Abstract
The assembly of antigen receptor genes by V(D)J recombination is initiated by the RAG1/RAG2 protein complex, which introduces double-strand breaks between recombination signal sequences and their coding DNA. Truncated forms of RAG1 and RAG2 are functional in vivo and have been used to study V(D)J cleavage, hybrid joint formation and transposition in vitro. Here we have characterized the activities of the full-length proteins. Unlike core RAG2, which supports robust transposition in vitro, full-length RAG2 blocks transposition of signal ends following V(D)J cleavage. Thus, one role of this non-catalytic domain may be to prevent transposition in developing lymphoid cells. Although full-length RAG1 and RAG2 proteins rarely form hybrid joints in vivo in the absence of non-homologous end-joining factors, we show that the full-length proteins alone can catalyze this reaction in vitro.
Keywords: hybrid joining/RAG1/RAG2/transposition/V(D)J recombination
Introduction
In order to generate a diverse repertoire of antigen receptors, immunoglobulin and T cell receptor genes are assembled from component gene segments by the process known as V(D)J recombination (reviewed in Bassing et al., 2002; Gellert, 2002). Each V, D and J gene segment is flanked by a recombination signal sequence (RSS), which is composed of a conserved heptamer and nonamer separated by a spacer whose length (12 or 23 base pairs), but not sequence, is conserved. Recombination of these gene segments is initiated by the RAG1 and RAG2 proteins. Together these proteins introduce a double-strand break between the coding DNA and the flanking RSS, resulting in the formation of blunt, 5′ phosphorylated signal ends and covalently sealed hairpin coding ends. In more detail, V(D)J cleavage is known to occur via a two-step reaction. RAG1 and RAG2 together bind to a pair of RSSs and introduce a nick at the 5′ end of each signal. They subsequently catalyze the attack by the resulting free 3′ hydroxyl groups on the opposing strands to generate a pair of hairpin coding ends and a pair of blunt signal ends. These ends are then resolved into precise signal joints and imprecise coding joints, with the help of DNA repair factors from the non-homologous end-joining (NHEJ) pathway, including Ku70/80, DNA-PKcs, XRCC4, Ligase IV and Artemis (reviewed in Bassing et al., 2002; Gellert, 2002; Schlissel, 2002).
Reconstitution of cleavage in vitro has usually been performed with catalytically active ‘core’ versions of the proteins (amino acids 384–1008 out of 1040 in RAG1 and amino acids 1–383 out of 527 in RAG2) that were more amenable to purification than the full-length proteins (Gellert, 2002). These proteins also support V(D)J recombination in vivo. While the purified core RAG proteins cannot, on their own, rejoin the broken ends to form the standard products of V(D)J recombination, they are able to mediate resolution of the broken DNA by two alternative pathways. In one pathway, the 3′ hydroxyl group of the signal end attacks a hairpinned coding end to form an open-and-shut joint or a hybrid joint (Melek et al., 1998). Hybrid joints, where the coding DNA originally adjacent to the 12RSS is joined to the 23RSS and vice versa, are observed in vivo, as are open-and-shut joints (Lewis et al., 1988; Morzycka-Wroblewska et al., 1988). In a second pathway, the core RAG proteins can carry out transposition in vitro, catalyzing the attack of the signal ends into an unrelated target sequence (Agrawal et al., 1998; Hiom et al., 1998; Melek and Gellert, 2000). RAG-mediated transposition has yet to be observed in vivo.
Although the C-terminus of RAG2 and the N-terminus of RAG1 appear to be dispensable for the basic recombination reaction in vivo and for V(D)J cleavage, transposition and hybrid joint formation in vitro, these regions are highly conserved throughout evolution (Litman et al., 1999; Peixoto et al., 2000 and references therein). Moreover, in vivo experiments have demonstrated differences between the behavior of full-length and core RAG proteins. First, the frequency of V(D)J recombination on exogenous or integrated substrates in fibroblast cells is lower with the core than full-length proteins (Sadofsky et al., 1993, 1994; Silver et al., 1993; Cuomo and Oettinger, 1994; Kirch et al., 1996). Secondly, recombination by the core RAG proteins in fibroblast cells leads to a greater accumulation of signal ends than is observed with full-length proteins (Steen et al., 1999). Thirdly, the absence of the N-terminus of RAG1 results in reduced D-to-J rearrangement, with differential effects observed on the assembly of endogenous T cell receptor and immunoglobulin genes (Roman et al., 1997; Noordzij et al., 2000; Santagata et al., 2000). Fourthly, in both pro-B cell lines and mice that express core RAG2, a mild reduction in DH-to-JH joining is seen, while VH-to-DJH joining is substantially reduced (Kirch et al., 1998; Liang et al., 2002; Akamatsu et al., 2003). Finally, both the full-length and core RAG proteins can form hybrid joints in cells expressing NHEJ factors. In the absence of NHEJ factors, only the core RAG proteins can efficiently initiate hybrid joining (Sekiguchi et al., 2001).
Given the differences between the behavior of the full-length and core RAG proteins in vivo, we sought to characterize the function of the full-length proteins in vitro. We describe here the expression and purification of the full-length RAG1 and RAG2 proteins, and the comparison of their performance in cleavage, transposition and hybrid joint formation. We find that hybrid joint formation occurs in vitro in the presence of full-length RAG proteins, suggesting that the substantial reduction in hybrid joining in NHEJ-deficient cells must require additional factors present in vivo. However, the C-terminal domain of RAG2 blocks the capture of unrelated target DNA when coding DNA is present, thereby preventing transposition. This observation suggests a means by which the RAG proteins avoid causing unwanted genomic instability.
Results
Expression and purification of full-length RAG1 and RAG2
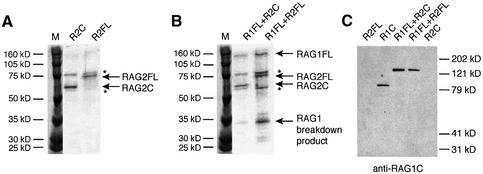
In order to compare the biochemical activities of the core and full-length RAG proteins, we used vaccinia virus vectors for expression of the full-length proteins in HeLa cells (see Materials and methods). Full-length RAG2 (RAG2FL), containing amino acids 1–527, was expressed with N-terminal FLAG (Hopp et al., 1988) and polyhistidine tags for purification purposes, and a C-terminal influenza hemagglutinin (HA) tag (Colman et al., 1987), which allows verification of the presence of the C-terminus. Core RAG2 (RAG2C), containing amino acids 1–383, and the same N- and C-terminal tags was expressed and purified in the same manner. The resulting proteins are shown in Figure 1A.
Fig. 1. Purification of full-length RAG1 and RAG2. (A and B) Coomassie-stained 10% SDS polyacrylamide gels with purified RAG proteins. Positions of RAG2C, RAG2FL and RAG1FL are indicated. Additional products present in all vaccinia preparations are marked with an asterisk. (C) Western blot stained with a polyclonal anti-RAG1 core antibody.
RAG1C, containing amino acids 384–1040, and an N-terminal polyhistidine tag was purified from Escherichia coli with high yield and activity. However, active RAG1FL (amino acids 1–1040) could not be obtained from E.coli and was therefore expressed with N-terminal FLAG and polyhistidine tags, as well as a C-terminal polyhistidine tag, using the vaccinia expression system. Coexpression with RAG2 (either core or full-length; see Figure 1B) enhanced the activity of RAG1FL (Figure 2B). An N-terminal break-down product of RAG1 (as determined by western analysis; data not shown), remains in the RAG1 preparations, but this product is too small to contain catalytic residues and therefore cannot contribute to the activity observed. The other bands visible on the gel (marked by asterisks) are present in all vaccinia preparations, and are not portions of the RAG proteins (Figure 1C; data not shown).
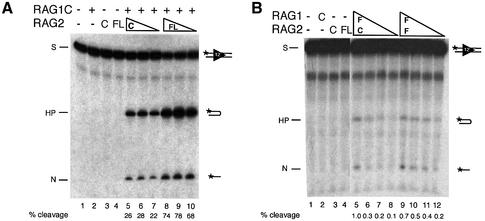
Fig. 2. Full-length RAG1 and RAG2 are active in cleavage at a single RSS. (A) Cleavage reactions were performed with decreasing amounts of RAG2C (40, 20 and 10 ng) and RAG2FL (20, 10 and 5 ng) in the presence of 150 ng RAG1C and resolved on a denaturing polyacrylamide gel. Positions of the substrate (S), nick (N) and hairpin (HP) are indicated. The percentage of the substrate cleaved (including both hairpin and nicked products) was determined by phosphoimager analysis and is indicated below the gel. (B) Cleavage reactions were performed as above with decreasing amounts of RAG1FL, copurified with either RAG2C or RAG2FL (∼80, 40, 20 and 10 ng of combined protein) as indicated.
Full-length proteins are active in single-site and coupled cleavage assays
When combined with RAG1C, RAG2FL was ∼5-fold more active per mole than RAG2C in cleaving an oligonucleotide containing a single RSS (Figure 2A, compare lanes 6 and 7 with 8 and 9). Both RAG2C and RAG2FL were titrated to determine the protein concentration that allows maximum cleavage. RAG2FL cleaves up to 80% of the substrate at its optimal concentration, whereas RAG2C cleaves only ∼30% of the substrate. It is unclear whether the increased cleavage is due to higher specific activity of the protein itself, or due to a higher percentage of active protein within the preparation. However, several preparations of RAG2FL were uniformly more active than preparations of RAG2C.
We also examined the ability of RAG1C/RAG2FL to cleave a pair of RSSs on a linearized plasmid substrate. The conditions of this assay permit cleavage only in the presence of both a 12RSS and a 23RSS, following the 12/23 rule (Eastman et al., 1996; van Gent et al., 1996). Once again, RAG1C/RAG2FL is able to cleave a higher percentage of the substrate than RAG1C/RAG2C (Figure 3A, compare lane 5 with 9). Although single-site cleavage events are evident, they still depend on the presence of a 12/23 RSS pair. No cleavage occurs on substrates that contain either two 12RSSs or a 12RSS alone (Figure 3C). Thus, in conjunction with RAG1, RAG2FL is more active than RAG2C and performs coupled cleavage following the dictates of 12/23 restriction.
Fig. 3. Full-length RAG1 and RAG2 are active in coupled cleavage and follow the 12/23 rule. (A) RAG2FL is more active in coupled cleavage than RAG2C. Lanes 5–7 contain 10 ng RAG2C and decreasing amounts of RAG1C (300, 150 and 50 ng). Lanes 8–10 contain 300 ng of RAG1C and decreasing amounts of RAG2FL (20, 10 and 5 ng). All reactions were performed on a substrate containing both 12 and 23 RSSs. Substrates and products resulting from coupled cleavage and single-site cleavage at either the 12RSS (filled triangles) or 23RSS (open triangles) are indicated. (B) RAG1FL is active in coupled cleavage. Lane 1 contains 100 ng RAG1C and 20 ng RAG2FL. Lane 2 contains ∼20 ng of copurified RAG1FL and RAG2FL. Lane 3 contains ∼40 ng of copurified RAG1FL and RAG2C. All reactions were performed on a substrate containing both 12 and 23 RSSs. All lanes are from the same exposure of the gel. Substrates and products resulting from coupled cleavage are indicated as above. (C) Cleavage by RAG1FL and RAG2FL is 12/23 restricted. Lanes 1, 3 and 5 contain 100 ng RAG1C and 20 ng RAG2FL. Lanes 2, 4 and 5 contain ∼80 ng of combined RAG1FL and RAG2FL. Reactions in lanes 1 and 2 were performed on a substrate containing a 12RSS and a 23RSS, lanes 3 and 4 on a substrate containing two 12RSSs, and lanes 5 and 6 on a substrate that contained only one 12RSS. Substrates and products resulting from coupled cleavage are indicated as above.
In the presence of RAG2C or RAG2FL, RAG1FL also cleaves an oligonucleotide containing a single RSS (Figure 2B) and mediates coupled cleavage on a plasmid substrate (Figure 3B), but its activity is substantially reduced compared with RAG1C. It is unclear whether lower activity is due to interference from the N-terminal fragment, a lower specific activity of the protein itself or a lower percentage of active protein within the preparation. Nonetheless, since any catalytic activity in the preparation must come from the full-length protein, the biochemical behavior of the full-length and core RAG proteins could be compared.
Full-length RAG proteins can mediate hybrid joint formation in vitro
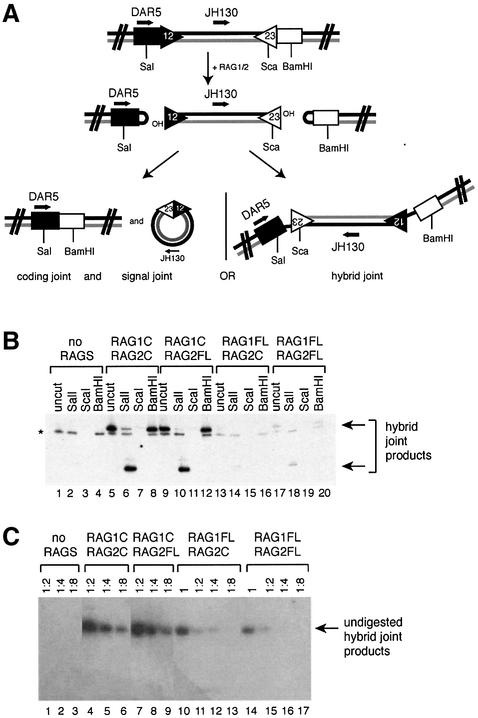
The verification that the full-length RAG1 and RAG2 proteins can carry out coupled cleavage allowed us to test their ability to mediate non-canonical resolutions of the cleavage complex. The observation that the full-length RAG proteins are much less effective than core RAGs at hybrid joint formation in vivo in an NHEJ-deficient background (Sekiguchi et al., 2001) led us to investigate whether the full-length proteins themselves are responsible for this inhibition. Previous work showed that the products of an in vitro coupled cleavage assay included hybrid joints along with the standard broken molecules (Melek et al., 1998). Thus, we used similar reaction conditions to those used for detecting cleavage of a pair of RSSs (Figure 3), but then subjected the reaction products to PCR using primers designed to specifically amplify hybrid joints (Figure 4A). The presence of hybrid joints was also confirmed by restriction digestion of the amplified products (see Figure 4).
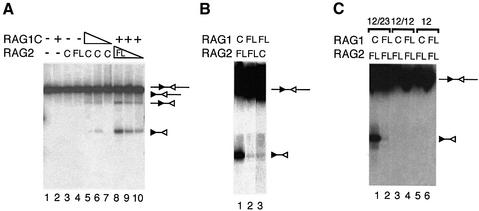
Fig. 4. Full-length RAG proteins can mediate hybrid joint formation. (A) Schematic diagram of the PCR assay used to detect hybrid joint formation. Positions of PCR primers, restriction sites and RSSs are indicated. (B) A plasmid substrate containing a 12RSS and 23RSS was incubated with RAG proteins under conditions that allow coupled cleavage (as described in Materials and methods), and the products were subjected to PCR with the primers DAR5 and JH130 as shown above. The PCR products were subjected to digestion with restriction enzymes to confirm the identity of the hybrid joint products, as shown. Digestion with SalI shortens the product, while ScaI cuts within the site recognized by the probe, preventing detection of a product. BamHI does not cut within the hybrid joint PCR product. Correct hybrid joint products are indicated with arrows. An asterisk denotes a PCR product not specific to the hybrid joint. (C) Two-fold serial dilutions of undigested hybrid joint products were amplified with PCR primers DAR5 and JH130.
In the presence of RAG1C, RAG2FL readily forms hybrid joints (Figure 4B, lanes 9–12). RAG1FL can also mediate hybrid joint formation, in the presence of either core or full-length RAG2 (Figure 4B, lanes 13–16 and 17–20). PCR performed on serial dilutions of the cleavage reaction indicates that RAG2FL is at least as active as RAG2C in hybrid joint formation. Given that the cleavage activity of RAG2FL is increased compared with RAG2C, it appears that hybrid joint formation is slightly reduced relative to cleavage in vitro in the presence of RAG2FL. However, this reduction in relative activity is estimated to be at most 2- to 4-fold, whereas the inhibition observed in vivo is ∼100-fold (Sekiguchi et al., 2001). The activity of RAG1FL is reduced ∼4- to 8-fold compared with RAG1C (Figure 4C), but RAG1FL still clearly forms hybrid joints. (This modest reduction is in keeping with the observed reduction in RAG1FL’s cleavage activity.) Thus, there is no significant inhibition of hybrid joint formation when the RAG1/2 complex includes full-length RAG1 or RAG2, either singly or together.
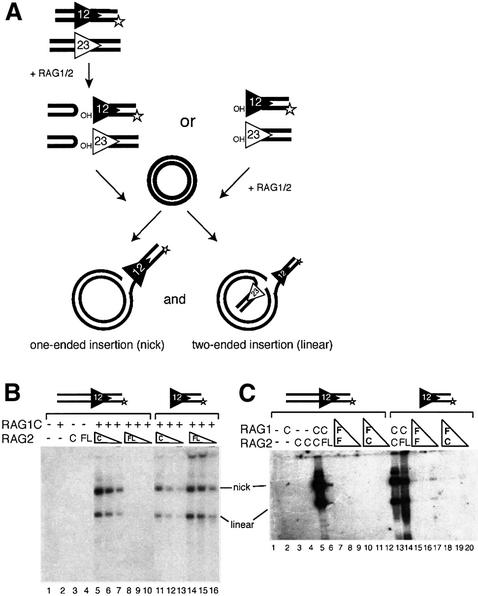
RAG2FL does not support efficient transposition following V(D)J cleavage
We next compared the ability of the full-length and core RAG proteins to transpose an RSS end derived from cleavage of a labeled ‘intact’ oligonucleotide donor (containing an RSS and flanking ‘coding’ DNA) into a plasmid substrate (see Figure 5A). Surprisingly, although it is more active in cleavage, RAG2FL with RAG1C is significantly less active than RAG2C with RAG1C in transposition of donor DNA arising from a cleavage reaction (Figure 5B, compare lanes 5–7 with 8–10, and C, compare lanes 5 and 6). The experiment was performed over a range of temperatures and concentrations of Mg2+ and HMG, but under all conditions tested, RAG2FL was less active than RAG2C (data not shown). Thus, the ‘non-core’ portion of RAG2 interferes with transposition in vitro.
Fig. 5. Full-length RAG proteins inhibit transposition of an intact substrate. (A) Schematic diagram of the transposition reaction. Intact and pre-cleaved substrates, as well as the two possible products of transposition (one-ended insertion resulting in a nicked plasmid or two-ended insertion resulting in a linearized plasmid) are shown. (B) Transposition reactions were performed with 20, 10 and 5 ng of RAG2C (lanes 5–7 and 11–13) and RAG2FL (lanes 8–10 and 14–16) in the presence of 150 ng core RAG1. Lanes 1–10 contain intact substrate, while lanes 11–16 contain pre-cleaved substrate. (C) Transposition reactions were performed with 80, 40 or 20 ng of RAG1FL copurified with RAG2FL (lanes 7–9 and 15–17) or RAG2C (lanes 10–12 and lanes 18–20). Lanes 1–12 contain intact substrate, while lanes 13–20 contain pre-cleaved substrate. Transposition with 150 ng RAG1C and 20 ng RAG2C (lanes 5 and 13) and 150 ng RAG1C and 20 ng RAG2FL (lanes 6 and 14) are also shown.
RAG2FL transposes a pre-cleaved donor
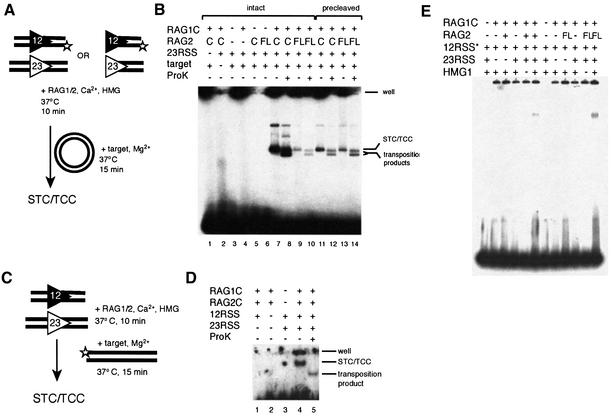
Transposition involves a series of steps: synaptic complex formation, RAG-catalyzed coupled cleavage of substrate DNA, the capture of the target DNA to form a target capture complex [target capture complex (TCC), composed of RAG proteins with donor and target DNA], and RAG-catalyzed strand transfer of the signal ended donor into the target DNA. In order to determine the step at which transposition is inhibited by RAG2FL, we looked at the ability of RAG2FL in conjunction with RAG1 to transpose a ‘pre-cleaved’ oligonucleotide donor, a duplex oligonucleotide that terminates in a blunt RSS with a 3′ hydroxyl, the same structure that is generated by V(D)J cleavage (see Figure 5A for schematic). Because RAG1C/RAG2FL exhibits robust cleavage activity and hence should have no difficulty generating the signal-ended donor, we again expected to see that RAG1C/RAG2FL was less active than RAG1C/RAG2C in transposition of a pre-cleaved substrate. Instead, we found that RAG1C/RAG2FL is more active in transposition with a pre-cleaved substrate than with an intact substrate (Figure 5B, compare lanes 14–16 with 8–10). Moreover, RAG1C/RAG2FL transposed the pre-cleaved donor more efficiently than did RAG1C/RAG2C (Figure 5B, compare lanes 14–16 with 11–13). This result may be due in part to the greater ability of RAG1C/RAG2FL to bind DNA with blunt signal ends, as RAG1C/RAG2FL forms a signal end complex (SEC) more readily than does RAG1C/RAG2C (Figure 6E).
Fig. 6. RAG2FL forms STC/TCC less efficiently than RAG2C. (A) Schematic diagram of the target capture assay, using a plasmid target and intact or pre-cleaved labeled oligonucleotide substrates. (B) RAG proteins were incubated with a labeled 12RSS, cold 23RSS and cold target plasmid for a short time to allow formation of a TCC and STC. One half of each reaction was treated with proteinase K and SDS to remove proteins and reveal transposition products. Lanes 7, 8, 11 and 13 contain RAG1C and RAG2C, lanes 9, 10, 13 and 14 contain RAG1C and RAG2FL. Lanes 1–10 contain intact substrate and lanes 11–14 contain pre-cleaved substrate. (C) Schematic diagram of the target capture assay using a labeled oligonucleotide target. (D) Core RAG proteins were incubated with cold 12RSS and 23RSS and a labeled oligonucleotide target for a short time to allow formation of TCC and STC. One half of each reaction was treated with proteinase K and SDS to remove proteins and reveal transposition products. The difference between the intensity of the bands with and without deproteination allows assessment of the percentage of TCC in the TCC/STC combined complex. (E) SEC formation. RAG proteins were incubated with a labeled 12RSS and cold 23RSS in the presence of HMG. Complexes were resolved on a 5% native polyacrylamide gel.
RAG2FL blocks target capture in the presence of coding DNA
The observation that a pre-cleaved substrate worked well as a donor, whereas an intact substrate did not, suggested that it was the presence of coding DNA from the intact substrate that posed a problem for RAG1C/RAG2FL. We therefore reasoned that RAG1C/RAG2FL might have difficulty capturing a plasmid target when coding DNA was present. We tested this idea by assessing the ability of the RAG proteins to generate a TCC. This complex is found in a mixture with the strand transfer complex (STC), in which the covalent links between the donor and the target have been made, but the proteins remained bound to the DNA. We used a labeled oligonucleotide substrate and unlabeled target plasmid to follow the formation of these complexes (see Materials and methods; Figure 6A) and used short incubation times to favor the formation of TCC over STC.
With an intact substrate, RAG1C/RAG2FL forms the STC/TCC mixed species much less efficiently than RAG1C/RAG2C (Figure 6B, compare lanes 9 and 10 with 7 and 8). Moreover, RAG1C/RAG2FL forms STC/TCC more readily with a pre-cleaved substrate than with an uncleaved substrate (Figure 6B, compare lanes 13 and 14 with 9 and 10). This is in sharp contrast to RAG1C/RAG2C where the amount of STC plus TCC generated from an intact donor significantly exceeds that from the pre-cleaved substrate (Figure 6B, compare lanes 7 and 8 with 11 and 12). In fact, the average ratio of STC plus TCC formed by RAG1C/RAG2C with intact compared with pre-cleaved substrates is ∼10, while for RAG1C/RAG2FL it is ∼0.9.
Does the lower level of STC/TCC formed with RAG1C/RAG2FL on an intact substrate reflect a decrease in target capture, decreased strand transfer, or both? The results in Figure 5 (lanes 14–16) already suggest that the strand transfer step is not appreciably inhibited. In order to distinguish between these possibilities more definitively, we needed to confirm that TCC was a substantial component of the STC/TCC band. The amount of STC present in the band of STC plus TCC can be determined by treating the reaction mixture with proteinase K to liberate the transposition product from bound protein. In the previous assay (Figure 6B, lanes 7, 9, 11 and 13), a large fraction of bound DNA did not enter the agarose gel, preventing direct quantification of the ratio of STC to TCC. To circumvent this problem, we used a labeled oligonucleotide target to assemble the STC and TCC complexes, in conjunction with unlabeled donor oligonucleotides and the RAG proteins (see Materials and methods; Figure 6C) (Neiditch et al., 2001). After forming STC/TCC under the same conditions used in Figure 6B, one half of the reaction was treated with SDS and proteinase K, while the other half was subjected to a mock treatment, and the resulting products were resolved on a native polyacrylamide gel. Consistent with previous observations (Neiditch et al., 2001; Tsai et al., 2002), ∼70% of the material in the STC plus TCC band is TCC, while 30% is the covalent STC transposition product (Figure 6D). (The results for RAG1C/RAG2C are presented here, because the amount of TCC formed by RAG1C/RAG2FL in this experiment was below the level of detection.) Because a large fraction of the STC plus TCC band is TCC, the decrease in STC plus TCC formation observed with RAG1C/RAG2FL (Figure 6B) must involve a decrease in target capture in the presence of coding DNA.
RAG1FL was also tested (together with RAG2C or RAG2FL) for its ability to mediate transposition (Figure 5C). Transposition was observed with a pre-cleaved donor, but not with an intact substrate. Thus, RAG1FL can catalyze transposition, but it is unclear whether the failure to detect transposition of the intact substrate reflects the low cleavage activity of RAG1FL or an active block of the later steps of transposition.
Discussion
The core versions of the RAG1 and RAG2 proteins contain the regions minimally required for V(D)J cleavage and recombination, but the high conservation among vertebrate species of the removed portions implies that they perform important functions. While a number of in vivo experiments have suggested roles for the non-core regions in the regulation of recombination and possibly in the handling of the DNA ends after cleavage, an in vitro comparison of the full-length and core RAG proteins was not possible until now due to difficulty in expressing the full-length versions. Here we show directly that full-length RAG2 fails to support efficient transposition of signal-ended donor DNA following V(D)J cleavage. A role for the RAG2 C-terminus in the inhibition of transposition has also been observed by Schatz and colleagues (C.-L.Tsai and D.G.Schatz, personal communication). However, full-length RAG1 and RAG2 are able to carry out hybrid joint formation in vitro.
The hybrid joints formed solely by the core RAG proteins in vitro are precise or nearly precise, resulting from RAG-mediated attack by the signal end on the hairpin coding end at or near the tip, with no further addition or loss of nucleotides, and they are joined only on the strand where the transesterification occurred (Melek et al., 1998). In vivo, core RAG proteins mediate formation of this same type of precise but incomplete joint, both in cells expressing all the NHEJ factors and in NHEJ-deficient cells (Bogue et al., 1997; Han et al., 1997; Sekiguchi et al., 2001). In addition, hybrid joints that have undergone extensive modification at the junction and are sealed on both ends are made in vivo in the presence of either core or full-length RAG proteins. However, these are generally dependent on NHEJ proteins and thus occur only in wild-type cells. In contrast, in NHEJ-deficient cells, the full-length proteins form very few hybrid joints, of which the majority contain large deletions and appear to have been repaired by the homologous end joining pathway (Sekiguchi et al., 2001). Although the in vivo results could have resulted from the full-length proteins blocking hybrid joint formation by the direct RAG-mediated pathway, our data now show that the full-length RAG proteins cannot be entirely responsible for this effect. Similar results have been obtained by Schatz and colleagues, although they observe a slightly greater reduction in hybrid joint formation by RAG2FL as compared with RAG2C (perhaps reflecting differences in the protein preparations). Thus, there may be factors in vivo (perhaps even a subset of the NHEJ proteins themselves) that, in conjunction with the full-length RAG proteins, prevent hybrid joint formation by this pathway.
Our results clearly demonstrate that RAG2FL blocks transposition of an intact substrate. Since, in the presence of RAG1, RAG2FL cleaves an intact substrate more robustly than RAG2C, the blocking of transposition by RAG2FL must occur after cleavage. In conjunction with RAG1, RAG2FL makes SEC but fails to form substantial amounts of TCC, indicating that target capture is the step at which transposition is blocked. Because RAG2FL does efficiently transpose a pre-cleaved substrate, it is clear that the protein itself can support target capture and strand transfer. Thus, the inhibition of target capture must be due to some effect of the coding DNA. One possibility is that after cleavage, coding DNA is bound more strongly by RAG2FL than by RAG2C. The continued presence of the coding DNA might then physically block target capture. It has been suggested that the target DNA and coding ends may occupy the same portion of the binding site of the RAG recombinase (Tsai et al., 2002). Our data support this hypothesis and suggest that the C-terminus of RAG2 either forms part of this binding site or influences its action. Although others have reported that a target can be captured when coding ends are still present, this form of TCC appears to be significantly less stable (Neiditch et al., 2001).
Based on their in vivo hybrid joining experiments, Sekiguchi et al. predicted that the full-length proteins would not only inhibit hybrid joining but would also inhibit transposition, as the basic biochemistry of the two reactions is the same (Sekiguchi et al., 2001). In both reactions, the hydroxyl group of the signal end attacks another DNA molecule: in one case, a hairpinned coding end, in the other, an unrelated target sequence. One explanation for why we find a block in transposition but not in hybrid joint formation is that transposition requires the capture and introduction of a new DNA molecule into the post-cleavage complex, whereas hybrid joining does not. Our data are consistent with a model in which RAG2FL holds on more tightly to coding ends after cleavage, inhibiting target capture, but holding them in a fashion that permits hybrid joint formation. While hybrid joining does not require capture of new DNA, it most likely requires a conformational change that allows exchange of signal and coding ends. Whatever conformational changes are required for hybrid joint formation, these must be largely permitted by the RAG proteins acting alone. The decrease in direct hybrid-joining seen with RAG2FL in vivo may indicate that in vivo conditions impose greater constraints on a post-cleavage complex formed with full-length proteins than with their core counterparts, such that the full-length RAG proteins are not positioned to directly catalyze hybrid joining. Alternatively, or in addition, the full-length RAG proteins may interact with cellular factors that block the full-length proteins from mediating a direct attack on the hairpin coding DNA.
Although several studies have examined the ability of RAG1 and RAG2 to mediate inter- and intramolecular transposition in vitro (Agrawal et al., 1998; Hiom et al., 1998; Melek and Gellert, 2000), no studies have been able to show evidence of transposition in vivo, using either core or full-length RAG proteins. It has been postulated that there are mechanisms in place in vivo that prevent transposition, since any transposition events in vivo could cause chromosomal translocations and genomic instability. The C-terminal tail of RAG2 may provide one layer of inhibition in vivo, but additional layers must also be involved.
Recent work has revealed that the RAG1 and RAG2 proteins are required for the completion of V(D)J recombination in vivo, even after the initial cleavage stage (Jones and Gellert, 2001; Neiditch et al., 2001; Tsai et al., 2002). It has been proposed that the RAG proteins have an architectural role, holding the cleaved ends in appropriate conformations and perhaps recruiting repair factors (Jones and Gellert, 2001; Neiditch et al., 2001; Tsai et al., 2002). In this work we have provided the first direct biochemical evidence for a post-cleavage property of the non-core portion of RAG2 that affects the resolution stage of the V(D)J recombination reaction. Thus, the RAG recombinase itself is involved in regulating the outcome of the recombination reaction and, at least in vitro, in preventing unwanted transposition events.
Materials and methods
DNA substrates
Oligonucleotide substrates VDJ100/101 (12RSS with flanking coding sequence), VDJ132/133 (23RSS with flanking coding DNA), VDJ104/106 (12RSS, pre-cleaved), YD24/VDJ134 (23RSS, pre-cleaved) and mm30t/mm30b (target for TCC assay) (Neiditch et al., 2001) were gel purified, annealed, and 5′-end labeled with [32P]ATP as described previously (Cuomo et al., 1996). Plasmid substrates pDVG42 (12/23), pPS12 (12/12), pPS14 (12 alone) and pPS15 (23 alone) were digested with AatII and gel-purified before use in coupled cleavage assays. Oligonucleotide sequences are as described in Cuomo et al. (1996) and Kim et al. (1999).
Vector and recombinant vaccinia virus construction
The NcoI–SmaI fragment from LG4.3, containing RAG2FL with a C-terminal HA tag, was subcloned into the pTM1-derived vector pTM3F9n, which contains FLAG and His9 tags positioned just upstream of the NcoI site and downstream of a bacteriophage T7 promoter, to create the plasmid pSRK6. The NcoI–SphI fragment from LG6, containing RAG1FL, was subcloned into pDRK534 (Kim et al., 1999) to create pSRK7. The NcoI–XhoI fragment from pSRK7, containing RAG1FL, was subcloned into pTM3F9n as above. Recombinant vaccinia viruses were generated using standard conditions and desired recombinants were isolated by XGPRT selection as described previously (Ausubel et al., 1989). Recombinant plaques were expanded to high titer using HeLa S3 cells.
Purification of proteins and western analysis
Recombinant RAG2C, recombinant RAG2FL, and RAG1FL, coexpressed with either core or full-length RAG2 from vaccinia infection of HeLa cells, were expressed and purified as described previously (McBlane et al., 1995). Recombinant RAG1C was purified from E.coli as described previously (Kim et al., 1999). Purified proteins were resolved on 10% polyacrylamide gels and either stained with Coomassie Blue or transferred to nitrocellulose membranes and subjected to western blot analysis (Ausubel et al., 1989) with anti-His antibodies (Santa Cruz Biotechnology; data not shown) or a chicken polyclonal anti-core RAG1 (a kind gift of Dr Nadja Patenge).
Cleavage assays
Standard oligonucleotide cleavage assays were performed essentially as described previously (Kim and Oettinger, 1998), except that samples were incubated for 2 h at 30°C.
Coupled cleavage assays were performed as described previously (van Gent et al., 1996) using the plasmid substrates described above. Samples were separated on a 1% TAE agarose gel, and gels were transferred to nitrocellulose membranes by Southern blot. Southern blots were hybridized overnight at a temperature of 68°C with a 0.9 kb probe derived from a BamHI–SalI digest of pDVG42 (representing the region between the RSSs) and 32P-labeled by random priming. Blots were visualized by autoradiography.
Hybrid joint assay
Hybrid joint assays were performed following the method described in Melek et al. (1998).
Transposition
In vitro transposition reactions were carried out as described previously (Kim et al., 1999).
STC/TCC capture
For detection of the STC and TCC transposition intermediates using labeled target DNA, initial binding reactions contained 25 mM K-morpholinepropanesulfonic acid (MOPS) (pH 7.0), 4 mM dithiothreitol, 75 mM potassium glutamate, 5 mM CaCl2, 100 µg of bovine serum albumin/ml, 80 ng of RAG1, 20 ng of RAG2, 50 ng of HMG1 and 0.02 pmol each of uncleaved 12RSS and 23RSS oligonucleotide donor (VDJ100/101 and VDJ132/133, respectively) in a final volume of 6.5 µl. Binding was allowed to proceed at 37°C for 10 min, after which MgCl2 (5 mM, final concentration), dimethyl sulfoxide (DMSO) (10%, final w/v ratio) and 0.1 pmol of labeled oligonucleotide target (mm30t/mm30b) were added (10 µl final reaction volume). Reaction mixtures were then incubated for an additional 15 min. The reactions were divided in half, with SDS (1%, final w/v ratio) and proteinase K (200 ng/ml final concentration) added to one half but not the other. After both samples were incubated for 15 min at 37°C, 2 µl of 50% glycerol were added to each and the samples were then loaded directly onto a 6% non-denaturing, DNA-retardation polyacrylamide gel (Novex) and run at 90 V for 90 min in 0.5× Tris–borate-EDTA at 4°C. Dried gels were visualized by phosphoimager analysis and autoradiography.
For STC/TCC capture with labeled donor DNA, reaction conditions are as described above except that a labeled 12RSS oligonucleotide was substituted for the unlabeled 12RSS, and 0.1 pmol of unlabeled pUC19 plasmid target was substituted as the target. Entire reaction mixtures were separated on a 1% agarose-ME gel, run at 80 V for 120 min in 1× Tris–acetate-EDTA at 4°C.
Electrophoretic mobility shift assay
Binding reactions were performed and resolved on native polyacrylamide gels as described previously (Mundy et al., 2002).
Acknowledgments
Acknowledgements
We thank Nadja Patenge for the kind gift of the polyclonal anti-RAG1 core antibody, and critical reading of the manuscript. We also thank Anne Clatworthy and Katrina Morshead for critical discussions. We are grateful to Chia-Lun Tsai and David Schatz for communicating results prior to publication. A.G.M. is a Howard Hughes Medical Institute Predoctoral Fellow. This work was supported by the National Institutes of Health grant RO1 GM48026 (M.A.O.).
References
- Agrawal A., Eastman,Q.M. and Schatz,D.G. (1998) Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature, 394, 744–751. [DOI] [PubMed] [Google Scholar]
- Akamatsu Y. et al. (2003) Deletion of the RAG2 C terminus leads to impaired lymphoid development in mice. Proc. Natl Acad. Sci. USA, 100, 1209–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (1989) Current Protocols in Molecular Biology. Greene Publishing Associates–Wiley Interscience, New York, NY.
- Bassing C.H., Swat,W. and Alt,F.W. (2002) The mechanism and regulation of chromosomal V(D)J recombination. Cell, 109, Suppl., S45–S55. [DOI] [PubMed] [Google Scholar]
- Bogue M.A., Wang,C., Zhu,C. and Roth,D.B. (1997) V(D)J recombination in Ku86-deficient mice: distinct effects on coding, signal and hybrid joint formation. Immunity, 7, 37–47. [DOI] [PubMed] [Google Scholar]
- Colman P.M., Laver,W.G., Varghese,J.N., Baker,A.T., Tulloch,P.A., Air,G.M. and Webster,R.G. (1987) Three-dimensional structure of a complex of antibody with influenza virus neuraminidase. Nature, 326, 358–363. [DOI] [PubMed] [Google Scholar]
- Cuomo C.A. and Oettinger,M.A. (1994) Analysis of regions of RAG-2 important for V(D)J recombination. Nucleic Acids Res., 22, 1810–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuomo C.A., Mundy,C.L. and Oettinger,M.A. (1996) DNA sequence and structure requirements for cleavage of V(D)J recombination signal sequences. Mol. Cell. Biol., 16, 5683–5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman Q.M., Leu,T.M. and Schatz,D.G. (1996) Initiation of V(D)J recombination in vitro obeying the 12/23 rule. Nature, 380, 85–88. [DOI] [PubMed] [Google Scholar]
- Gellert M. (2002) V(D)J recombination: rag proteins, repair factors and regulation. Annu. Rev. Biochem., 71, 101–132. [DOI] [PubMed] [Google Scholar]
- Han J.O., Steen,S.B. and Roth,D.B. (1997) Ku86 is not required for protection of signal ends or for formation of nonstandard V(D)J recombination products. Mol. Cell. Biol., 17, 2226–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiom K., Melek,M. and Gellert,M. (1998) DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell, 94, 463–470. [DOI] [PubMed] [Google Scholar]
- Hopp T.P., Prickett,K.S., Price,V., Libby,R.T., March,C.J., Cerretti,P., Urdal,D.L. and Conlon,P.J. (1988) A short polypeptide marker sequence useful for recombinant protein identification and purification. Biotechnology, 6, 1205–1210. [Google Scholar]
- Jones J.M. and Gellert,M. (2001) Intermediates in V(D)J recombination: a stable RAG1/2 complex sequesters cleaved RSS ends. Proc. Natl Acad. Sci. USA, 98, 12926–12931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.R. and Oettinger,M.A. (1998) Functional analysis of coordinated cleavage in V(D)J recombination. Mol. Cell. Biol., 18, 4679–4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.R., Dai,Y., Mundy,C.L., Yang,W. and Oettinger,M.A. (1999) Mutations of acidic residues in RAG1 define the active site of the V(D)J recombinase. Genes Dev., 13, 3070–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirch S.A., Sudarsanam,P. and Oettinger,M.A. (1996) Regions of RAG1 protein critical for V(D)J recombination. Eur. J. Immunol., 26, 886–891. [DOI] [PubMed] [Google Scholar]
- Kirch S.A., Rathbun,G.A. and Oettinger,M.A. (1998) Dual role of RAG2 in V(D)J recombination: catalysis and regulation of ordered Ig gene assembly. EMBO J., 17, 4881–4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis S.M., Hesse,J.E., Mizuuchi,K. and Gellert,M. (1988) Novel strand exchanges in V(D)J recombination. Cell, 55, 1099–1107. [DOI] [PubMed] [Google Scholar]
- Liang H.-E., Hsu,L.-Y., Cado,D., Cowell,L.G., Kelsoe,G. and Schlissel,M.S. (2002) The ‘dispensable’ portion of RAG2 is necessary for efficient V-to-DJ rearrangement during B and T cell development. Immunity, 17, 639–651. [DOI] [PubMed] [Google Scholar]
- Litman G.W., Anderson,M.K. and Rast,J.P. (1999) Evolution of antigen binding receptors. Annu. Rev. Immunol., 17, 109–147. [DOI] [PubMed] [Google Scholar]
- McBlane J.F., van Gent,D.C., Ramsden,D.A., Romeo,C., Cuomo,C.A., Gellert,M. and Oettinger,M.A. (1995) Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell, 83, 387–395. [DOI] [PubMed] [Google Scholar]
- Melek M. and Gellert,M. (2000) RAG1/2-mediated resolution of transposition intermediates: two pathways and possible consequences. Cell, 101, 625–633. [DOI] [PubMed] [Google Scholar]
- Melek M., Gellert,M. and van Gent,D.C. (1998) Rejoining of DNA by the RAG1 and RAG2 proteins. Science, 280, 301–303. [DOI] [PubMed] [Google Scholar]
- Morzycka-Wroblewska E., Lee,F.E. and Desiderio,S.V. (1988) Unusual immunoglobulin gene rearrangement leads to replacement of recombinational signal sequences. Science, 242, 261–263. [DOI] [PubMed] [Google Scholar]
- Mundy C.L., Patenge,N., Matthews,A.G. and Oettinger,M.A. (2002) Assembly of the RAG1/RAG2 synaptic complex. Mol. Cell. Biol., 22, 69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiditch M.B., Lee,G.S., Landree,M.A. and Roth,D.B. (2001) RAG transposase can capture and commit to target DNA before or after donor cleavage. Mol. Cell. Biol., 21, 4302–4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordzij J.G., Verkaik,N.S., Hartwig,N.G., de Groot,R., van Gent,D.C. and van Dongen,J.J. (2000) N-terminal truncated human RAG1 proteins can direct T-cell receptor but not immunoglobulin gene rearrangements. Blood, 96, 203–209. [PubMed] [Google Scholar]
- Peixoto B.R., Mikawa,Y. and Brenner,S. (2000) Characterization of the recombinase activating gene-1 and 2 locus in the Japanese pufferfish, Fugu rubripes. Gene, 246, 275–283. [DOI] [PubMed] [Google Scholar]
- Roman C.A., Cherry,S.R. and Baltimore,D. (1997) Complementation of V(D)J recombination deficiency in RAG-1–/– B cells reveals a requirement for novel elements in the N-terminus of RAG-1. Immunity, 7, 13–24. [DOI] [PubMed] [Google Scholar]
- Sadofsky M.J., Hesse,J.E., McBlane,J.F. and Gellert,M. (1993) Expression and V(D)J recombination activity of mutated RAG-1 proteins. Nucleic Acids Res., 21, 5644–5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadofsky M.J., Hesse,J.E. and Gellert,M. (1994) Definition of a core region of RAG-2 that is functional in V(D)J recombination. Nucleic Acids Res., 22, 1805–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santagata S., Gomez,C.A., Sobacchi,C., Bozzi,F., Abinun,M., Pasic,S., Cortes,P., Vezzoni,P. and Villa,A. (2000) N-terminal RAG1 frameshift mutations in Omenn’s syndrome: internal methionine usage leads to partial V(D)J recombination activity and reveals a fundamental role in vivo for the N-terminal domains. Proc. Natl Acad. Sci. USA, 97, 14572–14577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlissel M.S. (2002) Does artemis end the hunt for the hairpin-opening activity in V(D)J recombination? Cell, 109, 1–4. [DOI] [PubMed] [Google Scholar]
- Sekiguchi J.A., Whitlow,S. and Alt,F.W. (2001) Increased accumulation of hybrid V(D)J joins in cells expressing truncated versus full-length RAGs. Mol. Cell, 8, 1383–1390. [DOI] [PubMed] [Google Scholar]
- Silver D.P., Spanopoulou,E., Mulligan,R.C. and Baltimore,D. (1993) Dispensable sequence motifs in the RAG-1 and RAG-2 genes for plasmid V(D)J recombination. Proc. Natl Acad. Sci. USA, 90, 6100–6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen S.B., Han,J.O., Mundy,C., Oettinger,M.A. and Roth,D.B. (1999) Roles of the ‘dispensable’ portions of RAG-1 and RAG-2 in V(D)J recombination. Mol. Cell. Biol., 19, 3010–3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C.L., Drejer,A.H. and Schatz,D.G. (2002) Evidence of a critical architectural function for the RAG proteins in end processing, protection and joining in V(D)J recombination. Genes Dev., 16, 1934–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gent D.C., Ramsden,D.A. and Gellert,M. (1996) The RAG1 and RAG2 proteins establish the 12/23 rule in V(D)J recombination. Cell, 85, 107–113. [DOI] [PubMed] [Google Scholar]