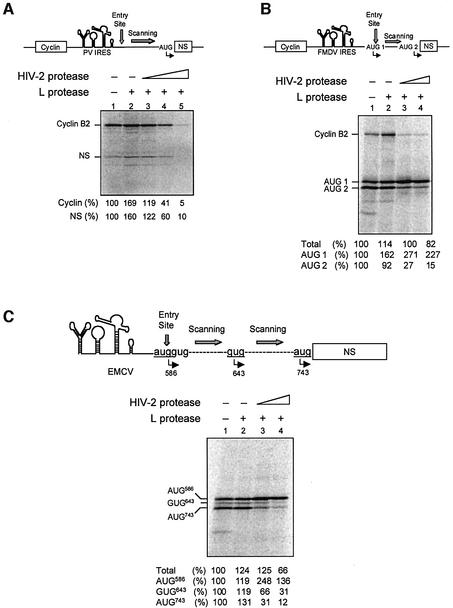
To address this, we first used the PV IRES in which the initiation codon is reached by scanning of the initiation complex from the entry site, 157 nt above. RRL was first pre-incubated with L protease in order to process endogenous eIF4GI into p100 and then further incubated with HIV-2 PR to convert p100 into Ch-1. Translation assays were programmed with bicistronic XL-PV under different conditions: no protease (Figure 8A, lane 1); L protease alone (lane 2); and L protease followed by HIV-2 PR (lanes 3–5). As expected, translation of both cistrons was stimulated by L protease treatment (lane 2), and translation of the first uncapped cistron was dramatically inhibited upon addition of HIV-2 PR. In sharp contrast with data obtained with the EMCV IRES, we also observed a drastic inhibition of translation of the second cistron (NS) driven by the PV IRES (Figure 8A, lane 5), showing that aa 635–681 of eIF4GI are essential to perform translation initiation with this IRES. As PV, but not EMCV IRES-driven translation, involves ribosomal scanning we have hypothesized that this region of eIF4GI could be required in the scanning process.
Fig. 8. Conversion of p100 into Ch-1 inhibits ribosomal scanning. RRL under full translation conditions was pre-incubated without (lanes 1) or with (lanes 2–5) 0.5 µl of L protease for 20 min at 30°C. Either buffer (lanes 1 and 2) or 3.5 ng/µl (lanes 3), 7 ng/µl (lanes 4) or 14 ng/µl (A, lane 5) of HIV-2 PR was added to the reaction. After 1 h at 30°C, Palinavir (10 µM final) was added and translation was programmed with (A) bicistronic XL–PV mRNA (10 ng/µl), (B) bicistronic XL–FMDV mRNA (10 ng/µl) or (C) monocistronic EMCV–PV (10 ng/µl). Samples were processed on SDS–PAGE, submitted to autoradiography and the relative intensities of the bands were quantified and the results are presented at the bottom of each panel. They are expressed as a percentage of total initiation events (Total), or as the relative translation efficiency for each individual initiation codon as indicated on the figure. A schematic diagram of the constructs used and their initiation sites are represented on top of each panel.
Therefore, we have used the FMDV IRES in which initiation occurs both at the first AUG (AUG 1) triplet at the 3′ landing site of the IRES but also at the next AUG (AUG 2) some 84 nt downstream (Belsham, 1992). The same experiment as in Figure 8A was repeated with bicistronic XL-FMDV (Figure 8B). In agreement with results obtained previously, the overall translation efficiency driven by the FMDV IRES was only slightly impaired by HIV-2 PR treatment (lane 4). However, this showed a rather surprising pattern as initiation at the downstream AUG, which is thought to be reached by linear scanning, was almost abolished upon treatment of the lysate with HIV-2 PR while initiation at the upstream AUG was enhanced under the same conditions (lanes 3 and 4). As cleavage of eIF4GI by the L protease alone (lane 2) did not alter the relative utilization of the two AUGs, this result suggests that removal of aa 635–681 from p100 disrupts ribosomal scanning.
To confirm this, a similar experiment was carried out with a chimeric construct (pEMCV-PV; see Figure 8C) harbouring the EMCV IRES (up to the oligopyrimidine tract) followed by the downstream sequences of the PV type 1 in which AUG586 was put in frame with AUG743 (numbering of the AUGs corresponds to the consensus genomic sequence of PV type 1). On such a construct, the body of the EMCV IRES promotes ribosomal entry some 25 nt downstream of the polypyrimidine-rich tract. However, the authentic initiation site found on the wild-type EMCV has been replaced by the AUG triplet found on PV type 1, which lies in a poor Kozak context (Kozak, 1989). As a result, some ribosomes bypass AUG586 and scan the region downstream to initiate either at GUG643 or AUG743.
EMCV-PV RNA was translated in the reticulocyte lysate under the conditions used for XL-FMDV; when no protease was added, three isoforms of the NS proteins were detected corresponding to initiation at AUG586, GUG643 and AUG743. It should be noted that, at the Mg2+ and K+ concentrations used, initiation events occurred at AUG586 and AUG743 with similar frequency. Pre-treatment of the lysate with L protease did not disrupt the relative utilization of the start codons (Figure 8C, lane 2) as described previously (Ohlmann and Jackson, 1999). Once more, addition of HIV-2 PR did not impair significantly overall translation efficiency (lane 4) but resulted in a drastic inhibition of translation arising from both GUG643 and AUG743 together with an increase in initiation at the proximal start site (lanes 3 and 4). Taken together, these results show that removal of eIF4GI635–681 does not affect, and even stimulates, initiation at the initial binding site but drastically inhibits ribosome scanning on the mRNA.
Discussion
In this work, we first showed that recombinant HIV-2 PR was able to cleave eIF4GI into two distinct C-terminal fragments named Ch-1 and Ch-2 (Figure 1). The initial cleavage site, generating Ch-1, is situated a few amino acids downstream of the picornaviral L and 2A proteases cleavage sites (Lamphear et al., 1995; see Figure 2B), at aa 678–679 and 681–682. The second cleavage site is located between aa 1086 and 1087 and further processes Ch-1 into Mh and Ch-2 (Figure 2A), corresponding to the central region and the very C-terminal part of eIF4GI, respectively (Figure 2B). Although HIV-1 PR and HIV-2 PR cleave eIF4GI at the same sites, HIV-2 PR generates a higher amount of Ch-1, suggesting a reduced preference of this enzyme for the cleavage site leading to Ch-2 (Figure 1). It should be noted that, as previously described for the HIV-1 PR (Ohlmann et al., 2002), eIF4GII was not substrate for the HIV-2 PR (data not shown).
Thus, by cleaving endogenous eIF4GI present in the RRL, HIV-2 PR provided us with an interesting tool to study the role of these regions of eIF4GI in translation initiation. One major advantage of using viral proteases is that it allows work with endogenous levels of eIF4GI rather than adding exogeneously expressed recombinant fragments.
Our data show that cap-dependent translation in the RRL was dramatically inhibited by HIV-2 PR (Figure 3A) due to loss of the N-terminal domain of eIF4GI containing the eIF4E-binding site. Surprisingly, translation of uncapped mRNAs was also inhibited upon cleavage of eIF4GI at a low dose of HIV-2 PR, and abolished using higher amounts of protease (Figure 3B). This was rather unexpected because Ch-1 (aa 682–1560) is very similar to the p100 fragment generated upon cleavage by L/2A proteases (aa 635/642–1560), which is able to promote and stimulate translation of uncapped mRNAs (Ohlmann et al., 1996; Borman et al., 1997; De Gregorio et al., 1998). No other initiation factor seems to be proteolysed at the doses of HIV-2 PR we used (Figure 4A), and translation of uncapped mRNA can be rescued by addition of recombinant p100 (Figure 4B), suggesting that the translation inhibition observed with HIV-2 PR is due to eIF4GI cleavage. In order to investigate in more detail the functional differences between p100 and Ch-1 fragment, we have performed the ‘clipping’ of p100 fragment into Ch-1 (Figure 5). As a result, translation of uncapped mRNA was drastically inhibited by the removal of aa 635–681 from p100.
In sharp contrast to uncapped mRNA, EMCV IRES-driven translation was moderately affected by addition of a low dose of HIV-2 PR, the level of inhibition varying between 30–50% (Figures 3–6). Thus, two additive mechanisms can be proposed to explain this slight inhibition. First, an intrinsic decreased ability of Ch-1 to promote initiation of IRES-driven translation supported by the fact that, unlike p100, Ch-1 is not completely associated with the ribosomal fraction (Figure 6B). Also, experiments performed with reconstituted lysate showed that Ch-1 alone promoted EMCV IRES-driven translation at a lower efficiency than the control level (Figure 6C). The second mechanism is that the slight decrease in EMCV IRES-driven translation could be the result of further processing of Ch-1 into Ch-2 and Mh fragments (Figure 3D). In that case, the Mh fragment (aa 682–1085) would exhibit reduced ability to support IRES-driven translation. This is in agreement with previous work showing that while the central region of eIF4GI (aa 613–1092) was sufficient to perform binding to the EMCV IRES (Pestova et al., 1996; Imataka and Sonenberg, 1997; Lomakin et al., 2000), this binding was less efficient than that of p100 under the same conditions, suggesting that the second binding site for eIF4A (situated in the C-terminal region of eIF4GI) plays an important modulatory role in this process. Taken together, these data show that Ch-1 retains the main part of the activity of intact eIF4GI on IRES-driven translation (Figure 6C) with further cleavage of Ch-1 into Mh resulting in additional loss of activity (Figure 3D).
As p100 and Ch-1 are identical except for the first 47 (in the case of L protease cleavage site) or 40 aa (in the case of 2A protease site) this prompted us to investigate the biochemical properties of this region. A peptide (named thereafter eIF4GI642–681) corresponding to the 40 aa located between the 2A and HIV-2 cleavage site was chemically synthesized and used in UV-crosslinking assays. Results show that eIF4GI642–681 was efficiently crosslinked to various RNA probes including capped, uncapped and IRES-containing RNAs (Figure 7). This suggests that eIF4GI642–681 corresponds to a domain with RNA-binding properties. This region is rich in arginine residues that may be involved in peptide–RNA interaction. Interestingly, the initiation factor eIF4B also contains an arginine motif located in the C-terminus region that also binds RNA in a non-specific manner (Methot et al., 1994).
As the results obtained so far suggest a role for eIF4GI642–681 in RNA binding, we have looked for a role for this small domain. Translation initiation can be broadly divided into two major steps: (i) attachment of the 43S complex to the mRNA and (ii) progression of the preinitiation complex on the mRNA molecule until the AUG initiation codon is decoded. The first step is mediated by a cap–eIF4E–eIF4GI interaction or alternatively by direct binding of eIF4GI onto an IRES element. Our data show that removal of eIF4GI642–681 by clipping p100 with the HIV-2 PR results in a drastic inhibition of uncapped RNA with little effect on EMCV IRES-driven translation. This suggests that Ch-1 is still able to promote internal ribosomal binding, which is consistent with a previous report showing that aa 722–969 of eIF4GI constitute the minimal domain required for EMCV translation (Lomakin et al., 2000). However, the inability of Ch-1 to promote uncapped RNA translation has remained puzzling. In fact, little is known on uncapped translation except that: (i) it is strongly stimulated by cleavage of eIF4GI by 2A/L proteases or addition of recombinant p100 (Ohlmann et al., 1995; Ziegler et al., 1995a,b; Borman et al., 1997; De Gregorio et al., 1998); (ii) ribosomal binding occurs at the 5′ uncapped end (De Gregorio et al., 1998); and (iii) following 48S formation, the complex scans along the 5′-UTR until it reaches the AUG codon (De Gregorio et al., 1998). Our data show that removal of the small RNA-binding domain (eIF4GI642–681) from p100 almost abolishes uncapped translation suggesting that this region is important for ribosomal binding or scanning or both. We first tested the hypothesis that inhibition of uncapped translation could reflect a default in ribosome scanning. Such an effect would have gone unrevealed with the sole use of the EMCV IRES RNA in which ribosomal binding takes place directly at the AUG codon (Kaminski et al., 1990).
Thus, we have studied translation of the PV IRES in which the AUG start codon is reached by the 43S complex after scanning some 157 nt from the entry site. Our results show that translation driven by the PV IRES was severely inhibited upon cleavage of eIF4GI by HIV-2 PR (Figure 8A). We also studied translation of the FMDV IRES, in which initiation occurs both at the entry site at the 3′ of the IRES (AUG 1) and at the next AUG triplet (AUG 2) some 84 nt downstream (Belsham, 1992). Initiation at the second AUG, which has been shown to be reached by the ribosomes upon scanning from the first initiation codon (Belsham, 1992), was significantly reduced after cleavage of eIF4GI by the HIV-2 PR (Figure 8B). These data were confirmed by the use of the EMCV-PV chimeric RNA described previously (Ohlmann and Jackson, 1999). On this RNA, initiation competent ribosomes are delivered to the AUG triplet at the 3′-binding site of the IRES (AUG586) as with wild-type EMCV; however, AUG586 lies in a poor surrounding context and, as a result, some entering ribosomes will scan 157 nt downstream to initiate translation at the next AUG triplet (AUG743). Data presented in Figure 8C showed that cleavage of eIF4GI by the HIV-2 PR drastically impaired initiation events at AUG743. Therefore, and in agreement with data obtained with the PV and FMDV-IRES, it strongly suggests that eIF4GI642–681 is critical for migration of the preinitiation complex along the mRNA molecule. Interestingly, both with XL-FMDV and EMCV-PV, we observed a significant increase of initiation events at the proximal AUG after addition of the HIV-2 PR. As in both constructs, and unlike XL-EMCV, the first AUG is in a suboptimal context, it may be possible that the inability of the 48S complex to perform scanning stalls the ribosome to the AUG, allowing more time for its recognition. A similar mechanism has been previously described in which introduction of a moderately stable RNA hairpin 14 nt downstream to an AUG codon lying in a suboptimal context slowed ribosomal scanning and resulted in efficient recognition of the AUG triplet (Kozak, 1990). Taken together, these results show that Ch-1 is able to promote ribosomal entry in an initiation competent manner on the FMDV and EMCV IRESs but cannot support scanning to the downstream initiation site.
One possible way to explain these data is that cleavage of eIF4GI by HIV-2 PR occurs at position 681 and could affect the helicase activity of eIF4F by disrupting the eIF4A-binding site. This would be consistent with a report mapping the upstream eIF4A-binding site on eIF4GI to aa 672–970 (Morino et al., 2000). However, in another report the eIF4A-binding site has been restricted to aa 722–949 (Lomakin et al., 2000). Moreover, the fact that translation driven by the EMCV IRES, which requires eIF4A for activity, was efficient in the HIV-2 PR-treated lysate suggests that the inhibition of ribosome scanning was not due to impairment of helicase activity (Lomakin et al., 2000).
Another hypothesis to explain an inhibition of the scanning mechanism could be an eventual degradation of initiation factors eIF1 or eIF1A upon incubation of RRL with HIV-2 PR, as these factors have been shown to be essential for scanning to occur (Pestova et al., 1998a). Our western blot analysis (Figure 4A) and the rescue experiments with p100 (Figure 4B) rule out this possibility. It may also be possible that inhibition of ribosome scanning is due to a defect in the recruitment of factors essential for scanning (yet unknown) or that the 40 aa domain is necessary to maintain the shape of the C-terminal part of eIF4GI in a suitable conformation for scanning along the mRNA.
However, we favour another model whereby the RNA-binding region of eIF4GI (eIF4GI642–681) is critical to maintain contact between the preinitiation complex and the mRNA during scanning. The fact that eIF4GI642–681 corresponds to a general RNA-binding domain is consistent with the model of scanning whereby the migrating ribosome moves along all genome-encoded cellular or viral mRNAs irrespective of their structure or primary nucleotide sequence. Such a model would extend the activity of the eukaryotic initiation factor eIF4GI beyond the initial stage of ribosomal attachment and suggests that the latter is also critical to guide the 40S subunit and associated factors through the mRNA molecule during scanning to the initiation codon.
Materials and methods
Proteases, antibodies and other reagents
Rabbit antiserum raised against the C-terminal portion of eIF4GI (aa 1139–1166), eIF4A and eIF4E were generous gifts of Dr S.J.Morley (University of Sussex, UK) as well as goat anti-serum raised against eIF4B. Rabbit antibodies against p48eIF3 and p66eIF3 were kindly donated by Dr P.Jalinot (ENS-Lyon, France). Goat antiserum against eIF3 and rabbit antiserum against eIF1A were kind gifts of Dr J.W.B.Hershey (UC Davis, CA). Rabbit antisera raised against eIF1 human protein was a kind donation from Dr M.Feitelson (Thomas Jefferson University, PA). Recombinant proteins: eIF4GI-eIF4E and p100 C-terminal fragment were a kind gift of Dr S.J.Morley.
Recombinant HIV-1 PR (a generous donation from Rhône Poulenc) and HIV-2 PR (from NIH AIDS reagent program; Rittenhouse et al., 1990) were dialysed in buffer A (20 mM MOPS pH 7.2, 25 mM KCl, 10 mM NaCl, 1.1 mM MgCl2, 0.1 mM EDTA). Preparation of in vitro translated L protease was carried out as described previously (Ohlmann et al., 1995).
The peptidomimetic Palinavir was provided by Biopharma, and E-64 was purchased from Sigma.
Plasmids
Standard procedures were used for restriction digestion, plasmid DNA construction and purification. mRNA was synthesized in vitro as described previously (Ohlmann and Jackson, 1999). The pDC-HCV was obtained from the pMLV-CB63 (Berlioz and Darlix, 1995), the pXL-FMDV, pXL-EMCV, pXL-PV and pEMCV-PV have been described previously (Ohlmann and Jackson, 1999). The cyclin B2 mRNA was generated by in vitro transcription after linearization of pXL-FMDV with BamHI. Natural capped globin mRNA was purchased from Gibco-BRL.
Synthesis of eIF4GI642–681
Forty amino acid peptide corresponding to the region of eIF4GI comprised between the 2A and HIV-2 PR cleavage sites and spanning from aa 642–681 was chemically synthesized by Eurogentec.
Isolation of ribosome pellet from RRL
RRL (200 µl) was incubated in the presence or absence of in vitro translated FMDV L (15 µl) or HIV-2 PR (7 µg/ml) for 60 min at 30°C. At the end of the incubation period, the protease inhibitor E-64 (Sigma; 500 µM final concentration) or Palinavir (10 µM final concentration) was added to the lysate and the mixture centrifuged for 60 min at 400 000 g in a Beckman TL100 centrifuge. The post-ribosomal supernatant was removed and the ribosomal pellet was washed three times in 200 µl of buffer A and resuspended in 40 µl of buffer A (one-fifth of the initial volume), frozen and kept at –80°C.
In vitro translation in the reticulocyte lysate
Capped and uncapped mRNAs, except XL-PV, were translated in nuclease-treated RRL (10 µl reactions; Promega) in the presence of 75 mM KCl, 0.5 mM MgCl2, 15 mM 2-aminopurine and 20 µM of each amino acid (except methionine). XL-PV mRNA was translated in RRL in the presence of 80 mM KCl, 5% amino acids mixture and 2.5% HeLa S10 extracts (kind gifts from Dr A.Borman). The mixture was incubated for 45 min at 30°C in the presence of 0.6 mCi/ml [35S]methionine. Translation products were then separated on 15% SDS–PAGE, the gel was dried and subjected to autoradiography for 12 h using Biomax films (Eastman Kodak Co.). The intensity of the bands was quantified using a STORM 850 PhosphorImager (Molecular Dynamics).
UV crosslinking
UV-crosslinking assays were performed by incubating [32P]UTP-labelled capped or uncapped cyclin B2 (nt 1–170) or EMCV (nt 260–834) RNA probe (75 000 c.p.m.) with different amounts of eIF4GI642–681 peptide (from 0.015 to 0.48 µg) in a final volume of 20 µl Buffer B (10 mM HEPES pH 7.9, 2 mM MgCl2, 35 mM KCl, 0.05 % NP-40). This mixture was incubated at 30°C for 15 min and submitted to irradiation at an energy level of 0.24 J/cm2 prior to digestion with RNase A during 30 min at 37°C. The RNA crosslinked proteins were resolved on a 20% SDS–polyacrylamide gels and subjected to autoradiography. Unlabelled-competitor luciferase RNA was purchased from Promega.
Protein sequencing
Baculovirus (1 µg) expressed eIF4GI–eIF4E was incubated with recombinant HIV-2 PR (47 ng/µl) for 1 h at 30°C and separated on 10% SDS–PAGE. The fragments corresponding to the 100, 55 and 50 kDa species were then isolated and subjected to trypsin digestion and mass spectrometry analysis (MS and MSMS, CEA of Grenoble, France).
Acknowledgments
Acknowledgements
The authors thank Rhône-Poulenc for donating recombinant HIV-1 PR, Dr S.J.Morley for eIF4A, eIF4B, eIF4E and eIF4GI antibodies, p100 and eIF4GI–eIF4E recombinant proteins, Dr P.Jalinot for the anti-eIF3p66 and eIF3p48 antibodies, Dr J.W.B.Hershey for the anti-eIF3 and eIF1A antibodies, Dr A.A.M.Thomas and Dr M.Feitelson for the anti-eIF1 antibodies, Dr R.J.Jackson and Caroline Gabus for plasmids, Biopharma for donating Palinavir and C.Daude for technical assistance. The following reagent was obtained through the AIDS Research and Reference Reagent Program, NIAID, NIH: HIV-2 PR from Bret Shirley and Mr Michael Cappola, Boehringer Ingelheim Pharmaceuticals, Inc. This work was supported by grants from the Agence Nationale de Recherche sur le SIDA (ANRS 2001-128) and the Association de la Recherche contre le Cancer (ARC 5466).
References
- Asano K., Clayton,J., Shalev,A. and Hinnebusch,A.G. (2000) A multifactor complex of eukaryotic initiation factors, eIF1, eIF2, eIF3, eIF5 and initiator tRNA(Met) is an important translation initiation intermediate in vivo. Genes Dev., 14, 2534–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsham G.J. (1992) Dual initiation sites of protein synthesis on foot-and-mouth disease virus RNA are selected following internal entry and scanning of ribosomes in vivo. EMBO J., 11, 1105–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsham G.J., McInerney,G.M. and Ross-Smith,N. (2000) Foot-and-mouth disease virus 3C protease induces cleavage of translation initiation factors eIF4A and eIF4G within infected cells. J. Virol., 74, 272–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlioz C. and Darlix,J.L. (1995) An internal ribosomal entry mechanism promotes translation of murine leukemia virus gag polyprotein precursors. J. Virol., 69, 2214–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borman A.M., Kirchweger,R., Ziegler,E., Rhoads,R.E., Skern,T. and Kean,K.M. (1997) elF4G and its proteolytic cleavage products: effect on initiation of protein synthesis from capped, uncapped and IRES-containing mRNAs. RNA, 3, 186–196. [PMC free article] [PubMed] [Google Scholar]
- De Gregorio E., Preiss,T. and Hentze,M.W. (1998) Translational activation of uncapped mRNAs by the central part of human eIF4G is 5′ end-dependent. RNA, 4, 828–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever T.E. (1999) Translation initiation: adept at adapting. Trends Biochem. Sci., 24, 398–403. [DOI] [PubMed] [Google Scholar]
- Etchison D., Milburn,S.C., Edery,I., Sonenberg,N. and Hershey,J.W. (1982) Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220 000-dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J. Biol. Chem., 257, 14806–14810. [PubMed] [Google Scholar]
- Gingras A.C., Gygi,S.P., Raught,B., Polakiewicz,R.D., Abraham,R.T., Hoekstra,M.F., Aebersold,R. and Sonenberg,N. (1999a) Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev., 13, 1422–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras A.C., Raught,B. and Sonenberg,N. (1999b) eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem., 68, 913–963. [DOI] [PubMed] [Google Scholar]
- Gradi A., Svitkin,Y.V., Imataka,H. and Sonenberg,N. (1998) Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc. Natl Acad. Sci. USA, 95, 11089–11094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray N.K. and Wickens,M. (1998) Control of translation initiation in animals. Annu. Rev. Cell. Dev. Biol., 14, 399–458. [DOI] [PubMed] [Google Scholar]
- Gunnery S. and Mathews,M.B. (1995) Functional mRNA can be generated by RNA polymerase III. Mol. Cell. Biol., 15, 3597–3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunnery S., Maivali,U. and Mathews,M.B. (1997) Translation of an uncapped mRNA involves scanning. J. Biol. Chem., 272, 21642–21646. [DOI] [PubMed] [Google Scholar]
- Hentze M.W. (1997) eIF4G: a multipurpose ribosome adapter? Science, 275, 500–501. [DOI] [PubMed] [Google Scholar]
- Imataka H. and Sonenberg,N. (1997) Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol. Cell. Biol., 17, 6940–6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imataka H., Gradi,A. and Sonenberg,N. (1998) A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J., 17, 7480–7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R.J. and Kaminski,A. (1995) Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA, 1, 985–1000. [PMC free article] [PubMed] [Google Scholar]
- Jackson R.J. and Wickens,M. (1997) Translational controls impinging on the 5′-untranslated region and initiation factor proteins. Curr. Opin. Genet. Dev., 7, 233–241. [DOI] [PubMed] [Google Scholar]
- Kaminski A., Howell,M.T. and Jackson,R.J. (1990) Initiation of encephalomyocarditis virus RNA translation: the authentic initiation site is not selected by a scanning mechanism. EMBO J., 9, 3753–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva V.G., Pestova,T.V., Hellen,C.U. and Shatsky,I.N. (1998) Translation eukaryotic initiation factor 4G recognizes a specific structural element within the internal ribosome entry site of encephalomyocarditis virus RNA. J. Biol. Chem., 273, 18599–18604. [DOI] [PubMed] [Google Scholar]
- Korneeva N.L., Lamphear,B.J., Hennigan,F.L. and Rhoads,R.E. (2000) Mutually cooperative binding of eukaryotic translation initiation factor (eIF) 3 and eIF4A to human eIF4G-1. J. Biol. Chem., 275, 41369–41376. [DOI] [PubMed] [Google Scholar]
- Korneeva N.L., Lamphear,B.J., Hennigan,F.L., Merrick,W.C. and Rhoads,R.E. (2001) Characterization of the two eIF4A-binding sites on human eIF4G-1. J. Biol. Chem., 276, 2872–2879. [DOI] [PubMed] [Google Scholar]
- Kozak M. (1989) The scanning model for translation: an update. J. Cell Biol., 108, 229–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. (1990) Downstream secondary structure facilitates recognition of initiator codons by eukaryotic ribosomes. Proc. Natl Acad. Sci. USA, 87, 8301–8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamarre D. et al. (1997) Antiviral properties of palinavir, a potent inhibitor of the human immunodeficiency virus type 1 protease. Antimicrob. Agents Chemother., 41, 965–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamphear B.J., Kirchweger,R., Skern,T. and Rhoads,R.E. (1995) Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J. Biol. Chem., 270, 21975–21983. [DOI] [PubMed] [Google Scholar]
- Lomakin I.B., Hellen,C.U. and Pestova,T.V. (2000) Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol. Cell. Biol., 20, 6019–6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez de Quinto S. and Martinez-Salas,E. (2000) Interaction of the eIF4G initiation factor with the aphthovirus IRES is essential for internal translation initiation in vivo. RNA, 6, 1380–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick W.C. (1992) Mechanism and regulation of eukaryotic protein synthesis. Microbiol. Rev., 56, 291–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Methot N., Pause,A., Hershey,J.W. and Sonenberg,N. (1994) The translation initiation factor eIF-4B contains an RNA-binding region that is distinct and independent from its ribonucleoprotein consensus sequence. Mol. Cell. Biol., 14, 2307–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morino S., Imataka,H., Svitkin,Y.V., Pestova,T.V. and Sonenberg,N. (2000) Eukaryotic translation initiation factor 4E (eIF4E) binding site and the middle one-third of eIF4GI constitute the core domain for cap-dependent translation and the C-terminal one-third functions as a modulatory region. Mol. Cell. Biol., 20, 468–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley S.J., Curtis,P.S. and Pain,V.M. (1997) eIF4G: translation’s mystery factor begins to yield its secrets. RNA, 3, 1085–1104. [PMC free article] [PubMed] [Google Scholar]
- Ohlmann T. and Jackson,R.J. (1999) The properties of chimeric picornavirus IRESes show that discrimination between internal translation initiation sites is influenced by the identity of the IRES and not just the context of the AUG codon. RNA, 5, 764–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlmann T., Rau,M., Morley,S.J. and Pain,V.M. (1995) Proteolytic cleavage of initiation factor eIF-4 γ in the reticulocyte lysate inhibits translation of capped mRNAs but enhances that of uncapped mRNAs. Nucleic Acids Res., 23, 334–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlmann T., Rau,M., Pain,V.M. and Morley,S.J. (1996) The C-terminal domain of eukaryotic protein synthesis initiation factor (eIF) 4G is sufficient to support cap-independent translation in the absence of eIF4E. EMBO J., 15, 1371–1382. [PMC free article] [PubMed] [Google Scholar]
- Ohlmann T., Prevot,D., Decimo,D., Roux,F., Garin,J., Morley,S.J. and Darlix,J.L. (2002) In vitro cleavage of eIF4GI but not eIF4GII by HIV-1 protease and its effects on translation in the rabbit reticulocyte lysate system. J. Mol. Biol., 318, 9–20. [DOI] [PubMed] [Google Scholar]
- Pestova T.V., Shatsky,I.N. and Hellen,C.U. (1996) Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol. Cell. Biol., 16, 6870–6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T.V., Borukhov,S.I. and Hellen,C.U. (1998a) Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature, 394, 854–859. [DOI] [PubMed] [Google Scholar]
- Pestova T.V., Shatsky,I.N., Fletcher,S.P., Jackson,R.J. and Hellen,C.U. (1998b) A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev., 12, 67–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T.V., Kolupaeva,V.G., Lomakin,I.B., Pilipenko,E.V., Shatsky,I.N., Agol,V.I. and Hellen,C.U. (2001) Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl Acad. Sci. USA, 98, 7029–7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiss T. and Hentze,M.W. (1999) From factors to mechanisms: translation and translational control in eukaryotes. Curr. Opin. Genet. Dev., 9, 515–521. [DOI] [PubMed] [Google Scholar]
- Pyronnet S., Imataka,H., Gingras,A.C., Fukunaga,R., Hunter,T. and Sonenberg,N. (1999) Human eukaryotic translation initiation factor 4G (eIF4G) recruits mnk1 to phosphorylate eIF4E. EMBO J., 18, 270–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rau M., Ohlmann,T., Morley,S.J. and Pain,V.M. (1996) A reevaluation of the cap-binding protein, eIF4E, as a rate-limiting factor for initiation of translation in reticulocyte lysate. J. Biol. Chem., 271, 8983–8990. [DOI] [PubMed] [Google Scholar]
- Rau M., Ohlmann,T., Pain,V.M. and Morley,S.J. (1998) A fractionated reticulocyte lysate system for studies on protein synthesis initiation factors. Methods Mol. Biol., 77, 211–226. [DOI] [PubMed] [Google Scholar]
- Rittenhouse J., Turon,M.C., Helfrich,R.J., Albrecht,K.S., Weigl,D., Simmer,R.L., Mordini,F., Erickson,J. and Kohlbrenner,W.E. (1990) Affinity purification of HIV-1 and HIV-2 proteases from recombinant E.coli strains using pepstatin–agarose. Biochem. Biophys. Res. Commun., 171, 60–66. [DOI] [PubMed] [Google Scholar]
- Sachs A.B. and Varani,G. (2000) Eukaryotic translation initiation: there are (at least) two sides to every story. Nat. Struct. Biol., 7, 356–361. [DOI] [PubMed] [Google Scholar]
- Saleh L., Rust,R.C., Fullkrug,R., Beck,E., Bassili,G., Ochs,K. and Niepmann,M. (2001) Functional interaction of translation initiation factor eIF4G with the foot-and-mouth disease virus internal ribosome entry site. J. Gen. Virol., 82, 757–763. [DOI] [PubMed] [Google Scholar]
- Thoma C. et al. (2001) Generation of stable mRNA fragments and translation of N-truncated proteins induced by antisense oligodeoxynucleotides. Mol. Cell, 8, 865–872. [DOI] [PubMed] [Google Scholar]
- Trachsel H., Erni,B., Schreier,M.H. and Staehelin,T. (1977) Initiation of mammalian protein synthesis. II. The assembly of the initiation complex with purified initiation factors. J. Mol. Biol., 116, 755–767. [DOI] [PubMed] [Google Scholar]
- Ventoso I., Blanco,R., Perales,C. and Carrasco,L. (2001) HIV-1 protease cleaves eukaryotic initiation factor 4G and inhibits cap-dependent translation. Proc. Natl Acad. Sci. USA, 23, 12966–12971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler E., Borman,A.M., Deliat,F.G., Liebig,H.D., Jugovic,D., Kean,K.M., Skern,T. and Kuechler,E. (1995a) Picornavirus 2A proteinase-mediated stimulation of internal initiation of translation is dependent on enzymatic activity and the cleavage products of cellular proteins. Virology, 213, 549–557. [DOI] [PubMed] [Google Scholar]
- Ziegler E., Borman,A.M., Kirchweger,R., Skern,T. and Kean,K.M. (1995b) Foot-and-mouth disease virus Lb proteinase can stimulate rhinovirus and enterovirus IRES-driven translation and cleave several proteins of cellular and viral origin. J. Virol., 69, 3465–3474. [DOI] [PMC free article] [PubMed] [Google Scholar]