Abstract
The RAG1 and RAG2 proteins perform critical DNA recognition and cleavage functions in V(D)J recombination, and also catalyze efficient DNA transposition in vitro. No transposition in vivo by the RAG proteins has been reported, suggesting regulation of the reaction by as yet unknown mechanisms. Here we report that RAG-mediated transposition is suppressed by physiological concentrations of the guanine nucleotide GTP, and by the full-length RAG2 protein. Both GTP and full-length RAG2 inhibit transposition by blocking the non-covalent ‘capture’ of target DNA, and both are capable of inhibiting RAG-mediated hybrid joint formation in vitro. We also observe that another intracellular signaling molecule, Ca2+, stimulates RAG-mediated transposition and is capable of activating transposition even in reactions containing full-length RAG2 and GTP. RAG-mediated transposition has been proposed to contribute to the chromosomal translocations that underlie the development of lymphoid malignancies, and our findings highlight regulatory mechanisms that might prevent such occurrences, and circumstances in which these regulatory mechanisms could be overcome.
Keywords: Ca2+/GTP/RAG1/RAG2/RAG-mediated transposition/V(D)J recombination
Introduction
In developing lymphocytes, germline gene coding segments are rearranged to assemble functional immunoglobulin genes and T cell receptor genes by a process known as V(D)J recombination (Fugmann et al., 2000a; Gellert, 2002). V(D)J recombination is targeted by specific DNA sequences, known as recombination signal sequences (RSSs), that flank the coding segments. RSSs consist of a highly conserved heptamer (consensus 5′-CAC AGTG) and an AT-rich nonamer (consensus 5′-ACA AAAACC) separated by a spacer of either 12 or 23 base pairs (referred to as the 12-RSS and 23-RSS). Initiation of V(D)J recombination requires two lymphocyte-specific proteins, encoded by the recombination activating genes, RAG1 and RAG2 (Schatz et al., 1989; Oettinger et al., 1990). The RAG proteins, in conjunction with high mobility group proteins HMG1 or 2, recruit a 12-RSS and a 23-RSS into a synaptic complex (Hiom and Gellert, 1998; Jones and Gellert, 2002) and then introduce DNA double-stranded breaks at the junctions of the RSSs and their flanking coding segments (McBlane et al., 1995). DNA cleavage results in two covalently sealed hairpin coding ends and two blunt signal ends with 5′ phosphate and 3′ hydroxyl groups at their termini.
The non-homologous end-joining machinery completes the V(D)J recombination reaction by processing and joining the coding ends to form the coding joint (CJ), and aligning and ligating the signal ends to form the signal joint (SJ) (Grawunder and Harfst, 2001). While CJ and SJ formation utilize a common DNA repair pathway, they are distinct in several regards. CJ formation is typically associated with nucleotide addition and/or deletion at the junction, whereas SJ formation is usually precise. CJ formation proceeds rapidly, whereas SJ formation is thought to be a slow process, as suggested by the long half-life of signal end fragments (Ramsden and Gellert, 1995; Livàk and Schatz, 1997).
Signal end fragments are abundant in developing lymphocytes (Roth et al., 1992; Schlissel et al., 1993) and are found in a complex with the RAG proteins (Agrawal and Schatz, 1997; Hiom and Gellert, 1998; Perkins et al., 2002). In vitro, signal end fragments can be inserted into a target DNA molecule in a transposition reaction catalyzed by the RAG proteins (Agrawal et al., 1998; Hiom et al., 1998). Mechanistically, this reaction closely resembles the reactions carried out by several bacterial transposases as well as retroviral integrase (Fugmann et al., 2000a). Paradoxically, although RAG-mediated transposition is quite efficient in vitro, such activity has not yet been reported in vivo. Several models have been proposed to explain how the deleterious effects of RAG-mediated transposition might be limited. First, it was found that high Mg2+ concentrations promote the reversal of transposition, a process known as disintegration (Melek and Gellert, 2000). Such a mechanism could help prevent stable insertion of a signal end fragment into a new location, but would not necessarily prevent DNA damage and instability at the site of insertion (Hiom et al., 1998; Roth and Craig, 1998). Secondly, there is evidence that the RAG proteins can capture target DNA prior to DNA cleavage, which would result in preferential integration of the transposable element near the site of excision, thereby restricting the regions of the genome that could be damaged (Neiditch et al., 2001). In vivo and in vitro, the RAG proteins are capable of catalyzing hybrid joint formation, a reaction with close mechanistic parallels to transposition (Melek et al., 1998). Strikingly, RAG-mediated hybrid joint formation in vivo is suppressed by the N-terminal region of RAG1 (aa 1–383) and the C-terminal region of RAG2 (aa 388–527) (Sekiguchi et al., 2001), regions of the RAG proteins that lie outside of the ‘core’ domains required for catalysis. Thus, a third mechanism to regulate transposition in vivo could be contributed by the non-essential regions of the RAG proteins (Sekiguchi et al., 2001). The first two mechanisms would limit, but not prevent, damage, while the third has yet to be tested directly.
Propagation of transposable elements results in alterations of the host genome and can be detrimental to both hosts and transposons. To reach a balance between the survival of transposable elements and their hosts, transposition needs to be tightly regulated (Labrador and Corces, 1997). A number of transposases have been shown to be regulated by nucleotides (Stellwagen and Craig, 1998). For example, the excision and insertion of the Tn7 transposable element is control by TnsC, an ATPase and an ATP-dependent DNA-binding protein (Peters and Craig, 2001); transposition of bacteriophage Mu is influenced by the MuB protein, an ATP-dependent activator of MuA transposase (Yamauchi and Baker, 1998); and the binding of GTP is necessary for the activity of Drosophila P element transposase (Kaufman and Rio, 1992). The striking parallels between the RAG proteins and other transposases (van Gent et al., 1996; Melek et al., 1998; Fugmann et al., 2000a) prompted us to ask whether nucleotides might also regulate transposition by RAG1/RAG2. Here we report that RAG-mediated transposition is strongly and selectively inhibited by physiological concentrations GTP. Importantly, GTP inhibits the reaction by impairing the ability of the RAG proteins to interact with target DNA, and does not interfere with the primary function of the RAG proteins, DNA cleavage. Furthermore, we find that the C-terminal region of the RAG2 protein inhibits both RAG-dependent hybrid joint formation and transposition in vitro, in accord with the in vivo observations and hypothesis of Sekiguchi et al. (2001).
Results
GTP selectively inhibits RAG-mediated transposition
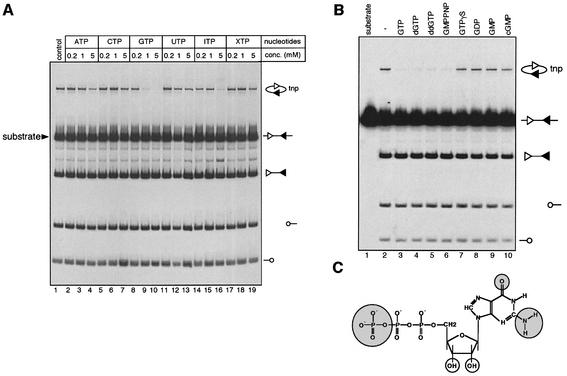
To investigate whether activities of the RAG proteins are regulated by nucleotide(s), we utilized a standard in vitro cleavage assay involving purified HMG2, truncated ‘core’ RAG1 and RAG2, and a linear body labeled DNA substrate containing an appropriate pair of RSSs. This assay allows simultaneous measurement of cleavage and transposition mediated by the RAG proteins (Agrawal et al., 1998). DNA cleavage by the RAG proteins results in the production of blunt signal end fragments and two hairpin coding ends, while intramolecular transposition of the signal end fragment results in circular DNA products, some of which can be visualized in this assay because they migrate at a distinct position above the input substrate during gel electrophoresis (Figure 1A).
Fig. 1. GTP is a specific inhibitor of RAG-mediated transposition. (A) The effect of nucleotides on the cleavage and transposition activities of the RAG1/RAG2 proteins was assessed using a body-labeled cleavage substrate (arrowhead) containing a 12-RSS and a 23-RSS in the presence of 10 mM MgCl2. Structures of the substrate and reaction products are indicated on the right, with the 12-RSS and 23-RSS depicted as open and filled triangles, respectively, and coding ends as open circles. The concentration of the nucleotides used is indicated above each lane. tnp, intramolecular transposition product. The reactions products were analyzed on a native 4% (acryamide:bis = 29:1) polyacryamide gel and quantitated using a PhosphorImager (Molecular Dynamics). (B) Effect of GTP derivatives (1 mM) on the activities of the RAG proteins was assessed using the same assay as in (A). (C) Structure of GTP with the functional groups critical for GTP inhibition highlighted in shaded circles and the functional groups that are dispensable highlighted in open circles.
Addition of various nucleotides to the reactions had no discernible effect on RAG-mediated DNA cleavage, but transposition was inhibited in a dose-dependent manner by GTP (lanes 8, 9 and 10). In contrast to other nucleotides such as ATP and ITP, which exhibited only moderate inhibitory effects at high concentration (5 mM, lanes 4 and 16, respectively), GTP inhibited RAG-mediated transposition at submillimolar concentrations (Figure 1A, lane 8, and see below), which closely mimic the average intracellular GTP concentration, estimated to be 468 ± 224 µM (Traut, 1994). To define further the functional groups of GTP critical for the inhibition of RAG-mediated transposition, GTP derivatives were assayed for their ability to inhibit transposition (Figure 1B). Deoxy GTP and dideoxy GTP exhibited a comparable inhibitory effect to that of GTP (Figure 1B, compare lanes 3 and 4 with 2), indicating that the 2′- and 3′-hydroxyl groups of the sugar moiety are not required. In contrast, changes in the triphosphate moiety (GDP, GMP and cGMP) strongly reduced inhibitory activity (Figure 1B, lanes 8–10), indicating that the γ phosphate plays a critical role. We also examined the effect of non-hydrolyzable GTP analogs, GMPPNP and GTPγS. GMPPNP is as potent an inhibitor of transposition as GTP, indicating that hydrolysis of the β–γ phosphate bond is not required. In contrast, GTPγS was not active at concentrations where GTP and GMPPNP strongly reduce RAG-mediated transposition. The reasons for this are not clear. One possibility is that substitution of a non-bridging oxygen with sulfur changes the way GTPγS interacts with the transposition machinery, as has been observed for some GTP-binding proteins (Cherfils et al., 1997).
GTP inhibits RAG-mediated transposition by blocking target capture
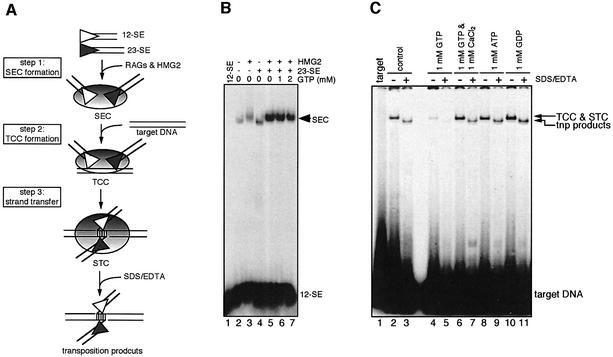
RAG-mediated transposition proceeds through a series of distinct DNA–protein complexes that can be reconstituted individually in vitro using purified RAG proteins, HMG2 and various DNA substrates (shown in Figure 2A). Because GTP appears to exert its function after DNA cleavage, we examined its effect on the formation of various post-cleavage DNA–RAG complexes. The assay for formation of the signal end complex (SEC) (step 1 in Figure 2A) was carried out by incubating the RAG proteins and HMG2 with a labeled 12-signal end substrate and an unlabeled 23-signal end substrate. SEC formation is strongly stimulated by the presence of both signal ends, the HMG2 protein and the RAG proteins (Figure 2B, compare lane 5 with 2, 3 and 4), and GTP has no effect on formation of this complex at concentrations where RAG-mediated transposition is strongly suppressed (compare lane 5 with 6 and 7). Therefore, GTP appears to act at a step downstream of SEC formation. We next assembled the target capture complex (TCC) and strand transfer complex (STC) from the preformed SEC and a labeled target oligonucleotide (Figure 2A, steps 2 and 3) (Neiditch et al., 2001; Tsai et al., 2002). Incubation of the SEC with target DNA resulted in the generation of a slowly migrating DNA–protein complex consisting of the TCC and the STC (Figure 2C, lane 2). To visualize the transposition products and estimate the relative contribution of the TCC and STC to the complex, we treated the reaction with SDS/EDTA, which separates non-covalently linked target DNA from signal ends. By comparing lanes 2 and 3, we estimate that ∼40% of the DNA–protein complex seen in lane 2 was derived from the STC. Addition of 1 mM GTP drastically reduced the formation of the DNA–protein complex (lane 4) and the generation of intermolecular transposition products (lane 5), suggesting that GTP inhibits RAG-mediated transposition by blocking the formation of a stable TCC. As expected, other nucleotides such as ATP and GDP do not have any inhibitory effect on the formation of the DNA–protein complexes (lanes 9 and 11, respectively) or transposition (lanes 10 and 12, respectively). We conclude that GTP inhibits transposition by selectively blocking the ability of the RAG proteins in the SEC to capture target DNA, although some inhibitory effect on strand transfer cannot be ruled out.
Fig. 2. Effect of GTP on the formation of post-cleavage complexes. (A) Schematic diagram of the DNA–RAG1/RAG2 complexes leading to transposition. (B) SEC formation reactions were carried out using a 5′ end-labeled 12-signal end oligonucleotide in a reaction buffer containing 4 mM MgCl2 in the presence or absence of HMG2 and unlabeled 23-signal end, and various amount of GTP, as indicated above the lanes. Reactions were analyzed on a native 6% (80:1) polyacrylamide gel. (C) TCC formation and intermolecular transposition reactions were carried out in a two-stage fashion. First, the SEC was formed as in (B) except that both the 12-signal end and 23-signal end were unlabeled, and reactions contained nucleotide (1 mM) or 1 mM CaCl2 as indicated above the lanes. Formation of the TCC and STC were initiated by adding 5′-end-labeled oligonucleotide target DNA. Deproteinized transposition products were visualized by treating the reaction with SDS/EDTA prior to loading on a native 6% (80:1) polyacrylamide gel.
Identification of a putative GTP-binding domain in RAG1
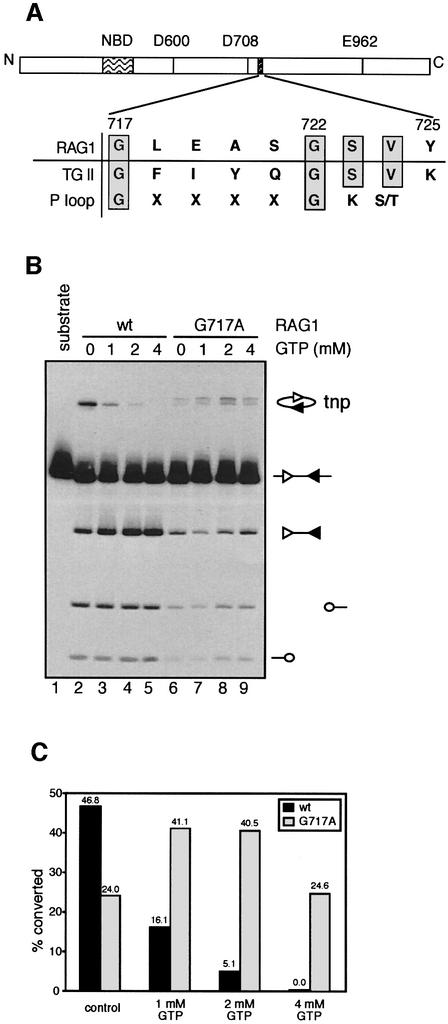
To explore further how GTP exerts it inhibitory effect, we searched for a GTP-binding domain in the amino acid sequences of RAG1, RAG2 and HMG2. None of these proteins contains a canonical GTP-binding domain such as is found in p21 ras or the α-subunit of trimeric G proteins. However, we found an evolutionary conserved region in RAG1 (aa 717–725) with some similarity to the GTP-binding domain of Gh/transglutaminase II (TGII) (Figure 3A; Iismaa et al., 2000). Interestingly, the GXXXXG motif in this region of RAG1 also resembles the consensus sequence of the P-loop motif that is found in many nucleotide-binding proteins, where it functions to coordinate phosphates of the bound nucleotide (Saraste et al., 1990).
Fig. 3. Identification of a putative GTP-binding domain. (A) Schematic diagram of the murine RAG1 protein depicting three active site residues (D600, D708 and E962), the nonamer-binding domain (NBD), and the putative GTP-binding domain (aa 717–725). A sequence alignment of RAG1, the GTP-binding domain of TGII and the consensus sequence of the P loop is shown, with identical residues shown in gray boxes. (B) Effect of GTP on the activities of the RAG1 mutant G717A was assessed using the assays described in Figure 1A in the presence of various concentrations of GTP as indicated above the lanes, except that the concentration of MgCl2 was 6 mM. (C) Quantitation of transposition in (B). Transposition activity was calculated as the ratio of the intensity of the transposition product to the sum of the intensities of the signal end fragment and the transposition product.
To determine whether this region of RAG1 plays a role in regulating the transposition activity of the RAG proteins, we mutated the amino acid residues shared between RAG1 and TGII. The purified mutant RAG proteins were then assayed for their cleavage and transposition activity in the presence or absence of GTP. Among the mutant RAG proteins we screened, substitution of glycine 717 with alanine (G717A) reduced cleavage activity, while transposition activity (calculated as the fraction of signal end DNA converted to transposition products) remained comparable with that of wild-type RAG1 (Figure 3B, compare lane 2 with 6). Strikingly, GTP did not inhibit transposition catalyzed by G717A RAG1, even at concentrations as high as 4 mM (Figure 3B and C). We observed that the transposition products generated by G717A migrated as a doublet in the gel. Restriction enzyme analyses of the transposition products revealed that the doublet arose from the use of a different spectrum of transposition target sites by G717A compared with wild-type RAG1 (data not shown). Hence, transposition catalyzed by G717A RAG1 is resistant to inhibition by GTP and exhibits altered target preferences. We conclude that G717 of RAG1 makes important contributions to the regulation of RAG-mediated transposition by GTP. Our results are consistent with the possibility that GTP exerts its effects by interacting with the region of RAG1 that includes aa 717–725, although extensive efforts to detect specific binding of GTP to RAG1, RAG2 or HMG2 have not been successful (data not shown). Further support for a role of this region of RAG1 in controlling transposition (and other post- cleavage activities of the RAG proteins) comes from our analyses of RAG1 proteins mutated at position 723 (Tsai et al., 2002). S723A and S723C RAG1 perform cleavage almost normally but exhibit a severe defect in transposition and hybrid joint formation. The inability of these mutants to perform transposition is due to a failure in target capture, which mimics the effect of GTP (Tsai et al., 2002).
Additionally, we found that substitution of the second conserved glycine with alanine (G722A) strongly reduced DNA cleavage (data not shown). In contrast, substitution of serine 721 (which is located in the non-conserved region of the P-loop motif) with alanine did not alter cleavage or transposition by the RAG proteins (data not shown). Overall, our results are consistent with the possibility that this region of RAG1 functions in a manner similar to that of a P-loop motif.
Full-length RAG2 exhibits a reduced transposition activity compared with core RAG2
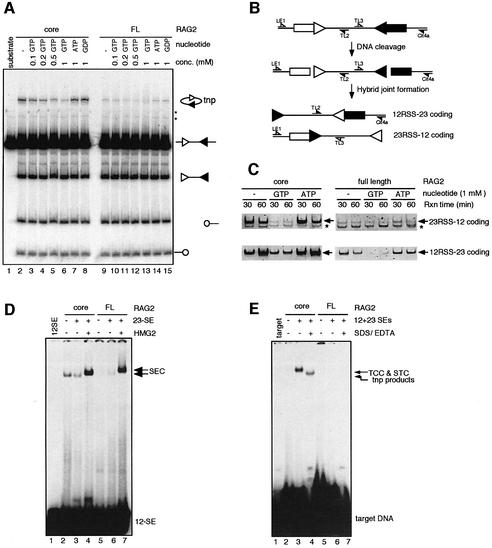
It has been shown that the ‘dispensable’ parts of the RAG proteins, namely the N-terminal region of RAG1 (aa 1–383) and C-terminal region of RAG2 (aa 388–527) play a critical role in vivo in suppressing RAG-mediated hybrid joint formation (Sekiguchi et al., 2001), a reaction in which signal ends attack and become joined to a different coding end (diagrammed in Figure 4B). The mechanistic similarity between RAG-mediated hybrid joint formation and transposition (Melek et al., 1998) prompted us to ask if these parts of the RAG proteins also suppress RAG-mediated transposition. We focused on the full-length RAG2 protein because full-length RAG1 has been difficult to analyze due to insolubility. We compared the transposition activity of the full-length and core RAG2 proteins (together with GST-core RAG1), and found that while the two proteins exhibited comparable cleavage activity, full-length RAG2 was much less active for transposition (Figure 4A, lanes 2 and 9). GTP inhibited transposition activity of both RAG2 proteins in a dose-dependent manner (compare lane 2 with 3–6, and lane 9 with 10–13). Other nucleotides such as ATP and GDP do not have any discernible inhibitory effect on RAG-mediated transposition (lanes 7, 8, 14 and 15). In keeping with the previous study (Sekiguchi et al., 2001), the full-length RAG2 protein exhibited a reduced activity in hybrid joint formation in vitro compared with the core RAG2 protein (Figure 4C). Hybrid joint formation by both core and full-length RAG2 is also inhibited by GTP (Figure 4C). Full-length RAG2 and 1 mM GTP each reduce hybrid joint formation by 5- to 10-fold, while the combination of the two results in a 20- to 50-fold reduction (as revealed by titration of the input substrate for the hybrid joint PCR; see Supplementary figure 1, available at The EMBO Journal Online). RAG2, including the C-terminal region, reduces transposition by impairing the ability of the RAG proteins to capture target DNA (Figure 4E, compare lane 3 with 6), but has no effect on formation of the SEC (Figure 4D, compare lane 4 with 7). Inhibition of transposition in vitro by the full-length RAG2 protein has also recently been observed by others (Elkin et al., 2003). Hence, GTP and the C-terminal region of RAG2 use similar mechanisms to inhibit RAG-mediated transposition, and both are able to inhibit hybrid joint formation.
Fig. 4. Full-length RAG2 exhibits reduced transposition and hybrid joint formation activities compared with core RAG2. (A) Standard in vitro 12/23 coupled cleavage reactions were carried out in the presence of 4 mM MgCl2 and various concentrations of nucleotides as indicated. The RAG proteins used in the reaction were partially purified from 293T cells co-expressing the GST-core RAG1 fusion protein and either core RAG2 (lanes 2–8) or full-length RAG2 (lanes 9–15). Interaction of these proteins with substrate DNA generates a faint smear at approximately the position of the transposition product (most easily seen in lanes 12 and 13). (B) Schematic diagram depicting the formation of hybrid joints, in which RSSs become linked to the opposite coding end. Half arrows represent PCR primers used for hybrid joint detection. (C) In vitro 12/23 coupled cleavage reactions were carried out in the presence of 1 mM GTP or ATP as indicated for 30 or 60 mins, and reaction products were used as templates for PCR amplification of hybrid joints. Prior to the analyses of hybrid joint formation by PCR, the templates were analyzed for the accumulation of cleavage products and were found to be comparable at all time points (data not shown). Asterisks indicate non-specific PCR products, while arrows indicate hybrid joint products. (D) The SEC formation assay was performed using MBP-core RAG1 together with either GST-core RAG2 (lanes 2–4) or GST-full-length RAG2 (lanes 5–7). (E) The assays for the TCC and STC formation and intermolecular transposition were carried out using the RAG proteins as indicated. The reaction conditions are identical to those described in Figure 2C.
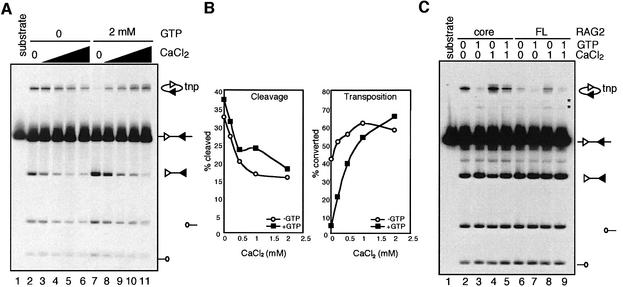
Ca2+ stimulates RAG-mediated transposition and overcomes GTP inhibition
Ca2+ supports RAG-mediated transposition in the absence of other divalent metal ions when precleaved RSSs are used (Hiom et al., 1998). The activities of TGII are regulated reciprocally by GTP and Ca2+, with GTP being an inhibitor and Ca2+ being an activator (Nakaoka et al., 1997). Identification of a RAG1 mutant resistant to GTP inhibition based on the sequence similarity between RAG1 and TGII, prompted us to ask whether Ca2+ can stimulate RAG-mediated transposition and antagonize GTP inhibition. As the CaCl2 concentration is increased, RAG-mediated cleavage decreases, but the efficiency of transposition increases (Figure 5A and B). More importantly, in the presence of 2 mM GTP, which efficiently suppresses RAG-mediated transposition, even low concentrations of CaCl2 readily overcome the GTP inhibition (Figure 5A, compare lane 7 with 8–11). Ca2+ exerts this effect by allowing the RAG proteins to capture the target DNA (Figure 2C, compare lanes 4 and 6). A similar relationship between GTP and Ca2+ was also demonstrated for full-length RAG2 (Figure 5C): Ca2+ stimulates transposition (lane 8) and can overcome inhibition by GTP (lane 9). Hence, as for TGII, GTP and Ca2+ exert reciprocal effects on RAG-mediated transposition. The minimal concentration of CaCl2 required for partial reversal of GTP inhibition is ∼100 µM (Figure 5A; data not shown). Since intracellular Ca2+ levels rarely exceed 5 µM (Alberts et al., 1994), it remains uncertain whether transient Ca2+ fluxes, such as those associated with B cell receptor signaling, could stimulate RAG-mediated transposition in developing lymphocytes.
Fig. 5. (A) The effect of CaCl2 on the activities of the RAG proteins and GTP inhibition of transposition was assayed using standard coupled cleavage assays (4 mM MgCl2) in the presence or absence of 2 mM GTP and increasing concentrations of CaCl2 (0.2, 0.5, 1 and 2 mM). (B) Quantitation of the results of (A). (C) The effect of 1 mM CaCl2 on transposition by core RAG2 and full-length RAG2 in the presence or absence of 1 mM GTP.
Discussion
Several types of genomic damage could be envisioned to arise from transposition mediated by RAG1/RAG2. Signal end fragments could act as insertional mutagens, and could disrupt the function of essential or tumor suppressor genes. In addition, the RAG proteins might act within the immediate product of insertion (the STC) to create chromosomal double-strand breaks and thereby contribute to chromosomal translocations (Roth and Craig, 1998; Melek and Gellert, 2000). Finally, movement of the RAG transposon, like many other transposons, might lead to more complex genome rearrangements (Kleckner et al., 1996).
It seems vital, therefore, that the transposase activity of the RAG proteins be tightly regulated. This is made all the more important by the apparently long half-life of the SEC inside normal lymphocytes (Ramsden and Gellert, 1995; Livàk and Schatz, 1997), and by the finding that SJs are not necessarily an inert, ‘safe’ product (Neiditch et al., 2002). Transposition is often regulated at the level of transposase expression (Labrador and Corces, 1997), but this is not feasible for RAG1/RAG2 because of its essential role in lymphocyte development. A mechanism that imposes inhibition at a post-cleavage step unique to transposition, and hence does not interfere with the recombinase functions of RAG1/RAG2, is appealing. Consistent with this idea, we find that both GTP and the C-terminal region of RAG2 specifically block the ability of the RAG proteins to interact with target DNA without compromising their DNA cleavage activity in vitro. A mechanism of this sort, which prevents productive interaction with target DNA, is clearly preferable to those previously articulated, which propose transposon disintegration (Melek and Gellert, 2000) or a constraint in the distance the transposon can jump (Neiditch et al., 2001).
Do the observations made in vitro reflect what takes place in vivo? In the absence of an assay for RAG-mediated transposition in vivo, this question cannot be definitively answered. Several factors, however, suggest that the observations reported here are relevant in vivo. First, GTP partially inhibits transposition at concentrations as low as 200 nM, and the concentration of GTP in the cell is ∼500 nM, a range in which GTP is an effective inhibitor. Secondly, GTP inhibition is highly selective and requires specific functional groups presented on GTP, as changes in hydrogen bond donors render GTP derivatives poor inhibitors. Thirdly, RAG1 contains a short region with sequence similarity to the GTP-binding domain of TGII, a protein that is reciprocally regulated by GTP and Ca2+. Mutation of RAG1 residues, predicted to be critical for regulating RAG-mediated transposition and/or for interacting with GTP, resulted in resistance to inhibition by GTP (G717A), a profound defect in transposition (S723A and S723C; Tsai et al., 2002) or aberrant cleavage activity (G722A). In contrast, mutation of a position predicted to be non-essential (S721) yielded a protein with no discernible defect in either DNA cleavage or transposition. These observations support the idea that GTP is a regulator of RAG-mediated transposition in vivo and that this effect is mediated by a specific GTP-binding domain in the RAG proteins (which includes aa 717–723 in RAG1). We note that this region of RAG1 lies close to the catalytic aspartate residue at aa 708 and immediately adjacent to a putative zinc-finger domain (aa 723–754; Rodgers et al., 1996) that has been suggested to interact with RAG2 (Aidinis et al., 2000). It is therefore easy to imagine that this region influences the structure of the active site and/or interactions of RAG1 with RAG2, and that GTP could influence one or both of these critical parameters by binding to this area. It is conceivable that this region of RAG1 makes direct contact with target DNA and that GTP interferes with this interaction. It is worth noting that, unlike many DNA transposases whose activities are positively regulated by nucleotide factors, transposition by the RAG proteins is negatively regulated by GTP.
A major motivation for our analysis of transposition by different forms of RAG2 was the previous finding that the C-terminal region of RAG2 (and the N-terminal region of RAG1) inhibit hybrid joint formation in vivo (Sekiguchi et al., 2001). These findings strongly predicted that the full-length RAG2 protein would inhibit hybrid joint formation in vitro, and led to the hypothesis that this region of RAG2 would also inhibit transposition (Sekiguchi et al., 2001). We have verified both of these predictions. Some hybrid joint formation is still observed in reactions containing full-length RAG2 and core RAG1 (Figure 4C; Supplementary figure 1), consistent with the in vivo results of Sekiguchi et al. (2001), but we cannot be sure that the inhibitory effect of the C-terminal domain of RAG2 is as strong in vitro as it is in vivo. Given the mechanistic similarities between transposition and hybrid joint formation (van Gent et al., 1996; Melek et al., 1998), and the ability of the C-terminal domain of RAG2 to inhibit hybrid joint formation in vivo and transposition in vitro, it seems plausible that this region of RAG2 also inhibits transposition in vivo.
The C-terminal region of RAG2 appears to be an independent folded domain (Liang et al., 2002) and plays a significant role in recombination of a subset of endogenous antigen receptor loci (Kirch et al., 1998; Liang et al., 2002), and importantly, enhances the formation of coding and signal joints while reducing the accumulation of signal end intermediates in transient V(D)J recombination assays (Steen et al., 1999). It has therefore been speculated that this portion of RAG2 contributes to the remodeling or disassembly of post-cleavage complexes prior to end joining (Steen et al., 1999; Gellert, 2002). Further support for this idea comes from our recent observation that the C-terminal region of RAG2 exacerbates the recombination defect of RAG1 mutants deficient in post-cleavage steps of the reaction (Tsai et al., 2002). Thus, it is plausible that the full-length RAG2 protein facilitates conformational changes or subunit reorganization in the signal end complex which block target capture.
The minimal Ca2+ concentration required to stimulate transposition by full-length RAG2 or in the presence of GTP in vitro (≈100 nM) are unlikely to be achieved in vivo. Thus, we view this observation as a cautionary note that the mechanisms that inhibit RAG-mediated transposition are reversible and that, under some circumstances, RAG-mediated transposition can be activated.
While GTP and the C-terminal region of RAG2 inhibit transposition at the same step, the capture of target DNA, it is not known whether they act by similar mechanisms. The effect of GTP appears modest in the context of full-length RAG2, and might be less obvious still if the full-length RAG1 protein were also used. Inhibition by GTP may be a failsafe, or backup, mechanism necessary to ensure a very low incidence of RAG-mediated transposition. Develop ing lymphocytes are abundant and highly proliferative, and undesired events, even rare, can lead to severe consequences.
It has been proposed that RAG1/RAG2 entered the early vertebrate genome as a transposable element, and that germline mobilization of this element was responsible for creating the primordial ‘split’ antigen receptor gene (Sakano et al., 1979; Oettinger et al., 1990; Thompson, 1995; Agrawal et al., 1998; Hiom et al., 1998). If correct, this model implies that the RAG transposon was active in vertebrate cells, at least during an early phase of vertebrate evolution, and it seems likely that the transposition activity would have needed to be suppressed once antigen receptor loci were established. Thus, it is possible that different inhibitory mechanisms evolved at different evolutionary stages. For example, the inhibitory effect of GTP may have predated entry into the vertebrate lineage and acted to control propagation of the RAG transposon. The C-terminal region of RAG2, which is dispensable for DNA cleavage, could have evolved to inhibit RAG-mediated transposition at a later stage, as the RAG1/RAG2 transposase was transforming into a recombinase. In this regard, it may be informative to compare the transposition activity of RAG proteins from different species.
Materials and methods
DNA substrates and nucleotides
The body-labeled coupled cleavage substrate was generated by PCR from pC317 (Agrawal et al., 1998). The 12-signal end and 23-signal end oligonucleotide substrates for signal end complex formation, target capture complex formation, and intermolecular transposition were identical to the 12-RSS and 23-RSS, but lack coding flank sequences (Fugmann et al., 2000b). The double-stranded oligonucleotide target DNA for intermolecular transposition was made by annealing 5′-end labeled CLT011 (5′-CGCTCGGTTGCCGCCGGGCGTACTATATTGA-3′) with CLT012 (5′-TCAATATAGTACGCCCGGCGGCAACCGAGCG-3′). Nucleotide triphosphates (ATP, CTP, GTP and UTP) used in the study were purchased from Amersham Pharmacia Biotech. Other nucleotides (ITP, XTP, GDP, GMP, cGMP, GTPγS and GMPPNP) were purchased from Sigma. GTP from other sources yielded identical results.
Proteins
The RAG1 protein used in the in vitro coupled cleavage reactions (Figures 1, 3 and 5A) and in the TCC formation reactions (Figures 2 and 4B) was the MBP-tagged core RAG1 protein purified from bacteria as described previously (Tsai et al., 2002). The RAG2 protein used in the coupled cleavage reaction (Figures 1, 3 and 5A) was the polyhistidine-myc-tagged core RAG2 protein purified from the murine cell line F2A1 as described previously (Tsai et al., 2002). The RAG2 protein used in the TCC formation reactions was GST-tagged core RAG2 (Figure 2) and GST tagged full-length RAG2 (Figure 4B) purified from 293T cells transfected with pEBG-2ΔC and pEBG-FLRAG2, respectively (Spanopoulou et al., 1996). The activities of various RAG2 proteins were initially measured by in vitro coupled cleavage reactions in the presence of the MBP-tagged core RAG1 protein. The RAG proteins used in Figures 4A and 5C were GST-tagged core RAG1 and His-myc tagged core RAG2 or His-myc tagged full-length RAG2 purified from 293T cells co-transfected with pEBG-1ΔN and pEBB-R2C or pEBB-R2FL by glutathione–agarose affinity chromatography. pEBB-R2C was made by subcloning a XbaI fragment from pMS216 into the pEBB vector (Sadofsky et al., 1994). Mutagenesis of RAG1 was carried out as described previously (Tsai et al., 2002).
DNA cleavage and intramolecular transposition
Reactions for in vitro 12/23 coupled transposition were carried out under standard conditions as described (Tsai et al., 2002), except that in reactions containing high concentrations of nucleotides (4–5 mM), MgCl2 concentrations were increased to 10 or 6 mM (Figures 1A and 3B, respectively). Briefly, reactions (25 µl final volume) contained 10 ng of body-labeled substrate, 50 ng of each RAG protein and 15 ng of HMG2. Reactions were incubated for 2 h at 37°C in a buffer containing 20 mM HEPES–Na pH 7.4, 75 mM NaCl, 4 mM MgCl2, 1 µM ZnSO4, 0.1 mg/ml BSA and 2 mM DTT.
Formation of the SEC, TCC and intermolecular transposition products
Reactions for SEC formation, TCC formation and intermolecular transposition were carried out as described previously (Tsai et al., 2002), except that 5.4 mM CaCl2 was replaced with 4 mM MgCl2. Briefly, reactions for SEC formation (12 µl) contained 0.05 pmol labeled 12-signal end oligonucleotide substrate, 0.05 pmol unlabeled 23-signal end oligonucleotide, 50 ng of MBP-core RAG1 and 50 ng of GST-core RAG2 (or 100 ng of GST-full-length RAG2) in 25 mM MOPS–NaOH (pH 7.0), 75 mM potassium acetate, 0.1 mg/ml BSA, 4 mM DTT and 4 mM MgCl2, and were performed at 37°C for 15 min.
Detection of hybrid joints in vitro
Coupled cleavage reactions were carried out as described above for 30 or 60 min, and 1/20th of the products from the reactions were used for PCR amplification of hybrid joints (24 cycles). The primers for amplifying 12-RSS-23-coding HJs were TL2 and Cit4a. The primers for amplifying 23-RSS-12-coding HJs were TL3 and LE1 (Leu et al., 1997). PCR products were resolved on a 6% polyacrylamide gel and visualized by staining with SYBR green.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We wish to dedicate this work to the memory of Paul Sigler. We thank Leon Ptaszek and Sebastian Fugmann for the GST-full-length RAG2 proteins. We thank members of the Schatz laboratory for numerous discussions and advice, and Vincenz Unger, Valerie Hand and David Hesslein for critical reading of the manuscript. We thank Marjorie Oettinger for sharing unpublished results. Oligonucleotide synthesis was performed by the W.M.Keck Foundation Biotechnology Resource Laboratory at Yale University. This work was supported by a grant to D.G.S. from the National Institutes of Health. D.G.S. is an investigator of the Howard Hughes Medical Institute.
References
- Agrawal A. and Schatz,D.G. (1997) RAG1 and RAG2 form a stable post-cleavage synaptic complex with DNA containing signal ends in V(D)J recombination. Cell, 89, 43–53. [DOI] [PubMed] [Google Scholar]
- Agrawal A., Eastman,Q.M. and Schatz,D.G. (1998) Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature, 394, 744–751. [DOI] [PubMed] [Google Scholar]
- Aidinis V., Dias,D.C., Gomez,C.A., Bhattacharyya,D., Spanopoulou,E. and Santagata,S. (2000) Definition of minimal domains of interaction within the recombination-activating genes 1 and 2 recombinase complex. J. Immunol., 164, 5826–5832. [DOI] [PubMed] [Google Scholar]
- Alberts B., Bray,D., Lewis,J., Raff,M., Roberts,K. and Watson,J.D. (1994) Molecular Biology of the Cell. Garland Publishing, Inc., New York, NY.
- Cherfils J., Menetrey,J., Le Bras,G., Janoueix-Lerosey,I., de Gunzburg,J., Garel,J.R. and Auzat,I. (1997) Crystal structures of the small G protein Rap2A in complex with its substrate GTP, with GDP and with GTPγS. EMBO J., 16, 5582–5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkin S.K., Matthews,A.G. and Oettinger,M.A. (2003) The C-terminal portion of RAG2 protects against transposition in vitro. EMBO J., 22, 1931–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugmann S.D., Lee,A.I., Shockett,P.E., Villey,I.J. and Schatz,D.G. (2000a) The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu. Rev. Immunol., 18, 495–527. [DOI] [PubMed] [Google Scholar]
- Fugmann S.D., Villey,I.J., Ptaszek,L.M. and Schatz,D.G. (2000b) Identification of two catalytic residues in RAG1 that define a single active site within the RAG1/RAG2 protein complex. Mol. Cell, 5, 97–107. [DOI] [PubMed] [Google Scholar]
- Gellert M. (2002) V(D)J recombination: rag proteins, repair factors, and regulation. Annu. Rev. Biochem., 71, 101–32. [DOI] [PubMed] [Google Scholar]
- Grawunder U. and Harfst,E. (2001) How to make ends meet in V(D)J recombination. Curr. Opin. Immunol., 13, 186–194. [DOI] [PubMed] [Google Scholar]
- Hiom K. and Gellert,M. (1998) Assembly of a 12/23 paired signal complex: A critical control point in V(D)J recombination. Mol. Cell, 1, 1011–1019. [DOI] [PubMed] [Google Scholar]
- Hiom K., Melek,M. and Gellert,M. (1998) DNA transposition by the RAG1 and RAG2 proteins: A possible source of oncogenic translocations. Cell, 94, 463–470. [DOI] [PubMed] [Google Scholar]
- Iismaa S.E., Wu,M.J., Nanda,N., Church,W.B. and Graham,R.M. (2000) GTP binding and signaling by Gh/transglutaminase II involves distinct residues in a unique GTP-binding pocket. J. Biol. Chem., 275, 18259–65. [DOI] [PubMed] [Google Scholar]
- Jones J.M. and Gellert,M. (2002) Ordered assembly of the V(D)J synaptic complex ensures accurate recombination. EMBO J., 21, 4162–4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman P.D. and Rio,D.C. (1992) P element transposition in vitro proceeds by a cut-and-paste mechanism and uses GTP as a cofactor. Cell, 69, 27–39. [DOI] [PubMed] [Google Scholar]
- Kirch S.A., Rathbun,G.A. and Oettinger,M.A. (1998) Dual role of RAG2 in V(D)J recombination—catalysis and regulation of ordered Ig gene assembly. EMBO J., 17, 4881–4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N., Chalmers,R.M., Kwon,D., Sakai,J. and Bolland,S. (1996) Tn10 and IS10 transposition and chromosome rearrangements: mechanism and regulation in vivo and in vitro. Curr. Top. Microbiol. Immunol., 204, 49–82. [DOI] [PubMed] [Google Scholar]
- Labrador M. and Corces,V.G. (1997) Transposable element-host interactions: regulation of insertion and excision. Annu. Rev. Genet., 31, 381–404. [DOI] [PubMed] [Google Scholar]
- Leu T.M.J., Eastman,Q.M. and Schatz,D.G. (1997) Coding joint formation in a cell free V(D)J recombination system. Immunity, 7, 303–314. [DOI] [PubMed] [Google Scholar]
- Liang H.E., Hsu,L.Y., Cado,D., Cowell,L.G., Kelsoe,G. and Schlissel,M.S. (2002) The ‘dispensable’ portion of RAG2 is necessary for efficient V-to-DJ rearrangement during B and T cell development. Immunity, 17, 639–651. [DOI] [PubMed] [Google Scholar]
- Livàk F. and Schatz,D.G. (1997) Identification of V(D)J recombination coding end intermediates in normal thymocytes. J. Mol. Biol., 267, 1–9. [DOI] [PubMed] [Google Scholar]
- McBlane J.F., van Gent,D.C., Ramsden,D.A., Romeo,C., Cuomo,C.A., Gellert,M. and Oettinger,M.A. (1995) Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell, 83, 387–395. [DOI] [PubMed] [Google Scholar]
- Melek M. and Gellert,M. (2000) RAG1/RAG2-mediated resolution of transposition intermediates: Two pathways and possible consequences. Cell, 101, 625–633. [DOI] [PubMed] [Google Scholar]
- Melek M., Gellert,M. and van Gent,D.C. (1998) Rejoining of DNA by the RAG1 and RAG2 proteins. Science, 280, 301–303. [DOI] [PubMed] [Google Scholar]
- Nakaoka H., Perez,D.M., Baek,K.J., Das,T., Husain,A., Misono,K., Im,M.J. and Graham,R.M. (1997) Gh: a GTP-binding protein with transglutaminase activity and receptor signaling function. Science, 264, 1593–1596. [DOI] [PubMed] [Google Scholar]
- Neiditch M.B., Lee,G.S., Landree,M.A. and Roth,D.B. (2001) RAG transposase can capture and commit to target DNA before or after donor cleavage. Mol. Cell. Biol., 21, 4302–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiditch N.B., Lee,G.S., Huye,L.E., Brandt,V.L. and Roth,D.B. (2002) The V(D)J recombinase efficiently cleaves and transposes signal joints. Mol. Cell, 9, 871–878. [DOI] [PubMed] [Google Scholar]
- Oettinger M.A., Schatz,D.G., Gorka,C. and Baltimore,D. (1990) RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science, 248, 1517–1523. [DOI] [PubMed] [Google Scholar]
- Perkins E.J., Nair,A., Cowley,D.O., Van Dyke,T., Chang,Y. and Ramsden,D.A. (2002) Sensing of intermediates in V(D)J recombination by ATM. Genes Dev., 16, 159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J.E. and Craig,N.L. (2001) Tn7: smarter than we thought. Nat. Rev. Mol. Cell Biol., 2, 806–814. [DOI] [PubMed] [Google Scholar]
- Ramsden D.A. and Gellert,M. (1995) Formation and resolution of double-strand break intermediates in V(D)J rearrangement. Genes Dev., 9, 2409–2420. [DOI] [PubMed] [Google Scholar]
- Rodgers K.K., Bu,Z., Fleming,K.G., Schatz,D.G., Engelman,D.M. and Coleman,J.E. (1996) A unique zinc-binding dimerization motif domain in RAG-1 includes the C3HC4 motif. J. Mol. Biol., 260, 70–84. [DOI] [PubMed] [Google Scholar]
- Roth D.B. and Craig,N.L. (1998) V(D)J recombination-a transposase goes to work. Cell, 94, 411–414. [DOI] [PubMed] [Google Scholar]
- Roth D.B., Nakajima,P.B., Menetski,J.P., Bosma,M.J. and Gellert,M. (1992) V(D)J recombination in mouse thymocytes: double-stranded breaks near T-cell receptor delta rearrangement signals. Cell, 69, 41–53. [DOI] [PubMed] [Google Scholar]
- Sadofsky M., Hesse,J. and Gellert,M. (1994) Definition of a core region of RAG-2 that is functional in V(D)J recombination. Nucleic Acids Res., 22, 1805–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakano H., Hüppi,K., Heinrich,G. and Tonegawa,S. (1979) Sequences at the somatic recombination sites of immunoglobulin light-chain genes. Nature, 280, 288–294. [DOI] [PubMed] [Google Scholar]
- Saraste M., Sibbald,P.R. and Wittinghofer,A. (1990) The P-loop—a common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci., 15, 430–434. [DOI] [PubMed] [Google Scholar]
- Schatz D.G., Oettinger,M.A. and Baltimore,D. (1989) The V(D)J recombination activating gene (RAG-1). Cell, 59, 1035–1048. [DOI] [PubMed] [Google Scholar]
- Schlissel M., Constantinescu,A., Morrow,T., Baxter,M. and Peng,A. (1993) Double-strand signal sequence breaks in V(D)J recombination are blunt, 5′-phosphorylated, RAG-dependent, and cell cycle regulated. Genes Dev., 7, 2520–2532. [DOI] [PubMed] [Google Scholar]
- Sekiguchi J., Whitlow,S. and Alt,F. (2001) Increased accumulation of hybrid V(D)J joins in cells expressing truncated versus full-length RAGs. Mol. Cell, 8, 1383–90. [DOI] [PubMed] [Google Scholar]
- Spanopoulou E., Zaitseva,F., Wang,F.-H., Santagata,S., Baltimore,D. and Panayotou,G. (1996) The homeodomain of Rag-1 reveals the parallel mechanisms of bacterial and V(D)J recombination. Cell, 87, 263–276. [DOI] [PubMed] [Google Scholar]
- Steen S.B., Han,J.O., Mundy,C., Oettinger,M.A. and Roth,D.B. (1999) Roles of the ‘dispensable’ portions of RAG-1 and RAG-2 in V(D)J recombination. Mol. Cell. Biol., 19, 3010–3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellwagen A.E. and Craig,N.L. (1998) Mobile DNA elements: controlling transposition with ATP-dependent molecular switches. Trends Biochem. Sci., 12, 486–490. [DOI] [PubMed] [Google Scholar]
- Thompson C.B. (1995) New insights into V(D)J recombination and its role in the evolution of the immune system. Immunity, 3, 531–539. [DOI] [PubMed] [Google Scholar]
- Traut T.W. (1994) Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem., 140, 1–22. [DOI] [PubMed] [Google Scholar]
- Tsai C.-L., Drejer,A.N. and Schatz,D.G. (2002) Evidence of a critical architectural function for the RAG proteins in end processing, protection, and joining in V(D)J recombination. Genes Dev., 16, 1934–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gent D.C., Mizuuchi,K. and Gellert,M. (1996) Similarities between initiation of V(D)J recombination and retroviral integration. Science, 271, 1592–1594. [DOI] [PubMed] [Google Scholar]
- Yamauchi M. and Baker,T.A. (1998) An ATP–ADP switch in mub controls progression of the mu transposition pathway. EMBO J., 17, 5509–5518. [DOI] [PMC free article] [PubMed] [Google Scholar]