Abstract
Proteorhodopsins, ubiquitous retinylidene photoactive proton pumps, were recently discovered in the cosmopolitan uncultured SAR86 bacterial group in oceanic surface waters. Two related proteorhodopsin families were found that absorb light with different absorption maxima, 525 nm (green) and 490 nm (blue), and their distribution was shown to be stratified with depth. Using structural modeling comparisons and mutagenesis, we report here on a single amino acid residue at position 105 that functions as a spectral tuning switch and accounts for most of the spectral difference between the two pigment families. Furthermore, looking at natural environments, we found novel proteorhodopsin gene clusters spanning the range of 540–505 nm and containing changes in the same identified key switch residue leading to changes in their absorption maxima. The results suggest a simultaneous diversification of green proteorhodopsin and the new key switch variant pigments. Our observations demonstrate that this single-residue switch mechanism is the major determinant of proteorhodopsin wavelength regulation in natural marine environments.
Keywords: diversity/rhodopsin/SAR86/spectral tuning/structure modeling
Introduction
Rhodopsins are currently known to belong to two distinct retinylidene protein families: the visual rhodopsins, found in eyes throughout the animal kingdom; and microbial rhodopsins, found in extreme haloarchaea, bacteria and eukaryotic microorganisms (Spudich et al., 2000). The two families share identical topologies, characterized by seven transmembrane helices, and are covalently linked to retinal. However, they do not share any significant similarities in their sequence and possibly have a different evolutionary origin (Spudich et al., 2000).
One of the most notable distinguishing properties of retinal among the various chromophores used in photosensory receptors is the large variation in its absorption spectrum depending on interaction with the apoprotein (‘spectral tuning’; Ottolenghi and Sheves, 1989; Birge, 1990). In rhodopsins, retinal is covalently attached to the ε-amino group of a lysine residue, forming a protonated retinylidene Schiff base. In methanol, a protonated retinylidene Schiff base exhibits a λmax of 440 nm. The protein microenvironment shifts λmax (the ‘opsin shift’; Yan et al., 1995) to longer wavelengths, e.g. to 487 nm in sensory rhodopsin II and to 568 nm in bacteriorhodopsin from Halobacterium salinarum. Spectral tuning in retinylidene proteins has been studied for decades (see Kochendoerfer et al., 1999; Lin and Sakmar, 1999; Nathans, 1999; Spudich et al., 2000, and references cited therein) but is still only partially understood.
The proteorhodopsins (PRs) are light-driven proton pumps (Béjà et al., 2000a; Dioumaev et al., 2002; Friedrich et al., 2002) belonging to the microbial rhodopsin superfamily, type 1 rhodopsins (Spudich et al., 2000), and associated (Béjà et al., 2000a) with the ubiquitous uncultured marine bacteria SAR86 group (Mullins et al., 1995; Béjà et al., 2000b; Eilers et al., 2000; Gonzalez et al., 2000; Rappé et al., 2000; Kelly and Chistoserdov, 2001; Suzuki et al., 2001; Zubkov et al., 2001; Pernthaler et al., 2002). Two related PR subgroups were found that absorb light with different absorption maxima, λmax 525 nm (green) and λmax 490 nm (blue). Their distribution was shown to be stratified with depth, with ‘green-absorbing’ pigments at the surface and ‘blue-absorbing’ pigments at deeper waters in accordance with light available at these depths (Béjà et al., 2001). The PR family is an ideal model system for investigating spectral tuning in microbial rhodopsins since both ‘green-absorbing’ and ‘blue-absorbing’ PR types share >78% of their amino acid residues (200 out of 247 are identical), while their visible absorption peaks differ by almost 40 nm. In contrast, the four types of H.salinarum rhodopsins show a comparable variation in color, but share <25–50% of their residues (Spudich et al., 2000).
As part of our effort to understand the global ecological implications of PR light harvesting and energy transduction to the ocean, we present here results on the identification and variation of a single amino acid which functions as a spectral tuning switch in these widespread pigments in natural environments.
Results and discussion
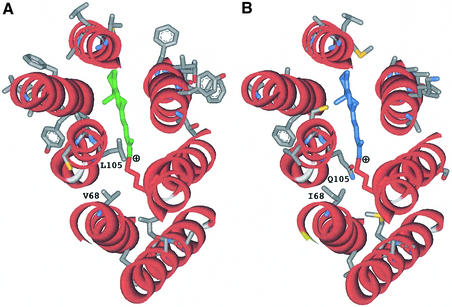
By comparing theoretical structural models, we identified candidates for the spectral difference between blue-absorbing and green-absorbing PRs (B-PRs and G-PRs, respectively). The models (Figure 1) were constructed by threading PR sequences on the 1.55 Å resolution structure of bacteriorhodopsin (Luecke et al., 1999; for details, see Béjà et al., 2000a). The eBAC31A08 (Béjà et al., 2000a; G-PR) and PalE6 (Béjà et al., 2001; B-PR) structural models were compared in order to find residue candidates accountable for the spectral difference between the two. As can be seen in Figure 1, almost all the residue changes between G-PRs and B-PRs are facing outward, in the opposite direction to the retinal chromophore, leaving only four polymorphic variable residues within the retinal binding pocket (residues in positions 40, 68, 105 and 106). Residues 40 and 106 are >6 Å from the Schiff base, whereas residues 68 and 105 are predicted to be positioned ∼5 Å from it; therefore positions 68 and 105 were chosen for mutation analysis.
Fig. 1. Structure modeling of PR (Béjà et al., 2000a) based on the 1.55 Å resolution structure of bacteriorhodopsin (Luecke et al., 1999). (A) Green-absorbing eBAC31A08 variant (G-PR); (B) blue-absorbing PalE6 variant (B-PR). Hydrophilic loops and amino acids in common between the two variants are omitted for clarity. Lys232 in helix G, Schiff base linked to retinal (marked green or blue), is labeled red. Structures were visualized using the ViewerLite 4.2 program (Accelrys Inc.).
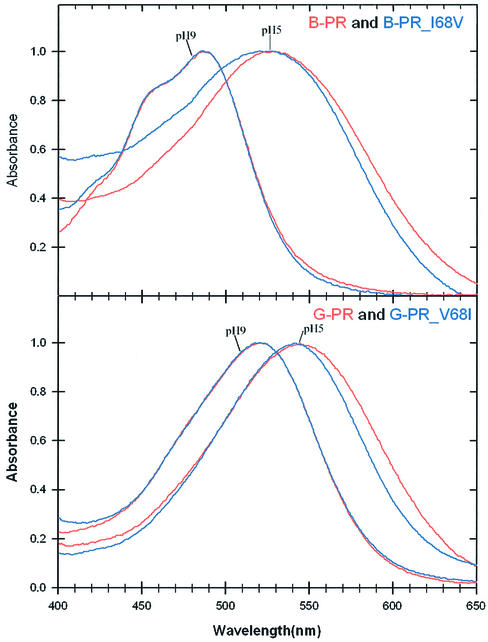
In general, the spectra of rhodopsins exhibit two forms in a proton-dependent equilibrium: an alkaline species in which the primary retinylidene Schiff base counterion (Asp97 in PRs) is unprotonated, and a red-shifted acidic species in which this residue is protonated. The PR alkaline species are active in light-driven proton ejection from the cell. To investigate the potential importance of residues 68 and 105 in PR wavelength absorption regulation, we mutated the PR pigments at these positions and compared the spectra of their acidic and alkaline forms. The residue at position 68 differs in B-PR and G-PR clades, being isoleucine in the former and valine in the latter. These residues where changed to valine in B-PR and isoleucine in G-PR. We changed these residues to valine in B-PR and isoleucine in G-PR. The results showed that the residue at position 68 has a negligible effect on the absorption spectra of both the acidic and alkaline forms (Figure 2).
Fig. 2. Acidic and alkaline absorption spectra of retinal-reconstituted position 68 mutant PRs in E.coli membranes. Spectra were generated by all-trans retinal addition to E.coli membranes containing apoproteins of wild-type and mutant PRs as follows: G-PR, wild-type PR from Monterey Bay (eBAC31A08 in Figure 4; Béjà et al., 2000a); B-PR, wild-type deep-water PR from the HOT station (palE6 in Figure 4; Béjà et al., 2001); BPR_V68I and GPR_V681, V68I and I68V mutants of G-PR and B-PR, respectively. Wild-type PR absorption spectra (in red) and mutant PR spectra (in blue) have been normalized to unity at their absorption maxima for comparison of the spectra.
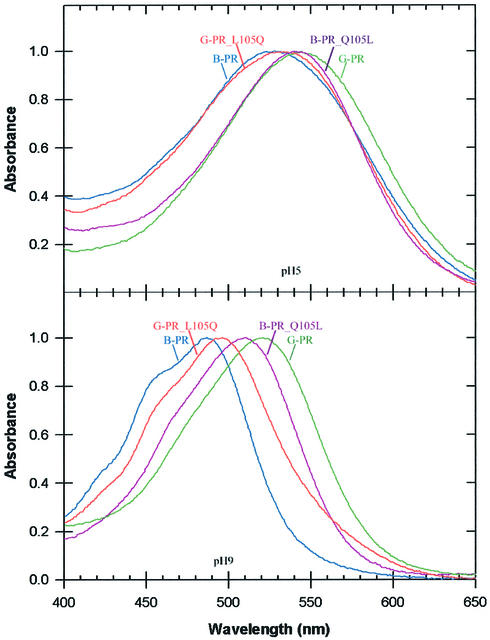
The difference between the two families at position 105 is a non-polar leucine residue (G-PR) versus a polar glutamine residue (B-PR). Therefore two PR protein mutants were constructed, in which the two residues at position 105 were interchanged (G-PR L105Q and B-PR Q105L), and their absorption spectra were compared. To assess the effect of the mutations, the mutant pigments were titrated to determine their alkaline and acidic spectra (W.Wang and J.L.Spudich, in preparation). The mutations at position 105 almost completely interconverted the absorption spectra of B-PR and G-PR (Figure 3). In the alkaline forms (pH 9.0), G-PR L105Q shifted to the blue and acquired vibrational fine structure like wild-type B-PR, and B-PR Q105L shifted to the red and lost the fine structure, exhibiting spectra similar to those of G-PR. In the acid forms (pH 5.0) the interconversion was essentially complete. The mutations at this site also shift the spectral transition pKa of G-PR and B-PR towards that of the other pigment (W.Wang, E.N.Spudich, O.A.Sineschekov, D.Man, O.Béjà and J.L.Spudich, in preparation). Therefore the Q–L difference between G-PR and B-PR pigments functions as a single-residue spectral tuning switch. The residue at position 105 is close to the protonated Schiff base nitrogen. A mutation at the equivalent position in bacteriorhodopsin (L93) was also shown to alter its absorption spectrum (Subramaniam et al., 1991). It is also notable that the corresponding microenvironment of the protonated retinylidene Schiff base has been found to be a site of wavelength regulation in human rhodopsin (Kochendoerfer et al., 1999).
Fig. 3. Acidic and alkaline absorption spectra of retinal-reconstituted position 105 mutant PRs in E.coli membranes. Spectra were generated and normalized as in Figure 2: G-PR, wild-type PR from Monterey Bay (eBAC31A08 in Figure 4; Béjà et al., 2000a); B-PR, deep-water PR from the HOT station (palE6 in Figure 4; Béjà et al., 2001); B-PR_ L105Q and G-PR_Q105L, L105Q and Q105L mutants of G-PR and B-PR, respectively.
The physical basis of the spectral tuning by variation at position 105 may derive from altered hydrogen bonding and subtle repositioning within the quaternary charge complex that appears largely to determine the energies of the ground and excited state in microbial rhodopsins (Kochendoerfer et al., 1999; Ren et al., 2001). The predicted location of the 105 residue sidechain near the protonated Schiff base supports a possible mechanism in which the polar glutamyl sidechain in clade II pigments (defined in Figure 4A) stabilizes the protonated Schiff base, thereby decreasing π-electron delocalization in the retinal and producing a higher energy gap between the ground and excited states, i.e. a blue shift. A similar mechanism has been suggested for the blue shift attributable to the introduction of Ser308 in the mouse green cone pigment (Sun et al., 1997). Moreover, a combined mutagenesis and resonance Raman spectroscopy analysis of human rod rhodopsin shows that the dielectric character and architecture of the chromophore-binding pocket are specifically altered by mutations in residues that compose the retinal-binding pocket. An increase in the number of dipolar sidechains near the protonated Schiff base of retinal was found to increase the ground–excited state energy gap via long-range dipole–dipole Coulomb interaction (Lin et al., 1998). Such interactions may explain the blue shift in PRs on substitution of the non-polar Leu with the polar Gln residue. A more detailed explanation of the Q–L switch mechanism in terms of quantum–mechanical/molecular–mechanical factors awaits atomic resolution coordinates from crystal structures of representatives from different clades.
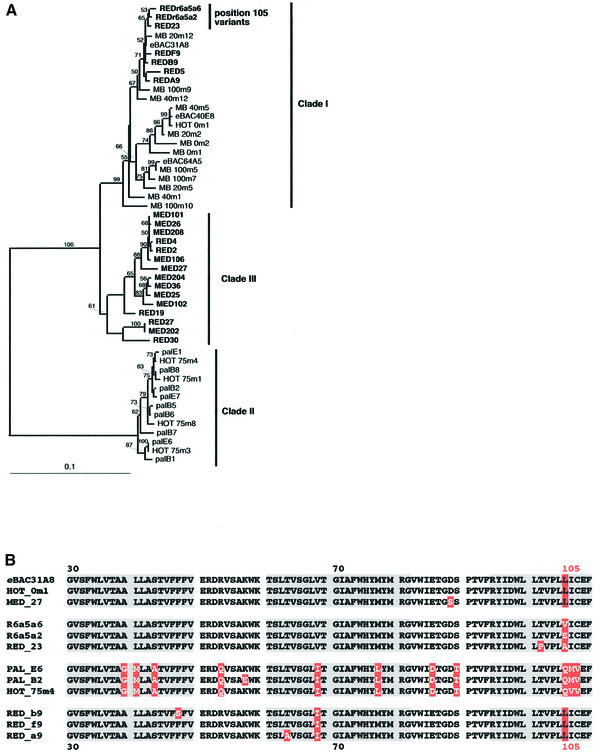
Fig. 4. Analysis of cloned PR genes from the Red Sea and the Mediterranean Sea. (A) DNA phylogenetic tree based on distance analysis of 740 nt positions by neighbor-joining using the Clustal X (1.81) program (Thompson et al., 1997). Bootstrap values >50% are indicated above the branches. Names in bold type indicate PR variants identified in this study. The scale bar represents number of substitutions per site. All previously reported PR variants have been incorporated in the tree: eBAC31A08–AF279106 (Béjà et al., 2000a) and AF349976–AF350003 (Béjà et al., 2001). (B) Multiple protein alignment of PRs from the different DNA-based groups shown in (A). Residue differences between the protein variants are marked in white on red; position 105 is marked above the alignment and is indicated in all sequences for clarity. Predicted transmembrane helices A, B and C are marked in gray.
To study the possible ecological in vivo significance of the in vitro analysis of residue 105 spectral tuning properties, we performed a search for more PR variants directly in their marine environment. Therefore we applied PR polymerase chain reaction (PCR) primers (Béjà et al., 2000a, 2001) to new environments.
The new PR sequences isolated from the Mediterranean Sea (Alborán Sea; MED clones in Figure 4A) and the Red Sea (RED clones) increased the previously known PR diversity, which was based on sequences isolated from Monterey Bay, California, the Hawaii Ocean Time (HOT) station and the Palmer station in Antarctica (Béjà et al., 2000a, 2001). The new sequences from the Mediterranean formed a new PR cluster (clade III in Figure 4A), based on DNA comparisons, and shared at least 88% DNA sequence identity with the Monterey Bay and shallow HOT cluster (clade I), and 69% with the Antarctic and deep HOT cluster (clade II). Nearly all DNA differences between clades I and III are synonymous substitutions; there is little difference between their predicted protein sequences. This finding might suggest that PR clade III is also encoded by SAR86-like bacteria, as a similar observation was recently reported for the GSAT (glutamate semialdehyde aminotransferase) gene from natural populations of marine Archaea sharing almost identical 16S rRNA (Béjà et al., 2002).
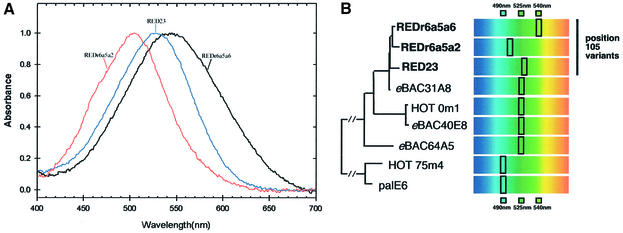
New sequences from the Red Sea station fell within clade I or were associated with the new clade III (Figure 4A). Some PRs from the Red Sea station that fall in clade I had isoleucine instead of valine at position 68, a change found in members from clade II (Figure 4B). As noted above, this modification in the pigments of clades I and II has no influence on the spectra, and therefore it remains to be determined whether it contributes to different properties in the Red Sea PR family. Other Red Sea variants, also integrated in clade I, had a residue change at position 105 (plus more residue changes compared with members of clade I) (Figure 4B), the same position implicated (this study) as the spectral tuning switch. Therefore, we expressed environmental Red Sea ‘position 105’ variants in Escherichia coli cells and examined their absorption spectra. Confirming that position 105 is a key residue for wavelength regulation, the Red Sea variants absorbed at different wavelengths ranging from 505 nm (REDr6a5a2, Glu at position 105) to 540 nm (REDr6a5a6, Val at position 105; Figure 5). These observations in natural samples strengthen our conclusion that 105 is the key residue position responsible for spectral tuning in PRs.
Fig. 5. pH-dependent absorption spectra of retinal-reconstituted PRs in E.coli membranes. (A) Spectra of deep Red Sea ‘position 105’ variants generated and normalized as in Figure 2 at pH 8. (B) Phylogeny of Red Sea ‘position 105’ PR variants (bold type) and schematic representation of their retinal-reconstituted form λmax in UT5600 E.coli membranes. PRs eBAC31A08, eBAC40E08 and eBAC65A05 were retrieved from surface waters in Monterey Bay (Béjà et al., 2000a, 2001), PR palE6 was collected from Antarctic waters, and HOT 0m1 and HOT 75m4 were collected from Hawaiian waters at 0 and 75 m, respectively (Béjà et al., 2001).
Low bootstrap confidence in the branching order among G-PRs and ‘position 105’ PRs (upper part of Figure 4B) suggests that G-PR and ‘position 105’ PR pigments are undergoing a rapid contemporaneous diversification of both PR mini-lineages similar to what has been suggested for marine cyanobacteria photosynthetic antenna pigments (Urbach et al., 1998; Ting et al., 2002). These natural PR variants may derive from different SAR86 strains or other planktonic microorganisms that adapt to different light conditions, or alternatively to the same bacterium armed with different adapted pigments that enable it to survive and harvest light energy in different niches. Since the identity of the bacteria harboring the PRs from clades II and III is unknown and it is not clear whether all SAR86 use this mechanism to harvest light (Fuhrman, 2002), further experiments are needed to distinguish these possibilities.
The ‘position 105’ Red Sea variants reported here were collected at a depth of 150 m, which in the Red Sea is still in the photic zone. A well-tuned pigment is expected to be more important at depths at which light is at a premium and hence to be more adapted to these conditions. Indeed, preliminary comparison with shallower samples from the same station indicates a greater abundance of ‘position 105’ variants in deeper samples (G.Sabehi, R.Massana, E.F.DeLong and O.Béjà, in preparation). It has recently been shown that multiplication of antenna genes is a major adaptation strategy to low-light conditions in a low-light-adapted cyanobacterium Prochlorococcus strain (Moore et al., 1998; Garczarek et al., 2000). Therefore the finding of the different ‘position 105’ PR variants at the bottom of the photic zone could be interpreted as the adaptation of SAR86 to minimal low-light conditions.
The variation in absorption maxima in highly similar (two to three residue changes) PR pigments resembles the variation in pigments in vertebrate color vision in which different isoforms of the same chromoprotein are adapted to different wavelengths (Nathans, 1999). The PR variants with blue to yellow–green absorption properties, which firmly belong to the ‘green’ clade of the PR tree (Figure 5B), confirm that the single spectral tuning switch residue identified in this study is broadly used in PRs in natural environments. This case illustrates the value of applying a combination of environmental genomics, theoretical structure prediction and site-directed mutagenesis to reveal the mode of action of natural selection in the environment.
Materials and methods
Bacterial strains
The E.coli outer-membrane-deficient strain UT5600 [ompT–] (Elish et al., 1988) was obtained from the E.coli Genetic Stock Center at Yale University (strain 7092).
Environmental DNA collection
Red Sea samples were collected from a depth of 150 m at a station in the northern Red Sea near the entrance to the Gulf of Suez (27.17°N, 34.22°E) in February 1999. Mediterranean samples were from the Alborán Sea (36.0°N, 4.25°W) collected in May 1998.
Site-directed mutagenesis
Mutants G-PR_V68I, B-PR_I68V, G-PR_L105Q and B-PR_Q105L were constructed by introducing mutations to G-PR (eBAC31A08 variant) or B-PR (palE6 variant) by using oligonucleotide-directed [V68InewF-5′-ctgtatctggtctcattactggtattgctttctggc-3′, V68InewR-5′-aaagcaataccagtaatg agaccagatacagttaatg-3′, I68VnewF-5′-ctgtatctggtctcgttactggtatagctttttg gc-3′, I68VnewR-5′-gctataccagtaacgagaccagatacagtaagtgaag-3′, Q105L sense2-5′-ggtgttccattactaatggttgagttctatc-3′, Q105Lanti2-5′-gatagaactca accattagtaatggaacacc-3′, L105QsenseG-5′-cagttcctctacaaatatgtgaattctac-3′, L105QantiG-5′-gtagaattcacatatttgtagaggaactg-3′, pBADNcoIsense-5′-ggagatatacatacCCATGG-3′ (NcoI site is underlined) and pBADPmeIanti-5′-ctggagaccGTTTAAAC-3′ (PmeI site is underlined)] site-specific mutagenesis via two-step PCR (Ho et al., 1989). The final PCR products were cloned into pBAD G-PR or pBAD B-PR clones by replacing the original inserts with NcoI and PmeI digestions. Point mutation identities were verified by sequencing the length of the PCR-generated inserts in the final constructed expression vector.
Environmental PR PCR amplification and expression
PRs were amplified by PCR directly from environmental samples using a modified PR forward primer R6sense-5′-ACCATGGGTAAATT ATTACTGATNTTAG-3′ (NcoI site is underlined), designed based on sequence retrieved from an Antarctic fosmid containing blue PR (J.R.de la Torre and E.F.DeLong, personal communication) and the original reverse PR primer (Béjà et al., 2000a), using independently two different high-fidelity proof-reading polymerase mixes (BIO-X-ACT™ from Bioline and TaKaRa Ex Taq™ from Takara Shuzo Co.; G.Sabehi, R.Massana, E.F.DeLong and O.Béjà, in preparation). PCR products were cloned using the QIAGEN® PCR cloning kit and unique RFLP groups were sequenced. Sequences were viewed using the Sequencher 4.1.2 program (Gene Codes Corporation) and selected sequences were expressed using the pBAD TOPO® expression kit (Invitrogen). Correctly oriented inserts were verified by NcoI digestion and religated to remove the signal sequence integrated in the commercial vector and to maintain the native signal sequence of the PR. To express PRs, O.N. cultures were diluted 1:100 and grown for 2 h at 37°C. Cells were induced with 0.2% l-arabinose for 3 h. All PCR products identical with samples handled or amplified previously in the laboratory were omitted from the analyses to avoid influence from possible contamination. The sequences reported in this study are deposited under DDBJ/EMBL/GenBank accession Nos AY210898–AY210919.
Absorption spectroscopy
Difference spectroscopy was used to measure the retinal-generated absorption in E.coli membranes. Ethanolic solutions of all-trans retinal were added to membrane suspensions and the course of pigment development monitored until completion (∼60 min) in an SLM Amico DW2000 spectrophotometer as described previously (Bieszke et al., 1999). To establish the absorption spectra of the alkaline and acidic forms of the pigments, the retinal addition was performed with membrane suspensions adjusted to a range of pH values between 4.0 and 9.0, and the regenerated absorption spectra were recorded. The transition between acid and alkaline forms in different proteorhodopsin pigments was found to exhibit pKa values in the range 6.5–8.0 (W.Wang, E.N.Spudich, O.A.Sineschekov, D.Man, O.Béjà and J.L.Spudich, in preparation).
Acknowledgments
Acknowledgements
We thank E.DeLong for encouragement and for sharing unpublished data, M.Sheves for help during initial stages of the research and G.Rhodes and N.Adir for help with molecular graphics programs. We thank B.Horwitz and B.Podbilewicz for helpful discussions and critical reading of this manuscript. This research was supported by the Human Frontiers Science Program P38/2002 (J.L.S. and O.B.), J.S.Frankford, E. and J.Bishop, J. and A.Taub and the New York Metropolitan research funds (O.B.), National Science Foundation grant 0091287 (J.L.S), National Institutes of Health R01 GM27750 (J.L.S.), the Robert A.Welch Foundation (J.L.S), EU project PICODIV EVK3-CT1999-00021 (R.M.) and the Israel Science Foundation 525/98 (A.P.).
References
- Béjà O. et al. (2000a) Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science, 289, 1902–1906. [DOI] [PubMed] [Google Scholar]
- Béjà O. et al. (2000b) Construction and analysis of bacterial artificial chromosome libraries from a marine microbial assemblage. Environ. Microbiol., 2, 516–529. [DOI] [PubMed] [Google Scholar]
- Béjà O., Spudich,E.N., Spudich,J.L., Leclerc,M. and DeLong,E.F. (2001) Proteorhodopsin phototrophy in the ocean. Nature, 411, 786–789. [DOI] [PubMed] [Google Scholar]
- Béjà O. et al. (2002) Comparative genomic analysis of archaeal genotypic variants in a single population and in two different oceanic provinces. Appl. Environ. Microbiol., 68, 335–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieszke J.A., Spudich,E.N., Scott,K.L., Borkovich,K.A. and Spudich,J.L. (1999) A eukaryotic protein, NOP-1, binds retinal to form an archaeal rhodopsin-like photochemically reactive pigment. Biochemistry, 38, 14138–14145. [DOI] [PubMed] [Google Scholar]
- Birge R.R. (1990) Nature of the primary photochemical events in rhodopsin and bacteriorhodopsin. Biochim. Biophys. Acta, 1016, 293–327. [DOI] [PubMed] [Google Scholar]
- Dioumaev A.K., Brown,L.S., Shih,J., Spudich,E.N., Spudich,J.L. and Lanyi,J.K. (2002) Proton transfers in the photochemical reaction cycle of proteorhodopsin. Biochemistry, 41, 5348–5358. [DOI] [PubMed] [Google Scholar]
- Eilers H., Pernthaler,J., Glockner,F.O. and Amann,R. (2000) Culturability and in situ abundance of pelagic bacteria from the North Sea. Appl. Environ. Microbiol., 66, 3044–3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elish M.E., Pierce,J.R. and Earhart,C.F. (1988) Biochemical analysis of spontaneous fepA mutants of Escherichia coli. J. Gen. Microbiol., 134, 1355–1364. [DOI] [PubMed] [Google Scholar]
- Friedrich T., Geibel,S., Kalmbach,R., Chizhov,I., Ataka,K., Heberle,J., Engelhard,M. and Bamberg,E. (2002) Proteorhodopsin is a light-driven proton pump with variable vectoriality. J. Mol. Biol., 321, 821–838. [DOI] [PubMed] [Google Scholar]
- Fuhrman J.A. (2002) Community structure and function in prokaryotic marine plankton. Antonie van Leeuwenhoek, 81, 521–527. [DOI] [PubMed] [Google Scholar]
- Garczarek L., Hess,W.R., Holtzendorff,J., van der Staay,G.W. and Partensky,F. (2000) Multiplication of antenna genes as a major adaptation to low light in a marine prokaryote. Proc. Natl Acad. Sci. USA, 97, 4098–4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez J.M., Simo,R., Massana,R., Covert,J.S., Casamayor,E.O., Pedrós-Alió,C. and Moran,M.A. (2000) Bacterial community structure associated with a dimethylsulfoniopropionate-producing North Atlantic algal bloom. Appl. Environ. Microbiol., 66, 4237–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho S.N., Hunt,H.D., Horton,R.M., Pullen,J.K. and Pease,L.R. (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene, 77, 51–59. [DOI] [PubMed] [Google Scholar]
- Kelly K.M. and Chistoserdov,A.Y. (2001) Phylogenetic analysis of the succession of bacterial communities in the Great South Bay (Long Island). FEMS Microbiol. Ecol., 35, 85–95. [DOI] [PubMed] [Google Scholar]
- Kochendoerfer G.G., Lin,S.W., Sakmar,T.P. and Mathies,R.A. (1999) How color visual pigments are tuned. Trends Biochem. Sci., 24, 300–305. [DOI] [PubMed] [Google Scholar]
- Lin S.W. and Sakmar,T.P. (1999) Colour tuning mechanisms of visual pigments. Novartis Found. Symp., 224, 124–135. [DOI] [PubMed] [Google Scholar]
- Lin S.W., Kochendoerfer,G.G., Carroll,K.S., Wang,D., Mathies,R.A. and Sakmar,T.P. (1998) Mechanisms of spectral tuning in blue cone visual pigments. Visible and Raman spectroscopy of blue-shifted rhodopsin mutants. J. Biol. Chem., 273, 24583–24591. [DOI] [PubMed] [Google Scholar]
- Luecke H., Schobert,B., Richter,H.T., Cartailler,J.P. and Lanyi,J.K. (1999) Structure of bacteriorhodopsin at 1.55 Å resolution. J. Mol. Biol., 291, 899–911. [DOI] [PubMed] [Google Scholar]
- Moore L.R., Rocap,G. and Chisholm,S.W. (1998) Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature, 393, 464–467. [DOI] [PubMed] [Google Scholar]
- Mullins T.D., Britcshgi,T.B., Krest,R.L. and Giovannoni,S.J. (1995) Genetic comparisons reveal the same unknown bacterial lineages in Atlantic and Pacific bacterioplankton communities. Limnol. Oceanogr., 40, 148–158. [Google Scholar]
- Nathans J. (1999) The evolution and physiology of human color vision: insights from molecular genetic studies of visual pigments. Neuron, 24, 299–312. [DOI] [PubMed] [Google Scholar]
- Ottolenghi M. and Sheves,M. (1989) Synthetic retinals as probes for the binding site and photoreactions in rhodopsins. J. Membr. Biol., 112, 193–212. [DOI] [PubMed] [Google Scholar]
- Pernthaler A., Pernthaler,J. and Amann,R. (2002) Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Microbiol., 68, 3094–3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappé M.S., Vergin,K. and Giovannoni,S.J. (2000) Phylogenetic comparisons of a coastal bacterioplankton community with its counterparts in open ocean and freshwater systems. FEMS Microbiol. Ecol., 33, 219–232. [DOI] [PubMed] [Google Scholar]
- Ren L., Martin,C.H., Wise,K.J., Gillespie,N.B., Luecke,H., Lanyi,J.K., Spudich,J.L. and Birge,R.R. (2001) Molecular mechanism of spectral tuning in sensory rhodopsin II. Biochemistry, 40, 13906–13914. [DOI] [PubMed] [Google Scholar]
- Spudich J.L., Yang,C.S., Jung,K.H. and Spudich,E.N. (2000) Retinylidene proteins: structures and functions from Archaea to humans. Annu. Rev. Cell. Dev. Biol., 16, 365–392. [DOI] [PubMed] [Google Scholar]
- Subramaniam S., Greenhalgh,D.A., Rath,P., Rothschild,K.J. and Khorana,H.G. (1991) Replacement of leucine-93 by alanine or threonine slows down the decay of the N and O intermediates in the photocycle of bacteriorhodopsin: implications for proton uptake and 13-cis-retinal–all-trans retinal reisomerization. Proc. Natl Acad. Sci. USA, 88, 6873–6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Macke,J.P. and Nathans,J. (1997) Mechanisms of spectral tuning in the mouse green cone pigment. Proc. Natl Acad. Sci. USA, 94, 8860–8865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M.T., Preston,C.M., Chavez,F.P. and DeLong,E.F. (2001) Quantitative mapping of bacterioplankton populations in seawater: field tests across an upwelling plume in Monterey Bay. Aquatic Microbiol. Ecol., 24, 117–127. [Google Scholar]
- Thompson J.D., Gibson,T.J., Plewniak,F., Jeanmougin,F. and Higgins,D.G. (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res., 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting C.S., Rocap,G., King,J. and Chisholm,S.W. (2002) Cyanobacterial photosynthesis in the oceans: the origins and significance of divergent light-harvesting strategies. Trends Microbiol., 10, 134–142. [DOI] [PubMed] [Google Scholar]
- Urbach E., Scanlan,D.J., Distel,D.L., Waterbury,J.B. and Chisholm,S.W. (1998) Rapid diversification of marine picophytoplankton with dissimilar light-harvesting structures inferred from sequences of Prochlorococcus and Synechococcus (Cyanobacteria). J. Mol. Evol., 46, 188–201. [DOI] [PubMed] [Google Scholar]
- Yan B., Spudich,J.L., Mazur,P., Vunnam,S., Derguini,F. and Nakanishi,K. (1995) Spectral tuning in bacteriorhodopsin in the absence of counterion and coplanarization effects. J. Biol. Chem., 270, 29668–29670. [DOI] [PubMed] [Google Scholar]
- Zubkov M.V., Fuchs,B.M., Burkill,P.H. and Amann,R. (2001) Comparison of cellular and biomass specific activities of dominant bacterioplankton groups in stratified waters of the Celtic Sea. Appl. Environ. Microbiol., 67, 5210–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]