Abstract
Glia cell missing (GCM) transcription factors form a small family of transcriptional regulators in metazoans. The prototypical Drosophila GCM protein directs the differentiation of neuron precursor cells into glia cells, whereas mammalian GCM proteins are involved in placenta and parathyroid development. GCM proteins share a highly conserved 150 amino acid residue region responsible for DNA binding, known as the GCM domain. Here we present the crystal structure of the GCM domain from murine GCMa bound to its octameric DNA target site at 2.85 Å resolution. The GCM domain exhibits a novel fold consisting of two domains tethered together by one of two structural Zn ions. We observe the novel use of a β-sheet in DNA recognition, whereby a five- stranded β-sheet protrudes into the major groove perpendicular to the DNA axis. The structure combined with mutational analysis of the target site and of DNA-contacting residues provides insight into DNA recognition by this new type of Zn-containing DNA-binding domain.
Keywords: DNA recognition/glia cells/transcription factor/X-ray crystal structure/Zn-binding
Introduction
GCM proteins form a small family of transcriptional regulators involved in fundamental developmental processes (Wegner and Riethmacher, 2001; Van de Bor and Giangrande, 2002). In Drosophila, where it was first identified, GCM directs the development of neuronal precursor cells into glial cells, acting as a master regulator of gliogenesis (Hosoya et al., 1995; Jones et al., 1995; Vincent et al., 1996). In contrast, neither of the two GCM homologs present in mammals appears to be involved in gliogenesis. Instead, GCMa regulates labyrinth formation in the developing placenta (Anson-Cartwright et al., 2000; Schreiber et al., 2000), while GCMb is involved in the development of the parathyroid gland (Gunther et al., 2000). Accordingly, inactivation of these genes leads to placental malfunction or parathyroid loss and hypoparathyroidism, respectively (Ding et al., 2001; Wegner and Riethmacher, 2001).
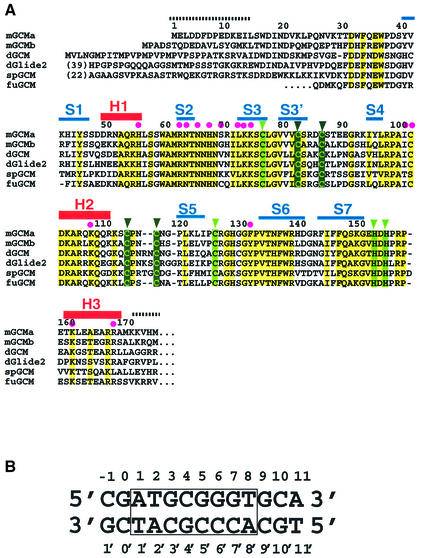
GCM homologs have also been identified in fish and sea urchins (Figure 1A), but no homologs have yet been detected in the sequenced genomes of fungi (Saccharomyces cerevisiae), plants (Arabidopsis thaliana) or nematodes (Caenorhabditis elegans).
Fig. 1. (A) Alignment of the GCM domains from mouse (mGCMa, mGCMb), Drosophila melanogaster (dGCM, dGlide2), sea urchin (spGCM) (Ransick et al., 2002) and the pufferfish fugu (fuGCM). Conserved residues and conservatively substituted residues are drawn on a yellow background. Secondary structure elements are shown above the mGCMa sequence. Regions indicated by broken lines are disordered and have not been included in the final model. Magenta dots indicate DNA-contacting residues; light green and dark green triangles indicate residues coordinating the first and second Zn ions, respectively. (B) Sequence of the 13mer DNA duplex present in crystal forms A and A′. The octameric target site is numbered from 1 to 8 (1′ to 8′ for the opposite strand) and boxed. Flanking base pairs upstream and downstream of the target site are numbered –1 to 0 and 9 to 11, respectively. (C) Stereo diagram of the final 2Fo – Fc electron density map contoured at 1.5σ. Strands S2 and S3 and the contacted DNA target site are shown. The figure was produced using the program BOBSCRIPT (Esnouf, 1999).
GCM transcription factors consist of ∼500 amino acid residues. The N-terminal moiety contains a DNA-binding domain of ∼150 residues. Sequence conservation is highest in this so-called GCM domain (Figure 1A). In contrast, the C-terminal moiety contains one or two transactivating regions and is only poorly conserved. In murine GCMb, an inhibitory region located between the two transactivating regions leads to decreased stability and lower transcriptional activity compared with other GCM transcription factors (Tuerk et al., 2000). GCM proteins bind their target sites as monomers. DNA selection experiments identified an 8 bp motif, 5′-ATGCGGGT-3′, as the optimal sequence; this is present with slight variations or in conserved form in potential target genes (Akiyama et al., 1996; Schreiber et al., 1997). As expected from their high degree of sequence similarity, the DNA-binding characteristics of different GCM homologs are very similar. Alanine mutations have identified a number of residues with critical roles in DNA recognition and stabilization of the GCM domain (Schreiber et al., 1998). Sequence conservation also indicated the importance of several conserved cysteine and histidine residues. EXAFS and microPixe analyses have demonstrated that most of these residues are involved in ligating two Zn ions required for the stability of the GCM domain (Cohen et al., 2002).
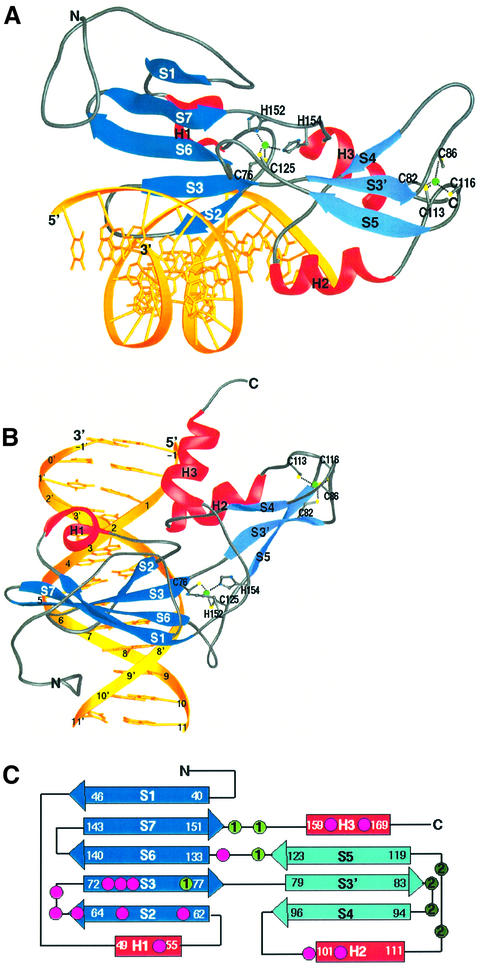
A detailed structural and functional analysis of the GCM domain has been hampered by the lack of a crystallographic structure. Here we present the crystal structure of the GCM domain of murine GCMa bound to a 13 bp DNA duplex containing its octameric target site (Figure 1B) at 2.85 Å resolution. Our results identify the GCM domain as a new class of Zn-containing DNA-binding domain with no similarity to any other DNA-binding domain. The GCM domain consists of a large and a small domain tethered together by one of the two Zn ions present in the structure (Figure 2). The large and the small domains comprise five- and three-stranded β-sheets, respectively, with three small helical segments packed against the same side of the two β-sheets. The GCM domain exercises a novel mode of sequence-specific DNA recognition, where the five-stranded β-pleated sheet inserts into the major groove of the DNA. Residues protruding from the edge strand of the β-pleated sheet and the following loop and strand contact the bases and backbone of both DNA strands, providing specificity for its DNA target site.
Fig. 2. Structure of the GCM domain. (A) Ribbon representation of the GCM domain bound to its cognate DNA. The β-sheets of the large and small domains are depicted in dark blue and light blue, respectively. Helices H1, H2 and H3 are shown in red, and the DNA is shown in yellow. The two Zn ions and their coordinating ligands are depicted. Figures 2A and B, 3B, 4A and 6 were produced using the program RIBBONS (Carson, 1991). (B) View of the GCM domain with the DNA axis running vertically. DNA bases are numbered according to Figure 1B. (C) Topology diagram of the GCM domain. DNA-contacting residues and the first and second Zn ion coordinating residues are marked as dots. The color code corresponds to Figures 1A and 2A.
Results and discussion
Overall structure
The GCM domain–DNA complex structure was solved by the multiple isomorphous replacement method using three iodinated DNA derivatives (Table I). The crystal contains one complex in the asymmetric unit. The current model contains 153 amino acid residues, 26 nucleotides, two Zn ions and four water molecules, and has been refined to a crystallographic R factor of 21.8% (Rfree = 28.3%) using all data from 20 to 2.85 Å. The final 2Fo – Fc electron density is well defined for the DNA, the polypeptide main chain and most of the protein side chains (Figure 1C). The highest mobility of the polypeptide chain is observed at the N- and C-terminal ends. N-terminal residues 1–13 are disordered and have not been included in the model. For the following residues 14–30 the main chain can be unambiguously followed but for most side chains the electron density is missing. The C-terminal residues 171–175 are also disordered.
Table I. Structure determination of the GCM domain–DNA complexa.
| Dataset | Crystal form A | Crystal form A′ |
|||
|---|---|---|---|---|---|
| Nat-1 | Nat-41 | IU16 | IU25 | IU34 | |
| Processing statistics | |||||
| Resolution range (Å) | 30–2.85 | 40.0–2.9 | 40–3.05 | 40–3.15 | 40–2.82 |
| Wavelength (Å) | 0.931 | 0.933 | 0.933 | 0.933 | 0.933 |
| Completeness (%) | 97.9 (95.6) | 91.9 (78.4) | 90.8 (72.7) | 91.3 (78.8) | 91.3 (74.6) |
| Multiplicity | 2.9 (2.4) | 4.1 (3.6) | 3.4 (2.7) | 3.4 (2.9) | 3.4 (2.7) |
| Rmeas (%)b | 4.9 (28.2) | 5.6 (27.4) | 8.6 (25.4) | 7.5 (19.5) | 7.4 (29.4) |
| I/σ (I) > 3 | 79.2 (34.8) | 81.8 (49.7) | 79.2 (46.4) | 82.0 (55.8) | 79.9 (42.2) |
| Phasing statistics | |||||
| No. of iodine atoms | 3 | 2 | 3 | ||
| Isomorphous difference (%) | 19.4 | 15.1 | 21.7 | ||
| Phasing power | |||||
| Isomorphous | – | 1.61 | 1.81 | 1.44 | |
| Anomalous | 0.94 | 0.85 | 0.84 | 0.73 | |
| Refinement | |||||
| Resolution range (Å) | 20–2.85 | ||||
| Rwork (%)c | 21.8 (5714 reflections) | ||||
| Rfree (%)c | 28.3 (516 reflections) | ||||
| Total no. of non-hydrogen atoms | 1810 | ||||
| No. of protein atoms | 1277 | ||||
| No. of DNA atoms | 527 | ||||
| No. of water molecules | 4 | ||||
| No. of Zn ions | 2 | ||||
| R.m.s.ds | |||||
| Bond lengths (Å) | 0.008 | ||||
| Bond angles (degrees) | 1.31 | ||||
aNat-1 (crystal form A) was used for the final refinement, whereas dataset Nat-41 (crystal form A′) was used as native data for the MIRAS phasing. For Nat-41, the anomalous signal of the Zn ions was included in the heavy-atom parameter refinement. Values in the highest resolution shell are given in parentheses.
bRmeas = ΣhklΣi|Ii(hkl) – <I(hkl)>|/ΣhklΣiIi(hkl), where Ii is the ith measurement of reflection I(hkl).
cRfree was calculated using 8.1% of the data. No σ cut-off was applied to the data.
The GCM domain has a roughly parallelepiped shape with dimensions of 60 × 30 × 30 Å. The longest dimension runs along the major groove at an angle of ∼45° with respect to the DNA axis (Figure 2B). The GCM domain can be divided into two domains. The large domain consists of an N-terminal extension, a five- stranded antiparallel β-sheet (strands S1, S2, S3, S6 and S7) and a short helix H1. Residues 31–39 of the N-terminal extension, helix H1 and the following linker residues 56–61 pack against the β-pleated sheet. Residues 31–39 and the linker residues 56–61 almost form the second layer of a β-barrel. However, only one main-chain hydrogen bond connects these two stretches of residues and therefore the β-barrel is only partially closed. The small domain contains a three-stranded β-pleated sheet (strands S3′, S4 and S5), helix H2 and the C-terminal helix H3. Helix H2 contains mostly polar residues and connects strand S4 with strand S5. A search for structurally similar proteins with the program DALI (Holm and Sander, 1993) did not find any high-scoring hits. The top hits matched the five-stranded β-sheet of the GCM domain with the seven-stranded β-sheet of bovine profilin (Cedergren-Zeppezauer et al., 1994) (Z score of 3.5) and with the six-stranded β-sheet formed by the C-terminal 100 residues of the mouse ap2 clathrin adaptor α-subunit (Traub et al., 1999) (Z score of 3.0). The overall similarities are low, as indicated by the Z scores, although the β-sheets in these two proteins share the same topology with the GCM domain, except for the insertion of the smaller domain between GCM domain strands S3 and S6 (Figure 2C). Despite the division of the GCM domain into two domains we do not consider them to form independent folding units. In fact, the two domains share a large hydrophobic interface and are probably unable to move independently with respect to each other. Furthermore, one of the two Zn coordination centers plays an important role in tethering the two domains together by coordinating Cys76, Cys125, His152 and His154. The residues following the two histidines fill a groove between the two domains and also contribute to connecting the two domains.
DNA recognition
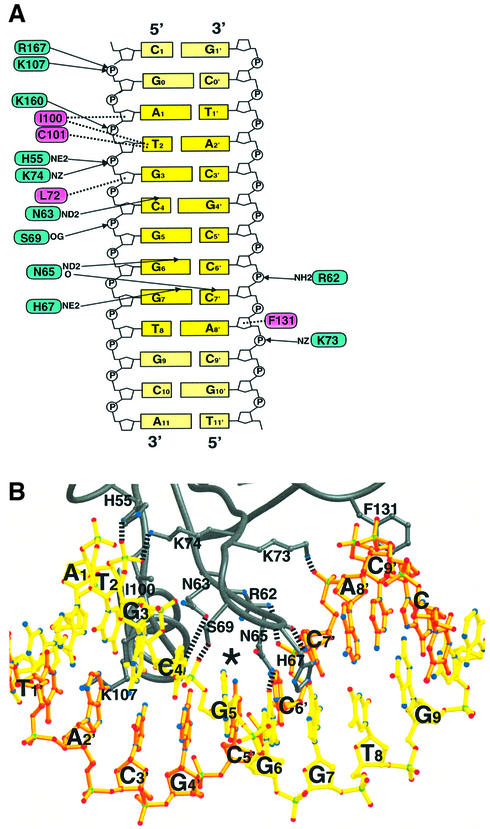
Both domains of the GCM domain are involved in DNA recognition, forming a clamp that seizes the substrate from two sides of the major groove (Figure 2A). The β-sheet of the large domain forms the upper jaw of the clamp, with its strands oriented orthogonally to the DNA axis (Figure 2A and B). At the edge of this sheet, the β-hairpin formed by strands S2 and S3 constitutes the most important recognition element within the GCM domain. This hairpin inserts into the major groove and contacts four backbone phosphates (positions 3, 5, 6′ and 8′) and three bases (Cyt4, Gua6 and Gua7) (Figure 3). Polar backbone contacts are made by residues Arg62, Ser69, Lys73 and Lys74; the last two residues point their side chains in opposite directions, bridging across the entire major groove to contact phosphates Gua3 and Ade8′ from complementary DNA strands. In addition, Leu72 forms a hydrophobic contact with the deoxyribose of Gua3 (Figures 1C and 3). Base-specific contacts are mediated by residues Asn63, Asn65 and His67 from strand S2 and the loop following it. The side chain OD1 and ND2 atoms of Asn63 point towards the exocyclic N4 atom of Cyt4 and the N7 atom of Gua3, respectively. However, both interatomic distances exceed 3.3 Å, which is too much to form direct hydrogen bonds. The ND2 atom of Asn65 forms a hydrogen bond with the exocyclic O6 of Gua6, while its backbone carbonyl contacts the exocyclic N4 of Cyt7′ from the complementary DNA strand. The side chain NE2 atom of His67 forms a hydrogen bond with the O6 of Gua7.
Fig. 3. DNA recognition by the GCM domain. (A) Protein–DNA interactions between the GCM domain and its DNA target site. Arrows and dotted lines indicate polar and hydrophobic interactions, respectively. Residues involved in polar and hydrophobic interactions are drawn on blue and magenta backgrounds, respectively. (B) Ribbon representation of the interactions between the GCM domain and its DNA target site Upper and lower strands as shown in Figure 1B are depicted in yellow and orange, respectively. Broken lines indicate polar interactions.
The lower jaw of the clamp is formed by helix H2 of the small domain. Within this helix, Lys107 contacts the phosphate group of Gua0, while at its N-terminus Ile100 and the backbone atoms of Cys101 form a hydrophobic barrier buttressing the exocyclic methyl group of Thy2. Cys101 is the only strongly conserved cysteine in the GCM domain that does not coordinate Zn (Figure 1A). Its sulfhydryl group points towards DNA bases Gua0 and Ade3, explaining mutagenesis results whereby Cys101 was shown to confer redox sensitivity to DNA binding (Schreiber et al., 1998). In addition to the two jaw regions, DNA binding also involves residues His55 and Lys160 from helices H1 and H3 and Phe131 in the linker between strands S5 and S6. His55 and Lys160 contact the phosphate groups of Gua3 and Thy2, respectively (Figure 3A), whereas Phe131 packs against the deoxyribose of Ade8′. Arg167 in helix H3 points towards the Gua0 phosphate. This is probably also an important contact, although in the crystal structure the Arg167 side chain is highly mobile and appears to be influenced by a phosphate group from a neighboring DNA strand in the crystal lattice. GCM domain residues contact both DNA strands, but it is worth noting that 12 residues contact one strand and only four residues (including Asn65) contact the other (Figure 3B). Almost all the DNA-contacting residues are conserved between different species (Figures 1A and 3). Subtle differences in the DNA-binding requirements of mGCMa and mGCMb (Tuerk et al., 2000) are probably not caused by differences in direct protein–DNA interactions but, rather, are indirect effects resulting from slight differences in the overall structure of both orthologs.
Conformation of the DNA
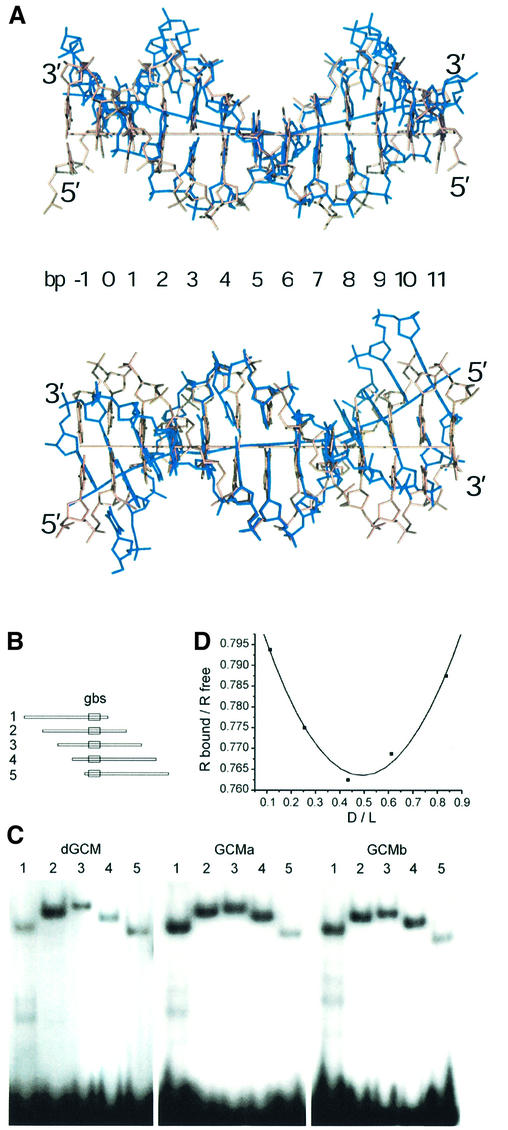
The overall conformation of the DNA in the GCM domain–DNA complex resembles B-form DNA, although its helical axis is highly distorted. These distortions consist of a central bend of ∼30° at bp 6 and two kinks of ∼25° between bp 2/3 and 7/8 (Figure 4A). These kinks direct the DNA axis in opposite directions, above and below the plane defined by the central bend. As a result the DNA axis has an S-like shape.
Fig. 4. DNA bending observed in the GCM domain–DNA complex. (A) Two orthogonal views of the 13mer DNA duplex in the GCM domain–DNA complex superimposed with canonical B-form DNA. Strands of the GCM-bound DNA are colored in blue. Helical axes were calculated using the program CURVES (Lavery and Sklenar, 1988). (B) The consensus GCM binding site (gbs) was inserted between the XbaI and SalI sites of pBEND2 (Kim et al., 1989) and retrieved with flanking sequences using the restriction enzymes BglII (1), XhoI (2), XmaI (3), Asp718 (4) and BamHI (5). This generates fragments of identical size with circular permutations of the same sequence and the GCM binding site at varying positions. (C) Circular permutation analyses of DNA bending by electrophoretic mobility shift assays with fragments 1–5 from (A) as probes and the GCM domains of Drosophila GCM (dGCM), mouse GCMa (GCMa) and mouse GCMb (GCMb) expressed in transiently transfected COS cells. (D) Calculation of bending angle for GCMa as described previously (Scaffidi and Bianchi, 2001). The mobility of the protein–DNA complexes (Rbound) was normalized to the mobility of the corresponding free probe (Rfree). The distance of the center of the GCM binding site from the 5′ end of the fragment was divided by the total length of the probe (flexure displacement D/L). The plotted points were interpolated with quadratic functions y = 0.207x2 – 0.203x + 0.813 (r2 = 0.987). The first- and second-order parameters are in close agreement and yield an estimate of 37° for the flexure angle. Similar calculations lead to flexure angles of 34° for Drosophila GCM and 35° for GCMb.
This overall curvature allows the DNA to form favorable hydrophobic and polar contacts with the protein. In the center of the binding site, the DNA curves around the five-stranded β-sheet that sticks into the major groove (Figure 4A, left panel). One important contact point is formed by the side chain and main chain carbonyl of residue Asn65 and bases Gua6 and Cyt7′. These interactions cause the base of Cyt7′ to rotate out of plane, leading to a considerable buckle and propeller twist of bp 7, which is propagated along the DNA duplex and contributes to the overall bend observed. A combination of polar and hydrophobic contacts is also responsible for the two kinks in opposite directions orthogonal to the central bend (Figure 4A). At one end of the duplex, one strand forms hydrophobic contacts with residues of helix H2 assisted through polar interactions with His55, Lys107 and Lys160 (see above) and leans towards the smaller domain, while at the other end the opposite strand passes through a cleft between the β-hairpin S2/S3 and the bulge between strands S5 and S6 with main contact points formed by Arg62, Lys73 and Phe131 protruding from the bulge (Figure 3B). The two kinks in opposite directions allow the 13mer DNA duplexes to pack continuously along the crystallographic b axis. However, even though the DNA stacks end to end, the polyphosphate backbone is discontinuous in the crystal. Adjacent DNA duplexes are rotated by ∼35° in opposite directions to the helical twist of the DNA. Therefore, the first base pair of each DNA duplex and the penultimate base pair of the neighboring duplex show the same twist angles.
In order to assess whether the observed DNA bending was due primarily to GCM domain binding or merely to crystal packing effects, we performed an electrophoretic mobility shift assay designed to measure the degree of DNA bending in solution. As probes, we used five DNA duplexes of identical length but with different permutations of the nucleotide sequence such that the GCM binding site was positioned differently within each probe (Figure 4B). Protein-induced DNA bending causes a probe with a centrally located binding site to be retarded more than one with a binding site near the end, and the magnitude of this effect can be used to estimate the bending angle (Scaffidi and Bianchi, 2001). When we performed the assay with the GCM domain of murine GCMa, the degree of retardation of the five probes differed significantly, corresponding to an estimated bending angle of 37° (Figure 4C and D). Similar bending angles were also obtained when the assay was performed with the GCM domains of murine GCMb and the Drosophila homolog dGCM. Therefore the solution studies also support a considerable bending of the DNA upon binding of the GCM domain. Thus the considerable deformation of DNA observed in our structure appears to be due primarily to the binding of the GCM domain, with at most only a minor contribution from the crystal packing.
Specificity of the DNA recognition
Experiments on DNA binding of mouse and Drosophila GCM domains to consensus and mutated DNA recognition sequences identified bp 2, 3, 6 and 7 as the strongest determinants of specificity (Schreiber et al., 1998). In accordance, we observe important hydrophobic contacts to Thy2 (Ile100, Cys102) and hydrogen bonds to Gua6, Cyt7′ (Asn65) and Gua7 (His67). The importance of bp 3 is less obvious from the crystal structure as Asn63 only indirectly contacts Gua3. However, changing Gua3 into Thy3 in bp 3 completely abolishes GCM binding (Schreiber et al., 1998). The sequence-dependent conformation of the bound DNA, which is often referred to as ‘indirect readout’, might specify this base pair. Indeed, at this position we see strong deviations of the DNA from the canonical B-form: the DNA is bent between bp 2 and 3 (see above), which accounts for a roll angle of 13° between them. In addition, bp 2 shows a strong buckle of ∼10° with Thy2 leaning towards Gua3.
To investigate the indirect recognition of bp 3 we also replaced guanine by adenine, cytosine and uracil. All these mutations lead to stronger GCM binding compared with the initial M3 mutant site (Figure 5C). Our results correlate well with the conformational mobility of dinucleotide steps deduced from the comparison of DNA duplex crystal structures (El Hassan and Calladine, 1996). This analysis identified TG/CA (present in the consensus GCM binding site) and TA/TA steps (3A site) as particularly flexible and often found in ‘hinges’ in DNA duplexes, whereas TT/AA steps (as present in the M3 site) are very rigid. Our results suggest that only certain base pairs are flexible enough to allow the pronounced roll between bp 2 and 3. The exocyclic 5-methyl group of thymine appears particularly unfavorable. Changing thymine into uracil (3U site) restores ∼50% of the wild-type DNA-binding affinity either because removing the 5-methyl group allows more conformational flexibility (El Hassan and Calladine, 1996) or because it prevents a clash with the adjacent 5-methyl group of Thy2.
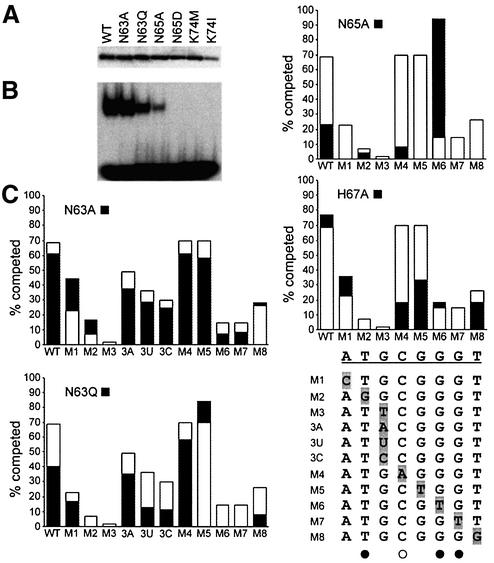
Fig. 5. DNA-binding properties of mutant GCM domains. (A) Expres sion of T7-epitope tagged wild-type (WT) and mutant (N63A, N63Q, N65A, N65D, K74M, K74I) GCM domains was verified by western blot of nuclear extracts from transfected COS cells with a monoclonal antibody against the tag. (B) Electrophoretic mobility shift assay with the consensus GCM binding site as probe and extracts from transfected COS cells expressing the wild-type and mutant GCM domains. Equal amounts of each GCM domain were used. (C) Comparative DNA- binding analysis of wild-type GCMa and GCM protein mutants by competition analyses. Electrophoretic mobility shift assays were performed with the consensus GCM binding site as probe and extracts expressing the wild-type and mutant GCM protein in the absence and presence of increasing amounts of competitor (5-, 10-, 25-, 50- and 100-fold molar excess). Oligonucleotides containing the consensus GCM binding site (WT) and its variants (M1–M8, 3A, 3U, 3C) were used as competitors. Conditions were such that in the absence of competitor, 20–30% of the radioactively labeled probe was in complex with the GCM domain. The competitor-dependent reduction of probe in the complex was determined by phosphoimager analysis. The graph summarizes the relative level of competition obtained with a 10-fold excess of each competitor (WT, M1–M8, 3A, 3U, 3C) for wild-type GCMa (open bars) and GCM mutants (black bars). WT and mutant target sites (M1–M8, 3A, 3U, 3C) are listed. Directly and indirectly contacted bases as observed in the crystal structure are marked with filled and open circles, respectively.
To gain further insight into GCM domain DNA recognition we mutated a number of residues of the DNA-contacting β-hairpin. We mutated three residues involved in base-specific contacts (mutations N63A, N63Q; N65A, N65D; H67A) and one residue contacting the DNA backbone (K74I, K74M, K74A). Expression of the mutated proteins in transiently transfected COS cells was verified by western blots, and their ability to bind to the consensus and mutated DNA target sites was tested by electrophoretic mobility shift assays (Figure 5A and B); DNA binding of the H76A and K74A mutants was analyzed earlier (Schreiber et al., 1998). Our results show distinct roles for Asn63 and Asn65 in site-specific DNA recognition. Mutant protein N63A binds with slightly lower affinity, which agrees with the crystal structure where Asn63 does not form direct hydrogen bonds with DNA bases. In contrast, mutant N65A shows greatly reduced DNA affinity because it can no longer contact Gua6 and Cyt7′. DNA binding is completely abolished in the N65D mutant, probably because the mutation introduces a carboxy group that points towards the Gua6 O6 atom. Our experiments also show the importance of the polar contact formed between Lys74 and the DNA backbone. Changing this residue into a leucine, methionine or alanine residue completely abolishes DNA binding (Figure 5B; Schreiber et al., 1998).
We also performed a series of competitive binding assays in which we assessed the ability of nine different DNA probes, comprising either the natural target site sequence or eight mutated variants (M1–M8), to displace wild-type and mutant GCM domains from the target site (Figure 5C). We observed considerable changes in the site specificity of the N63Q and N65A mutants. Mutant protein N63Q shows reduced binding affinity for the wild-type DNA sequence (Figure 5B) and instead preferentially binds DNA sites M4 and M5, whereas mutant protein N65A preferentially binds to the M6 site (Figure 5C). The crystal structure suggests that the slightly longer glutamine side chain of the N63Q mutant could fill a cavity in the major groove (indicated by an asterisk in Figure 3B), which would allow the N63Q mutant to form favorable interactions with the A–T and T–A base pairs of the M4 and M5 sites. However, the glutamine side chain probably does not form direct interactions with bp 3 as mutant N63Q (like N65A and the wild type) does not clearly distinguish between guanine, adenine, uracil and cytosine in bp 3 (Figure 5C). For the N65A mutant, model building suggests that the alanine CB atom forms a hydrophobic contact with the exocyclic methyl group of Thy6 in the M6 site, which could compensate for the loss of the polar interaction between Asn65 and the Gua6 O6. The H67A mutant shows similar DNA binding to the wild type but a strongly reduced binding to sites M4 and M5 not directly contacted by His67 (Figure 5C). This suggests that different DNA-contacting residues influence each other, probably because point mutations affect not only specific contacts but also the conformation of the entire S2/S3 β-hairpin.
Zn coordination in the GCM domain
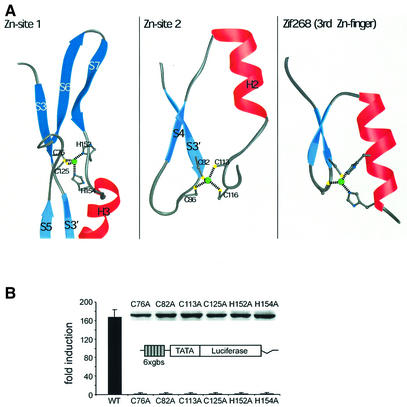
The GCM domain contains two tetrahedrally coordinated Zn ions. The first is coordinated by two cysteines (Cys76, Cys125) and two histidines (His152, His154) in the interface of the two domains (Figure 6A). Apart from Cys76, which protrudes from strand S3 of the large domain, the other three residues lie in linker regions joining the two domains: Cys125 in the loop between strands S5 and S6, and His152 and His154 in the linker between strand S7 and helix H3. Thus, the first Zn-site is an important coordination center, which tightly connects the large and small domains.
Fig. 6. Structural and functional roles of the Zn ions in the GCM domain. (A) Topology of the two Zn-sites observed in the GCM domain. Note the similar topology of Zn-site 2 (center) and classical Cys2His2 Zn-fingers (right) as present in Zif268 (Elrod-Erickson et al., 1996). (B) Transcriptional activities of mutant GCM proteins. A luciferase reporter plasmid carrying six tandemly arranged GCM-binding sites (6× gbs luc) was transfected into 293 cells together with pCMV5 expression plasmids for wild-type mGCMa (WT) or various mutant versions [Cys76 to Ala (C76A), Cys82 to Ala (C82A), Cys113 to Ala (C113A), Cys125 to Ala (C125A), His152 to Ala (H152A) and His154 to Ala (H154A)]. Luciferase activities in extracts from transfected cells were determined in three independent experiments, each performed in duplicate. Data are presented as fold inductions, which were calculated for each reporter plasmid by comparing luciferase activities with values from cells transfected with reporter plasmid and empty pCMV5 expression plasmid. Expression of wild-type mGCMa and its mutants in transfected cells was confirmed by western blot analysis using a polyclonal antiserum against mGCMa (see inset).
The second Zn ion is coordinated by four cysteines at the DNA-distal end of the small domain, connecting the S3′/S4 loop (Cys82, Cys86) with the H2/S5 loop (Cys113, Cys116). The sequence signature of this binding site, C-X3-C-X26-C-X2-C, resembles that of a classical Zn-finger, C-X2–4-C-X12-H-X3-H (Berg and Shi, 1996). Indeed, its topology is similar to the Zn-finger ββα topology, as observed, for example, in the protein Zif268 (Elrod-Erickson et al., 1996), although the third and fourth ligands (Cys113, Cys116) do not protrude from a helix but rather from the subsequent loop (Figure 6A). In classical Zn-fingers, the Zn-site is directly involved in DNA binding as it coordinates the recognition helix that contacts the DNA in the major groove. In contrast, the second Zn-site in the GCM domain is ∼20 Å away from the DNA backbone and does not participate directly in DNA binding.
We have previously shown that the Zn ions in the GCM domain could only be removed by the strong Zn chelator 1,10-phenanthroline under heat-denaturing conditions, a procedure that abrogates DNA-binding activity. However, these experiments did not distinguish between the two Zn-sites (Cohen et al., 2002). In contrast, alanine mutations of the Zn-coordinating cysteine residues show different roles of the Zn ions, which are consistent with our crystallographic results. Mutations of Cys76 and Cys125 coordinating the first Zn ion exhibited a complete loss of DNA binding, confirming their important roles in tethering the two domains together (Schreiber et al., 1998). Accordingly, the Drosophila melanogaster loss-of-function mutant glide/gcmN7-4 carries a point mutation of Cys93 (corresponding to mGCMa Cys76) to Ser96, which abolishes DNA binding and transcriptional activation (Miller et al., 1998). Alanine mutants of the cysteine residues coordinating the second Zn-site can still bind to DNA but show an altered electrophoretic mobility, indicating an altered structure of the GCM domain (Schreiber et al., 1998). Despite these differences, both Zn ions appear to stabilize the GCM domain. Changing any of the eight Zn-coordinating residues into an alanine reduces the amount of protein produced in transiently transfected cells (Schreiber et al., 1998), suggesting a significant decrease in protein stability.
We also analyzed the importance of both Zn-binding sites for the transactivation capacity of mGCMa by expressing mGCMa wild-type and mutant proteins in human 293 cells together with a luciferase reporter gene containing six GCM-binding sites (gbs) (Tuerk et al., 2000). Alanine mutations of all Zn-coordinating residues in the first (Cys76, Cys125, His152, His154) and second (Cys82, Cys113) Zn-sites lead to a complete loss of transcriptional activity compared with the wild-type protein (Figure 6B). Interestingly, we do not observe any differences in the transactivation capacity of mutants changing the first and second Zn-sites despite their different DNA-binding activities (see above). Western blots confirmed that all mutant proteins are expressed during transfection. Furthermore, increasing the amount of transfected expression plasmid did not restore the trans-activation capacity of the mutants (Figure 6B). Therefore, reduced expression or stability of the mutant proteins does not explain the reduced transcriptional activity. Instead, our results suggest that transactivating and DNA-binding domains of GCM interact and that the transactivating domain ‘senses’ structural disturbances of the DNA-binding domain introduced by the mutations. Analysis of the Drosophila gcm regulatory region (Ragone et al., 2003) and of the putative regulatory region of the repo gene (Akiyama et al., 1996) also indicates complex promoter structures containing clusters of GCM protein-binding sites. In addition, high levels of gcm expression can depend on lineage-specific partners like the transcription factor Prospero in Drosophila (Akiyama-Oda et al., 2000; Freeman and Doe, 2001; Ragone et al., 2001). Therefore, it is also conceivable that the structural disturbances introduced in the mutant protein affect molecular interactions between adjacently bound GCM molecules or other interacting factors.
Comparison with other DNA-binding proteins
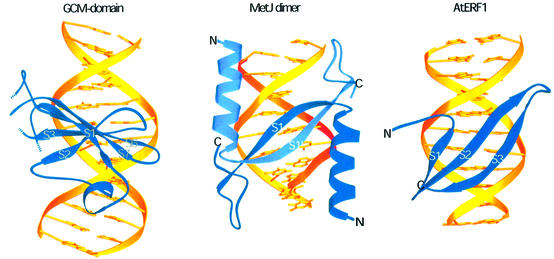
A number of other DNA-binding domains use β-strands to recognize their target sites in the major groove of the DNA (Tateno et al., 1997). In proteins of the MetJ/Arc family (Somers and Phillips, 1992; Raumann et al., 1994) and in the plasmid-encoded transcriptional repressor CopG (Gomis-Ruth et al., 1998), a two-stranded antiparallel β-sheet inserts into the major groove with the plane of the two-stranded sheet reposing on the edges of the bases (Figure 7). The two-stranded β-sheet is formed by two repressor monomers, each donating one strand. A related recognition element has been observed in the structure of the I-PpoI homing endonuclease, the DNA-binding domain of AtERF1 and the DNA-binding domain of the integrase from transposon Tn916, where a three-stranded antiparallel β-sheet protrudes into the major groove (Allen et al., 1998; Flick et al., 1998; Wojciak et al., 1999). However, a detailed inspection reveals that only two strands at a time are inserted into the major groove, whereas one strand stays closer to the polyphosphate backbone. Therefore, DNA recognition by three-stranded β-sheets resembles DNA recognition by the MetJ/Arc family (Figure 7).
Fig. 7. Comparison with other DNA-binding domains. Different use of β-sheets for DNA recognition in the major groove by the GCM domain compared with the bacterial repressor MetJ (Somers and Phillips, 1992) and the A.thaliana transcription factor AtERF1 (Allen et al., 1998).
In the GCM domain the use of the β-sheet for base-specific DNA recognition is very different. The β-sheet is rotated by 90° with respect to those in the examples cited above. Therefore, only one edge of the five-stranded antiparallel β-sheet sticks into the major groove, with the plane of the β-sheet running parallel to the DNA bases. To our knowledge such use of a β-sheet for DNA recognition has not been observed previously.
Relatively few DNA-binding domains use β-sheets for sequence-specific recognition in the major groove, in contrast to the abundant use of α-helices. As one explanation, it has been suggested that β-sheets evolve new specificities more slowly, as changes of single amino acids often affect the overall structure, whereas α-helices are relatively tolerant to point mutations (Connolly et al., 2000). The GCM domain appears to be particularly sensitive in this respect. Because the GCM domain β-sheet is inserted perpendicular to the DNA, only a few bases are recognized directly and additional specificity has to be provided by the small domain (see above). All point mutations that change the five-stranded β-sheet, the DNA contact region of the small domain and the relative orientation of the two domains to each other are likely to affect DNA binding. These constraints may have prevented the GCM domain from becoming such a ubiquitous DNA-binding domain as the Zn-finger or the helix–turn– helix superfamilies.
Materials and methods
Protein purification and crystallization
The GCM domain (residues 1–174) of mGCMa was expressed in Escherichia coli and purified as described previously (Cohen et al., 2002). DNA oligonucleotides were chemically synthesized and purified by anion-exchange chromatography following established procedures (Cramer and Müller, 1997). Iodo- and bromo-substituted DNA oligonucleotides were protected from light during the purification and co-crystallization. Purified GCM domain protein was concentrated to 13 mg/ml in 200 mM NaCl, 20 mM Tris pH 7.9 and 10 mM dithiothreitol. For co-crystallization, protein and DNA duplexes were mixed in a molar ratio of 1:1.2. DNA duplexes were tested that contained the consensus target site 5′-ATGCGGGT-3′ and varied from 8 to 19 bp in length, yielding several different crystal forms. The best crystals were obtained with a 13mer blunt-ended DNA duplex using 16–20% PEG 6000 as precipitant. Two related crystal forms, A and A′, were obtained using 100 mM MES pH 6.0 or 100 mM sodium citrate–citric acid pH 5.0 as buffers. Both forms belong to space group P21 and diffract to ∼2.8 Å resolution at the high-brilliance undulator beamlines of the ESRF. For crystal form A, the cell dimensions are a = 41.8 Å, b = 52.9 Å, c = 63.0 Å and β = 103.2, whereas for crystal form A′ the dimensions are a = 41.7 Å, b = 54.1 Å, c = 61.1 Å and β = 99.4. For both crystal forms, diffraction was strongest along the b* axis, displaying streaks at ∼3.4 Å reflecting the end-to-end stacking of DNA duplexes along the b axis. Native and iodo-DNA derivative crystals grew as thin plates to a maximum size of 150 × 150 × 20 µm. In contrast, crystals containing bromosubstituted DNA oligonucleotides were too small to allow any usable data to be collected. Crystals were stepwise soaked in cryoprotectant solution (25% glycerol final), mounted in cryoloops (Hampton) and flash cooled either using a nitrogen gas stream at 100 K or by simply dipping the crystals into liquid nitrogen. Diffraction data for native and derivative crystals were collected on ESRF beamlines ID14-4 and ID14-3 using a MarCCD X-ray detector.
Structure solution and refinement
Diffraction images were processed using the program XDS (Kabsch, 1988) or the HKL program package (Otwinowski and Minor, 1997). The best reproducible crystal form A′ was used to solve the structure by MIRAS using three iodosubstituted DNA derivatives. The quality of native and derivative datasets is summarized in Table I. Iodine sites were located using the program SOLVE (Terwilliger and Berendzen, 1999). Heavy atom parameters were refined using the program SHARP (de La Fortelle and Bricogne, 1997), yielding an overall figure of merit of 0.50 (0.33 for the highest resolution shell). Finally, the program RESOLVE (Terwilliger, 2000) was used for solvent flattening of the initial electron density map calculated with the program SHARP, which led to an overall figure of merit of 0.60 and 0.41 for the highest resolution shell.
The solvent-flattened electron density map allowed the construction of an initial model containing the DNA and ∼70% of the polypeptide chain using program O (Jones et al., 1991). At this stage not all the sheet-forming strands were continuously connected and in some regions the sequence assignment remained ambiguous. Programs REFMAC (CCP4, 1994) and CNS (Brünger et al., 1998) were both used during the refinement (using the same set of reflections for the free R value). In the early stages of the refinement, the experimental phases were kept as additional restraints. Phase combination using σA-weighted electron density maps allowed the stepwise completion of the model. During the refinement process the partially refined model was transferred from crystal form A′ to crystal form A, which showed a slightly lower overall temperature factor. The refinement was completed in crystal form A to a final crystallographic R factor of 21.8% (Rfree = 28.3%) using all data between 20 and 2.85 Å without any σ cut-off. The final model shows excellent geometry with no residues in the disallowed regions of the Ramachandran plot as evaluated by the program PROCHECK (Laskowski et al., 1993).
Generation of GCM proteins for the DNA-binding experiments
The expression plasmids for amino acids 31–190 of Drosophila GCM and the N-terminal 184 amino acids of mouse GCMb have been described previously (Schreiber et al., 1998; Tuerk et al., 2000). The N-terminal 174 amino acids of mouse mGCMa (Schreiber et al., 1998) were fused in-frame to a T7 epitope (Novagen) and inserted into the eukaryotic expression vector pCMV5. Using this plasmid as template, the following mutations were introduced by site-directed mutagenesis into the GCM domain of mouse GCMa: Asn63 to Ala (N63A); Asn63 to Gln (N63Q); Asn65 to Ala (N65A); Asn65 to Asp (N65D); Lys74 to Met (L74M); Lys74 to Ile (K74I). All expression cassettes were verified by DNA sequencing. For production of GCM proteins, COS cells [maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum (FCS)] were transfected with 10 µg of expression plasmid per 10 cm plate using DEAE–dextran (500 µg/ml) followed by chloroquine treatment. COS cells were harvested 48 h after transfection and extracts were prepared as described previously (Schreiber et al., 1998). Protein expression was detected by SDS–PAGE followed by western blotting using a monoclonal antibody against the T7 epitope (Novagen; 1:5000 dilution), horseradish peroxidase-coupled anti-mouse IgG antibodies and the reagents of the ECL detection system (Amersham).
Electrophoretic mobility shift assays
COS cell extracts expressing the various GCM proteins were incubated with 0.5 ng of 32P-labeled double-stranded oligonucleotides containing wild-type or mutant GCM binding sites (see Figure 5C for sequences) or with 32P-labeled DNA fragments retrieved from pBEND2-gbs by various restriction enzymes. Reaction conditions were as described previously (Schreiber et al., 1998). For competition experiments, unlabeled oligonucleotides carrying various versions of the GCM binding site were added in 5-, 10-, 25-, 50- and 100-fold molar excess. Samples were loaded onto native 5% polyacrylamide gels and electrophoresis was performed in 0.5× TBE (45 mM Tris, 45 mM boric acid and 1 mM EDTA pH 8.3) at 120 V for 1.5 h. Gels were dried and exposed for autoradiography. For determination of competition efficiencies, the relative amount of probe complexed to GCM proteins was quantified using a phosphoimager. Values obtained for a specific GCM protein with increasing amounts of the same competitor were fitted as described previously (Wegner et al., 1993), and the resulting equation was used to determine the amount of complex competed with a 10-fold molar excess. This served as a measure for the competition efficiency of the respective DNA.
Luciferase assays
The mGCMa mutations Cys76 to Ala (C76A), Cys82 to Ala (C82A), Cys113 to Ala (C113A), Cys125 to Ala (C125A), His152 to Ala (H152A) and His154 to Ala (H154A) have been analyzed previously for their DNA-binding ability in the context of the GCM domain (Schreiber et al., 1998). Here they were introduced into the complete open reading frame of mGCMa in the context of the eukaryotic expression vector pCMV5. To analyze the transactivation potential of mutant mGCMa proteins, human 293 cells (maintained in DMEM supplemented with 10% FCS) were transfected by the the calcium phosphate technique as described previously (Tuerk et al., 2000) with 2 µg of the 6× gbs luc reporter plasmid and 2 µg of pCMV5 expression plasmid per 60 mm plate. In control transfections, empty pCMV5 vector was used. Cells were harvested 48 h after transfection, and extracts were assayed for luciferase activity. Expression of all mutant mGCMa proteins was verified on western blots of extracts from transfected cells using the previously described rabbit antiserum against mGCMa (1:3000 dilution) (Tuerk et al., 2000) and horseradish peroxidase-coupled anti-rabbit IgG antibodies.
Accession code
The coordinates of the GCM–DNA complex have been deposited in the Protein Data Bank under accession code 1ODH.
Acknowledgments
Acknowledgements
We thank members of the ESRF/EMBL Joint Structural Biology Group (JSBG), in particular Steffi Arzt, Gordon Leonard, Sean McSweeney, Raimond Ravelli, William Shepard and Andrew Thompson, for support on various ESRF beamlines. We also thank Carlo Petosa for comments on the manuscript. M.W. acknowledges support from the Deutsche Forschungsgemeinschaft (SFB473).
References
- Akiyama Y., Hosoya,T., Poole,A.M. and Hotta,Y. (1996) The gcm-motif: a novel DNA-binding motif conserved in Drosophila and mammals. Proc. Natl Acad. Sci. USA, 93, 14912–14916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama-Oda Y., Hotta,Y., Tsukita,S. and Oda,H. (2000) Mechanism of glia-neuron cell-fate switch in the Drosophila thoracic neuroblast 6-4 lineage. Development, 127, 3513–3522. [DOI] [PubMed] [Google Scholar]
- Allen M.D., Yamasaki,K., Ohme-Takagi,M., Tateno,M. and Suzuki,M. (1998) A novel mode of DNA recognition by a β-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. EMBO J., 17, 5484–5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anson-Cartwright L., Dawson,K., Holmyard,D., Fisher,S.J., Lazzarini,R.A. and Cross,J.C. (2000) The glial cells missing-1 protein is essential for branching morphogenesis in the chorioallantoic placenta. Nat. Genet., 25, 311–314. [DOI] [PubMed] [Google Scholar]
- Berg J.M. and Shi,Y. (1996) The galvanization of biology: a growing appreciation for the roles of zinc. Science, 271, 1081–1085. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software for macromolecular structure determination. Acta Crystallogr. C, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Carson M. (1991) Ribbons 2.0. J. Appl. Crystallogr., 24, 958–961. [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–776. [DOI] [PubMed] [Google Scholar]
- Cedergren-Zeppezauer E.S., Goonesekere,N.C., Rozycki,M.D., Myslik,J.C., Dauter,Z., Lindberg,U. and Schutt,C.E. (1994) Crystallization and structure determination of bovine profilin at 2.0 Å resolution. J. Mol. Biol., 240, 459–475. [DOI] [PubMed] [Google Scholar]
- Cohen S.X., Moulin,M., Schilling,O., Meyer-Klaucke,M., Schreiber,J., Wegner,M. and Müller,C.W. (2002) The GCM domain is a Zn-coordinating DNA-binding domain. FEBS Lett., 528, 95–100. [DOI] [PubMed] [Google Scholar]
- Connolly K.M., Ilangovan,U., Wojciak,J.M., Iwahara,M. and Clubb,R.T. (2000) Major groove recognition by three-stranded β-sheets: affinity determinants and conserved structural features. J. Mol. Biol., 300, 841–856. [DOI] [PubMed] [Google Scholar]
- Cramer P. and Müller,C.W. (1997) Engineering of diffraction-quality crystals of the NF-κB P52 homodimer:DNA complex. FEBS Lett., 405, 373–377. [DOI] [PubMed] [Google Scholar]
- de La Fortelle E. and Bricogne,G. (1997) Maximum-likelihood heavy-atom parameter refinement for the multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol., 276, 472–494. [DOI] [PubMed] [Google Scholar]
- Ding C., Buckingham,B. and Levine,M.A. (2001) Familial isolated hypoparathyroidism caused by a mutation in the gene for the transcription factor GCMB. J. Clin. Invest., 108, 1215–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hassan M.A. and Calladine,C.R. (1996) Propeller-twisting of base-pairs and the conformational mobility of dinucleotide steps in DNA. J. Mol. Biol., 259, 95–103. [DOI] [PubMed] [Google Scholar]
- Elrod-Erickson M., Rould,M.A., Nekludova,L. and Pabo,C.O. (1996) Zif268 protein–DNA complex refined at 1.6 Å: a model system for understanding zinc finger–DNA interactions. Structure, 4, 1171–1180. [DOI] [PubMed] [Google Scholar]
- Esnouf R.M. (1999) Further additions to MolScript version 1.4, including reading and contouring of electron-density maps. Acta Crystallogr. D, 55, 938–940. [DOI] [PubMed] [Google Scholar]
- Flick K.E., Jurica,M.S., Monnat,R.J.,Jr and Stoddard,B.L. (1998) DNA binding and cleavage by the nuclear intron-encoded homing endonuclease I-PpoI. Nature, 394, 96–101. [DOI] [PubMed] [Google Scholar]
- Freeman M.R. and Doe,C.Q. (2001) Asymmetric Prospero localization is required to generate mixed neuronal/glial lineages in the Drosophila CNS. Development, 128, 4103–4112. [DOI] [PubMed] [Google Scholar]
- Gomis-Ruth F.X. et al. (1998) The structure of plasmid-encoded transcriptional repressor CopG unliganded and bound to its operator. EMBO J., 17, 7404–7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther T. et al. (2000) Genetic ablation of parathyroid glands reveals another source of parathyroid hormone. Nature, 406, 199–203. [DOI] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1993) Protein structure comparison by alignment of distance matrices. J. Mol. Biol., 233, 123–138. [DOI] [PubMed] [Google Scholar]
- Hosoya T., Takizawa,K., Nitta,K. and Hotta,Y. (1995) Glial cells missing: a binary switch between neuronal and glial determination in Drosophila. Cell, 82, 1025–1036. [DOI] [PubMed] [Google Scholar]
- Jones B.W., Fetter,R.D., Tear,G. and Goodman,C.S. (1995) Glial cells missing: a genetic switch that controls glial versus neuronal fate. Cell, 82, 1013–1023. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zhou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kabsch W. (1988) Evaluation of single-crystal X-ray diffraction data from a position-sensitive detector. J. Appl. Crystallogr., 21, 916–924. [Google Scholar]
- Kim J., Zwieb,C., Wu,C. and Adhya,S. (1989) Bending of DNA by gene-regulatory proteins: construction and use of a DNA bending vector. Gene, 85, 15–23. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Lavery R. and Sklenar,H. (1988) The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J. Biomol. Struct. Dynam., 6, 63–91. [DOI] [PubMed] [Google Scholar]
- Miller A.A., Bernardoni,R. and Giangrande,A. (1998) Positive autoregulation of the glial promoting factor glide/gcm. EMBO J., 17, 6316–6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Ragone G., Bernardoni,R. and Giangrande,A. (2001) A novel mode of asymmetric division identifies the fly neuroglioblast 6-4T. Dev. Biol, 235, 74–85. [DOI] [PubMed] [Google Scholar]
- Ragone G. et al. (2003) Transcriptional regulation of glial cell specification. Dev. Biol., 255, 138–150. [DOI] [PubMed] [Google Scholar]
- Ransick A., Rast,J.P., Minokawa,T., Calestani,C. and Davidson,E.H. (2002) New early zygotic regulators expressed in endomesoderm of sea urchin embryos discovered by differential array hybridization. Dev. Biol., 246, 132–147. [DOI] [PubMed] [Google Scholar]
- Raumann B.E., Rould,M.A., Pabo,C.O. and Sauer,R.T. (1994) DNA recognition by β-sheets in the Arc repressor-operator crystal structure. Nature, 367, 754–757. [DOI] [PubMed] [Google Scholar]
- Scaffidi P. and Bianchi,M.E. (2001) Spatially precise DNA bending is an essential activity of the sox2 transcription factor. J. Biol. Chem., 276, 47296–47302. [DOI] [PubMed] [Google Scholar]
- Schreiber J., Sock,E. and Wegner,M. (1997) The regulator of early gliogenesis glial cells missing is a transcription factor with a novel type of DNA-binding domain. Proc. Natl Acad. Sci. USA, 94, 4739–4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber J., Enderich,J. and Wegner,M. (1998) Structural requirements for DNA binding of GCM proteins. Nucleic Acids Res., 26, 2337–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber J., Riethmacher-Sonnenberg,E., Riethmacher,D., Tuerk,E.E., Enderich,J., Bosl,M.R. and Wegner,M. (2000) Placental failure in mice lacking the mammalian homolog of glial cells missing, GCMa. Mol. Cell. Biol., 20, 2466–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers W.S. and Phillips,S.E. (1992) Crystal structure of the met repressor-operator complex at 2.8 Å resolution reveals DNA recognition by β-strands. Nature, 359, 387–393. [DOI] [PubMed] [Google Scholar]
- Tateno M., Yamasaki,K., Amano,N., Kakinuma,J., Koike,H., Allen,M.D. and Suzuki,M. (1997) DNA recognition by β-sheets. Biopolymers, 44, 335–359. [DOI] [PubMed] [Google Scholar]
- Terwilliger T.C. (2000) Maximum-likelihood density modification. Acta Crystallogr. D, 56, 965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T.C. and Berendzen,J. (1999) Automated MAD and MIR structure solution. Acta Crystallogr. D, 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub L.M., Downs,M.A., Westrich,J.L. and Fremont,D.H. (1999) Crystal structure of the α appendage of AP-2 reveals a recruitment platform for clathrin-coat assembly. Proc. Natl Acad. Sci. USA, 96, 8907–8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerk E.E., Schreiber,J. and Wegner,M. (2000) Protein stability and domain topology determine the transcriptional activity of the mammalian glial cells missing homolog, GCMb. J. Biol. Chem., 275, 4774–4782. [DOI] [PubMed] [Google Scholar]
- Van de Bor V. and Giangrande,A. (2002) Glide/gcm: at the crossroads between neurons and glia. Curr. Opin. Genet. Dev., 12, 465–472. [DOI] [PubMed] [Google Scholar]
- Vincent S., Vonesch,J.L. and Giangrande,A. (1996) Glide directs glial fate commitment and cell fate switch between neurones and glia. Development, 122, 131–139. [DOI] [PubMed] [Google Scholar]
- Wegner M. and Riethmacher,D. (2001) Chronicles of a switch hunt: gcm genes in development. Trends Genet., 17, 286–290. [DOI] [PubMed] [Google Scholar]
- Wegner M., Drolet,D.W. and Rosenfeld,M.G. (1993) Regulation of JC virus by the POU-domain transcription factor Tst-1: implications for progressive multifocal leukoencephalopathy. Proc. Natl Acad. Sci. USA, 90, 4743–4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojciak J.M., Connolly,K.M. and Clubb,R.T. (1999) NMR structure of the Tn916 integrase–DNA complex. Nat. Struct. Biol., 6, 366–373. [DOI] [PubMed] [Google Scholar]