Abstract
Biogenesis of functional spliceosomal small nuclear RNAs (snRNAs) includes the post-transcriptional covalent modification of numerous internal nucleotides. We have recently demonstrated that synthesis of 2′-O-methylated nucleotides and pseudouridines in the RNA polymerase II-synthesized Sm snRNAs is directed by sequence-specific guide RNAs. Here, we provide evidence supporting the notion that modification of Sm snRNAs occurs in nucleoplasmic Cajal bodies (CBs), where modification guide RNAs accumulate. We show that short fragments of Sm snRNAs are correctly and efficiently modified when targeted to CBs, but not when these same fragments are targeted to the nucleolus. We also demonstrate that internal modification of the U2 snRNA occurs exclusively after nuclear import of the newly assembled Sm snRNP from the cytoplasm. Finally, we show that p80 coilin, the CB marker protein, is not required for snRNA modification. In coilin knockout cells, Sm snRNAs and their modification guide RNAs colocalize in residual CBs, which do not stockpile fibrillarin and fail to recruit the U3 small nucleolar RNA.
Keywords: Cajal body/p80 coilin/RNA modification/scaRNAs/spliceosomal snRNAs
Introduction
The U1, U2, U4, U5 and U6 small nuclear RNAs (snRNAs) are key components of the spliceosome, a huge ribonucleoprotein (RNP) responsible for the removal of precursor mRNA (pre-mRNA) introns. In addition to selecting the correct splice sites, the snRNAs are most likely the catalysts of the splicing reaction itself (Villa et al., 2002). The spliceosomal snRNAs contain many post-transcriptionally modified nucleotides. Altogether, the human U1, U2, U4, U5 and U6 snRNAs carry 21 pseudouridines and 13 2′-O-methyl groups (Massenet et al., 1998). Although unmodified U1, U4, U5 and U6 snRNAs can support pre-mRNA splicing in vitro (Massenet et al., 1998), modification of the U2 RNA is essential for assembly of the active spliceosome (Yu et al., 1998). The modified nucleotides are confined to snRNA regions involved in formation of RNA–RNA or RNA–protein interactions that are crucial for spliceosome function, strongly suggesting that they have beneficial effects on the efficacy and/or fidelity of pre-mRNA splicing.
Biogenesis of spliceosomal snRNAs is a complex process that is accompanied by complicated intracellular trafficking of maturing snRNAs (Will and Lührmann, 2001). The newly synthesized precursor snRNAs (pre-snRNAs) undergo nucleolytic processing, 5′-terminal cap formation, internal nucleotide modification and RNP assembly. Maturation of the RNA polymerase (pol) III-transcribed U6 snRNA occurs, at least partially, in the nucleolus, where synthesis of the eight 2′-O-methyl groups and three pseudouridines of U6 is directed by small nucleolar (sno)RNAs (Tycowski et al., 1998; Ganot et al., 1999). The U6 modification guide snoRNAs are structurally and functionally indistinguishable from snoRNAs directing modification of the 18S, 5.8S and 28S rRNAs (Kiss, 2001; Filipowicz and Pogacic, 2002; Terns and Terns, 2002). In fact, one 2′-O-methylation guide snoRNA was found to function in both U6 and 28S modification (Tycowski et al., 1998). The 2′-O-methylation guide snoRNAs possess conserved C (UGAUGA) and D (CUGA) box motifs, whereas the pseudouridylation guide snoRNAs fold into a consensus ‘hairpin–hinge– hairpin–tail’ secondary structure and contain so-called H (AnAnnA) and ACA boxes. Both classes of guide snoRNAs select the correct substrate nucleotides by forming transient base-pairing interactions with their target RNAs (Kiss, 2001; Filipowicz and Pogacic, 2002; Terns and Terns, 2002).
In contrast to U6, maturation of the RNA pol II-synthesized U1, U2, U4 and U5 snRNAs has a cytoplasmic phase. Shortly after synthesis, the pre-snRNAs that carry 3′-terminal extra nucleotides and a 5′ 7-methylguanosine primary cap are exported to the cytoplasm where seven Sm core proteins bind to the Sm structural motif of the snRNAs. Packaging into Sm snRNPs is a prerequisite for trimming off the 3′-terminal trailer, for hypermethylation of the primary cap to 2,2,7-trimethylguanosine (TMG) and for import of the newly assembled snRNPs into the nucleus (Mattaj, 1986).
While many aspects of snRNA biogenesis have been deciphered, it remains unclear where and when internal modifications are introduced into Sm snRNAs during their biogenesis. In the nucleus, snRNPs accumulate mainly in the interchromatin granule clusters, also known as speckles (Lamond and Earnshaw, 1998; Sleeman and Lamond, 1999b; Lewis and Tollervey, 2000). A smaller fraction of snRNPs accumulates in the nucleoplasmic Cajal bodies (CBs) (Matera, 1999; Dundr and Misteli, 2001; Ogg and Lamond, 2002) and under certain conditions, in the nucleolus (Lyon et al., 1997; Sleeman and Lamond, 1999a; Lange and Gerbi, 2000; Yu et al., 2001; Gerbi and Lange, 2002). Modification studies in microinjected Xenopus oocytes led to the conclusion that both 2′-O-methylation and pseudouridylation of the U2 snRNA take place in the nucleolus (Yu et al., 2001). More recently, we have identified eight distinct 2′-O-methylation and pseudouridylation guide RNAs that direct modification of the U1, U2, U4 and U5 snRNAs (Jády and Kiss, 2001; Darzacq et al., 2002; Kiss et al., 2002). In situ localization studies demonstrated that the Sm snRNA modification guide RNAs accumulate exclusively in CBs and, therefore, they were called small Cajal body-specific RNAs (scaRNAs).
In addition to the spliceosomal snRNPs, CBs have long been known to accumulate U3, U8 and U14 snoRNAs along with their associated proteins (e.g. fibrillarin and Nopp140). Furthermore, CBs are highly enriched in the survival of motor neurons (SMN) protein complex as well as several basal transcription factors. However, the cellular function(s) of this subnuclear organelle has remained speculative (Matera, 1999; Gall, 2000; Dundr and Misteli, 2001; Ogg and Lamond, 2002). Our finding that guide RNAs directing snRNA modification accumulate in CBs, raised the possibility that post-transcriptional modification of Sm snRNAs might take place in this compartment (Kiss, 2001, 2002; Darzacq et al., 2002). In this study, we provide experimental evidence supporting the notion that internal modification of Sm spliceosomal snRNAs takes place in CBs following import of the newly assembled snRNPs from the cytoplasm.
Results
The cytoplasmic precursor of U2 snRNA lacks internal modifications
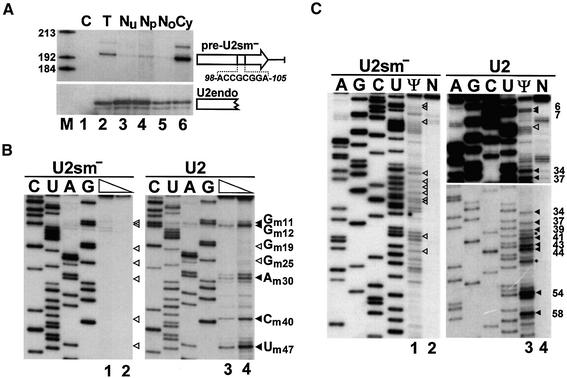
Previous microinjection experiments in Xenopus oocytes indicated that internal modification of the U2 snRNA takes place in the nucleus, but left open the question whether it occurs before or after the cytoplasmic phase of U2 biogenesis (Yu et al., 2001). Therefore, we first examined whether the cytoplasmic pre-U2 snRNA contains modified nucleotides. To this end, a mutant version of the human U2 snRNA, U2sm–, which carried an altered Sm binding site, was transiently expressed in simian COS-7 cells (Figure 1A). Consistent with the fact that the Sm site is crucial for 3′ end processing and re-entry of Sm snRNAs into the nucleus (Mattaj, 1986; Will and Lührmann, 2001), subcellular fractionation followed by RNase protection analysis demonstrated that the pre-U2sm– RNA accumulated mainly in the cytoplasm of transfected cells (lane 6) and carried an unprocessed 3′-terminal tail of ∼11 nucleotides (Ares et al., 1985). The endogenous U2 snRNA showed a predominant nucleoplasmic accumulation (lane 4), providing a good internal control for cellular fractionation.
Fig. 1. Modification of Sm mutant pre-U2 snRNA. (A) Human pre-U2 snRNA with an altered Sm binding site accumulates in the cytoplasm. Simian COS-7 cells were transfected with the pU2sm– expression construct. RNAs isolated from transfected cells (T) or from the nuclear (Nu), nucleoplasmic (Np), nucleolar (No) or cytoplasmic (Cy) fractions of transfected cells were analysed by RNase A/T1 mapping. Protected RNAs corresponding to the 3′ end-extended precursor of U2sm– and the endogenous U2 snRNA (U2endo) are indicated. Note that the exposure time of the upper panel was about 50 times longer than that of the lower panel. Lane C, control mapping with Escherichia coli tRNA. Lane M, molecular markers. (B) Primer extension mapping of 2′-O-methylated nucleotides. RNAs isolated from the cytoplasmic (lanes 1 and 2) or nuclear (lanes 3 and 4) fraction of COS-7 cells expressing the pre-U2sm– RNA were annealed with terminally labelled oligonucleotides complementary to the mutant (lanes 1 and 2) or wild-type (lanes 3 and 4) U2 RNA and incubated with AMV reverse transcriptase in the presence of 1 mM (lanes 1 and 3) or 0.004 mM (lanes 2 and 4) dNTPs. The extended products were fractionated on a 6% denaturing polyacrylamide gel. Lanes C, U, A and G represent dideoxy sequencing reactions performed on the pU2sm– and pU2 plasmids. Stop signals corresponding to 2′-O-methylated nucleotides and expected positions of 2′-O-methylated nucleotides are indicated by closed and open arrowheads, respectively. (C) Mapping of pseudouridine residues in the U2sm– RNA. Cytoplasmic (lanes 1 and 2) and nuclear (lanes 3 and 4) RNAs, either treated (Ψ) or non-treated (N) with CMC, were analysed by primer extension. The observed and expected positions of pseudouridines are indicated by closed and open arrowheads, respectively. Asterisks indicate stops generated by 2′-O-methylated nucleotides. For other details, see the legend to (B).
The 2′-O-methylation status of the cytoplasmic pre-U2sm– RNA was determined by primer-extension analysis. This procedure is based on the observation that, at low dNTP concentrations, reverse transcriptase pauses one nucleotide before and/or at the ribose-methylated nucleotide (Maden et al., 1995). When cytoplasmic RNAs were analysed with a primer complementary to the mutant Sm motif of the U2sm– RNA, no stops were observed at high (Figure 1B, lane 1) or low (lane 2) dNTP concentrations. Mapping of the endogenous U2 snRNA with a primer specific for wild-type U2, however, detected most of the known 2′-O-methylated nucleotides in human U2 snRNA (Massenet et al., 1998; Figure 1B, lanes 3 and 4). It is unclear why the Gm19 and Gm25 modified residues failed to arrest reverse transcriptase. Pseudouridylation of the pre-U2sm– RNA and the endogenous U2 snRNA was investigated by the N3-1-cyclohexyl-3-(2-morpholino ethyl)carbodiimide methyl-p-toluenesulfonate (CMC)-modification primer extension procedure (Bakin and Ofengand, 1993; Figure 1C). CMC specifically reacts with pseudouridines and arrests reverse transcriptase one nucleotide prior to the pseudouridylation site. While mapping of the wild-type U2 snRNA clearly detected most of the known pseudouridine residues in mammalian U2 snRNAs (lane 3), no pseudouridine was detected in the pre-U2sm– RNA (lane 1). Since the RNA-guided modification reactions seem be independent of the secondary structure of the substrate RNA, it is unlikely that the altered Sm motif of pre-U2sm– could generate a structure inaccessible for the modification machinery. In summary, our results not only confirm the idea that modification of U2 snRNA is a nuclear event that depends upon a functional Sm site (Yu et al., 2001), but extend the observation to reveal that cytoplasmic pre-U2sm– RNAs are unmodified.
To test whether the Sm-dependent modification of the U2 snRNA could occur in the nucleus prior to cytoplasmic export, we assessed the modification state of the endogenous HeLa pre-U2 snRNA. Taking advantage of the fact that the human pre-U2 snRNA carries an 11 nucleotide 3′-terminal tail (Ares et al., 1985), we utilized a primer complementary to this region to specifically analyse the pre-U2 snRNA (Figure 2). After CMC modification of HeLa cytoplasmic RNAs, primer extension analysis failed to detect any pseudouridines in pre-U2 snRNA (lane 1). Likewise, primer extension in the presence of low dNTP concentrations demonstrated that the pre-U2 snRNA contains no 2′-O-methylated nucleotides (lanes 3 and 4). We conclude that internal modifications are introduced into the U2 snRNA after its return to the nucleus.
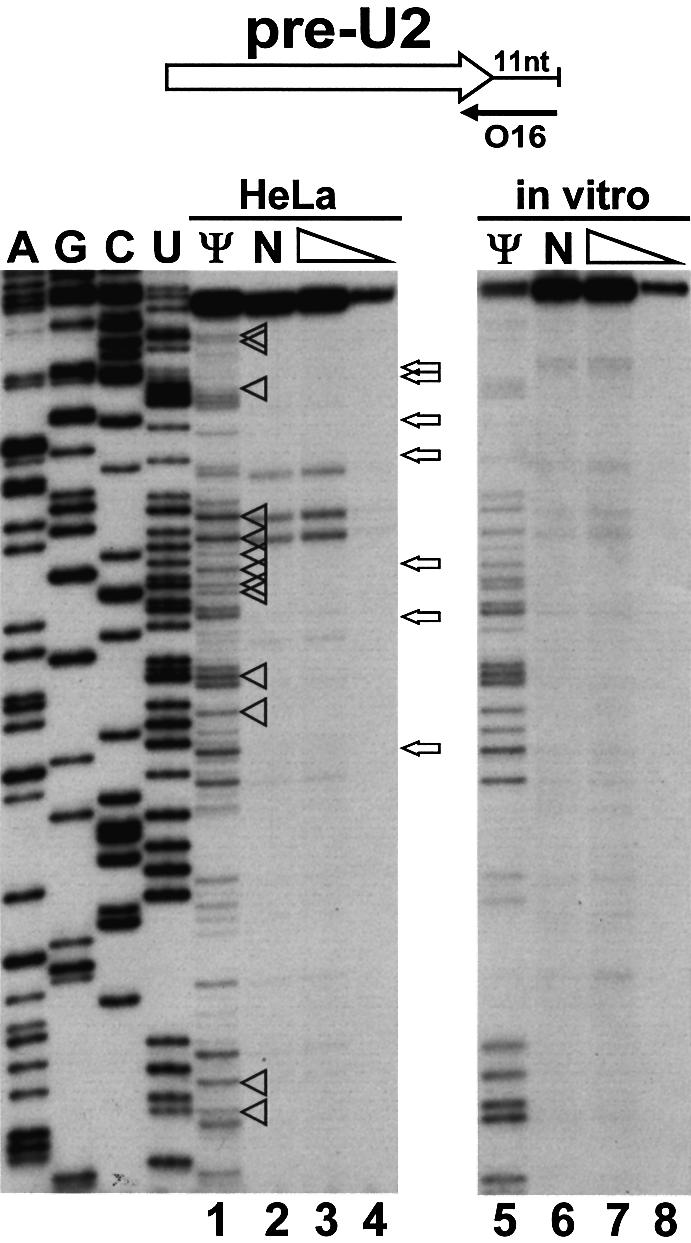
Fig. 2. Mapping of 2′-O-methylated nucleotides and pseudouridines in human pre-U2 snRNA. A terminally labelled oligonucleotide (O16) was annealed to HeLa cytoplasmic RNA or in vitro transcribed pre-U2 snRNA. RNA samples used for pseudouridylation mapping had been treated with CMC and alkali buffer (lanes 1 and 5) or only with alkali buffer (lanes 2 and 6). Primer extension was performed with AMV reverse transcriptase in the presence of 1 mM (lanes 1, 2, 3, 5, 6 and 7) or 0.004 mM (lanes 4 and 8) dNTPs. Lanes A, G, C and U, dideoxy sequencing reactions. The expected positions of 2′-O-methylated nucleotides and pseudouridines are indicated by open arrows and arrowheads, respectively. For other details, see the legend to Figure 1.
Nucleolar localization is not sufficient for modification of U5 snRNA in mammalian cells
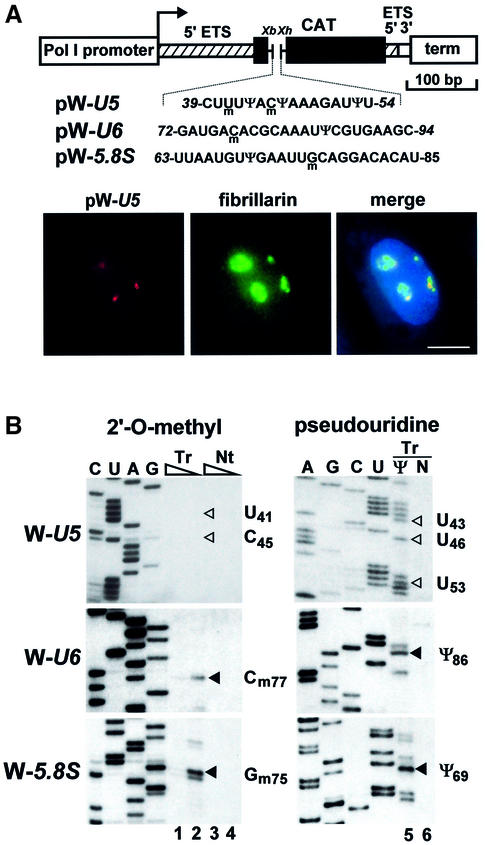
Having established that U2 and probably all pol II-specific snRNAs are modified in the nucleus following nuclear re-entry, we attempted to define the subnuclear locale where snRNA modification takes place. To date, two nuclear organelles, the nucleolus and the CB, have been implicated in modification of Sm-class snRNAs. Whereas microinjection experiments suggested that modification of U2 (Yu et al., 2001) and perhaps U4 and U5 snRNAs (Gerbi and Lange, 2002) takes place in the nucleolus of Xenopus oocytes, in situ localization studies in mammalian cells revealed a CB-specific accumulation for guide RNAs directing 2′-O-methylation and pseudouridylation of Sm snRNAs (Darzacq et al., 2002; Kiss et al., 2002). We have previously demonstrated that 2′-O-methylation of C45 and pseudouridylation of U46 residues in the U5 snRNA are directed by the U85 composite box C/D-H/ACA guide RNA (Jády and Kiss, 2001). Although in situ hybridization detects the U85 RNA only in CBs (Darzacq et al., 2002), in principle, a small fraction of U85 might be present in the nucleolus as well. Expression of short test RNAs within ribosomal minigene transcripts has proved to be a powerful technique to study nucleolar RNA modification activities in vivo (Ganot et al., 1997a, 1999). To detect U85 modification activities within the nucleolus, a short fragment of the human U5 snRNA (C39 to U54), encompassing the C45 and U46 modification sites, was inserted into the pW mouse ribosomal minigene (Hadjiolova et al., 1994; Figure 3A). This fragment of U5 is an excellent substrate for the U85 modification RNP both in vivo and in vitro (Jády and Kiss, 2001). Upon transfection into mouse cells, both the RNA pol I-directed transcription of the pW-U5 construct and accumulation of the W-U5 transcript were expected to occur in the nucleolus (Hadjiolova et al., 1994; Moss and Stefanovsky, 1995; Ganot et al., 1999). In situ hybridization with a pW-U5-specific fluorescent oligonucleotide probe showed that the W-U5 RNA accumulated in distinct regions of the nucleus (Figure 3A). Co-expression of a GFP-tagged version of human fibrillarin, an abundant nucleolar protein (Lapeyre et al., 1990), demonstrated that the W-U5 RNA accumulated within the nucleolus. This conclusion was further strengthened by cell fractionation experiments (data not shown). Primer extension mapping failed to detect any 2′-O-methylated nucleotides or pseudouridines in the U5-specific tag of the nucleolar W-U5 RNA (Figure 3B). In contrast, when short fragments of the U6 snRNA (G72 to C94) or the 5.8S rRNA (U63 to U85) were expressed within the nucleolar W-U6 and W-5.8 transcripts (Figure 3A), all modified nucleotides reported for these regions of mammalian U6 (Cm77 and Ψ86) and 5.8S (Gm75 and Ψ69) RNAs were faithfully synthesized (Figure 3B).
Fig. 3. Nucleolar expression and modification of mouse ribosomal minigene transcripts tagged with human U5, U6 and 5.8S-specific sequences. (A) Schematic structure of the pW-U5, pW-U6 and pW-5.8S expression constructs and in situ localization of the expressed W-U5 RNA in mouse L929 cells. The mouse pol I promoter and terminator (term), fragments derived from the 5′ (hatched boxes) and 3′ (open box) external transcribed spacers (ETS) of the mouse rRNA gene, and a fragment of the chloramphenicol acetyltransferase (CAT) gene are shown. Insertion of synthetic DNAs into the XbaI (Xb) and XhoI (Xh) sites of pW resulted in pW-U5, pW-U6 and pW-5.8S. Nucleotides 2′-O-methylated (m) and pseudouridylated (Ψ) in the human U5, U6 and 5.8S RNAs are indicated. Mouse cells transfected with the pW-U5 expression construct were hybridized with a fluorescent oligonucleotide probe specific for the W-U5 transcript. The nucleolus was visualized by expression of GFP-tagged fibrillarin. Nuclear DNA was stained with DAPI (blue). Bar, 10 µm. (B) Mapping of 2′-O-methylated nucleotides and pseudouridines. Mouse L929 cells were transfected with the pW-U5, pW-U6 or pW-5.8S expression construct as indicated. RNAs isolated from transfected (Tr) or non-transfected (Nt) cells were analysed by primer extension in the presence of 1 mM (lanes 1, 3, 5 and 6) or 0.004 mM (lanes 2 and 4) dNTPs. RNA analysed on lane 5 was treated with CMC. Lanes C, U, G and A represent dideoxy sequencing reactions performed on the pW-U5, pW-U6 or pW-5.8S plasmid. Stops representing modified nucleotides are indicated by closed arrowheads. The expected positions of 2′-O-methylated nucleotides and pseudouridines in the W-U5 transcript are indicated by open arrowheads.
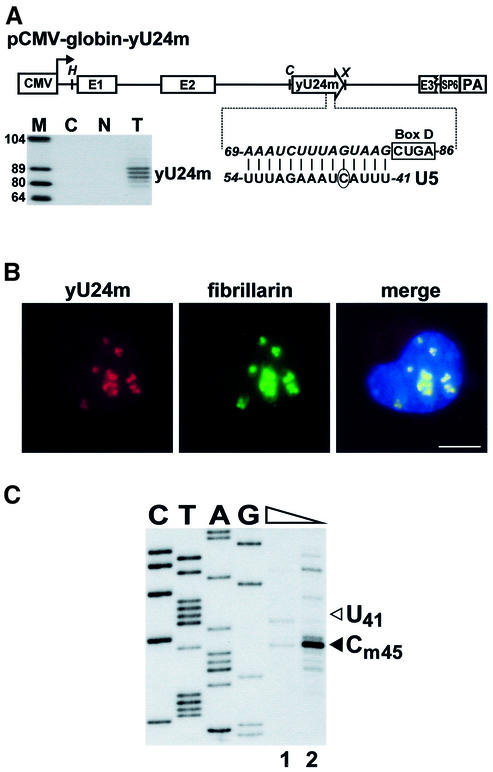
To exclude the formal possibility that the W-U5 transcript, in contrast to the W-U6 and W-5.8S RNAs, folds into a secondary structure that prevents its modification, we attempted to restore the modification of W-U5 by co-expressing an artificial 2′-O-methylation guide snoRNA (Figure 4A). The downstream target recognition element of the yeast U24 snoRNA was replaced for a sequence predicted to direct 2′-O-methylation of the C45 residue in the U5 snRNA. The mutant U24 snoRNA (yU24m) was inserted into the pCMV-globin expression construct (Darzacq et al., 2002) and co-transfected with the pW-U5 ribosomal minigene into mouse cells. Expression of yU24m was verified by RNase A/T1 mapping (Figure 4A, lane T). In situ hybridization of mouse cells co-expressing the yU24m snoRNA and fibrillarin–GFP constructs demonstrated that yU24m specifically accumulated in the nucleolus (Figure 4B). Modification mapping of the W-U5 transcript revealed that expression of the yU24m snoRNA restored 2′-O-methylation of W-U5 at the C45 residue (Figure 4C). We conclude that modification activities responsible for 2′-O-methylation and pseudouridylation of the U5 snRNA are absent from the nucleolus of cultured mouse cells.
Fig. 4. Restoration of 2′-O-methylation of the W-U5 RNA. (A) Schematic structure of the pCMV-globin-yU24m expression construct and expression of the yU24m snoRNA. The promoters of the cytomegalovirus (CMV) and the SP6 RNA polymerase, the exons of the human β-globin gene (E1–E3), the polyadenylation region of the bovine growth hormone gene (PA) are shown. The coding region of a mutant version of the yeast U24 snoRNA (yU24m) is represented by an open arrow. The sequence of the D box and the altered target recognition region of the yU24m snoRNA (in italics) positioning the C45 residue (circled) in the U5 snRNA for 2′-O-methylation are shown. Relevant restriction sites are indicated (H, HindIII; C, ClaI; X, XhoI). RNase A/T1 mapping of RNAs obtained from mouse L929 cells either transfected (T) or non-transfected (N) with the pCMV-globin-yU24m construct. The protected RNA triplet likely reflects a heterogeneity of the 3′ terminus of the expressed yU24m snoRNA (Ganot et al., 1997b). Lane C, control mapping with E.coli tRNA. Lane M, size markers. (B) In situ localization of yU24m RNA. Mouse L929 cells co-transfected with pW-U5, pCMV-globin-yU24m and pfibrillarin-GFP were probed with a fluorescent oligonucleotide complementary to the yU24m RNA. Bar, 10 µm. (C) Primer extension mapping of 2′-O-methylated nucleotides. RNA isolated from mouse cells co-expressing the yU24m and W-U5 RNAs was mapped with a primer specific for the W-U5 RNA. For other details, see the legend to Figure 1.
U5-specific sequences are correctly modified when targeted to CBs
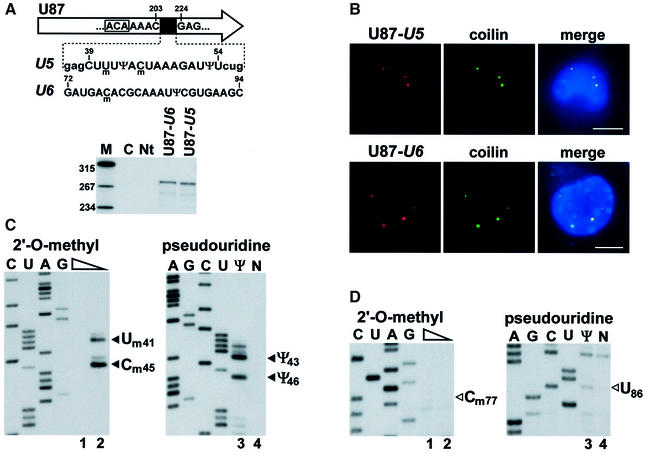
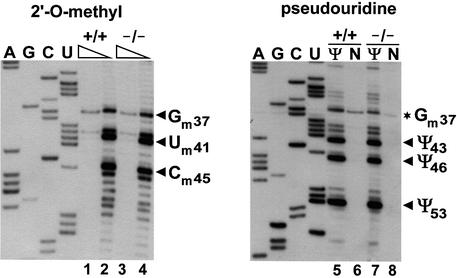
The finding that guide RNAs responsible for modification of the U1, U2, U4 and U5 snRNAs specifically localize to CBs suggested that post-transcriptional modification of Sm snRNAs takes place in this subnuclear organelle (Darzacq et al., 2002; Kiss et al., 2002). To provide experimental support for this idea, we tested whether modification guide RNPs accumulating in CBs are functionally active. To this end, the C39–U54 fragment of the U5 snRNA and, as a control, the G72–C93 fragment of the U6 snRNA were inserted into the coding region of the U87 scaRNA (Figure 5A). The U87-U5 and U87-U6 genes were cloned into the pCMV-globin expression construct (see Figure 4A) and transfected into mouse cells. Expression of the chimeric U87-U5 and U87-U6 RNAs was confirmed by RNase mapping (Figure 5A). Fluorescent in situ hybridization of the transfected cells with U87-U5- and U87-U6-specific oligonucleotide probes demonstrated that both chimeric RNAs colocalized with a transiently expressed red fluorescent protein (RFP)-tagged coilin, a molecular marker of the CB (Andrade et al., 1991; Raska et al., 1991; Figure 5B). Modification mapping revealed that both 2′-O-methylated nucleotides (Um41 and Cm45) and both pseudouridines (Ψ43 and Ψ46) present in this region of the human U5 snRNA were faithfully synthesized in the U5 tag of the U87-U5 RNA (Figure 5C). Likewise, when short fragments of the U2 snRNA (G31–U53 and U49–A66) were expressed in the U87 scaRNA and targeted to CBs, all known modified nucleotides were synthesized (data not shown). In contrast, no 2′-O-methyl groups and pseudouridines were detected in the U6 tag of the U87-U6 scaRNA (Figure 5D), indicating that factors mediating the synthesis of Cm77 and Ψ86 in U6 snRNA are not present or are inactive in the CB.
Fig. 5. Modification of a U5-specific sequence in the CB. (A) Expression of U87 scaRNA tagged with U5- or U6-specific sequences. A fragment of the human U87 scaRNA gene between C203 and G224 was replaced for short regions of the U5 or U6 snRNAs. The ACA motif of U87 is boxed. For a detailed structure of the U87 RNA, see figure 1 in Darzacq et al. (2002). Nucleotides facilitating cloning are in lower case letters. Pseudouridines (Ψ) and 2′-O-methylated nucleotides (m) are shown. The U87-U5 and U87-U6 genes were inserted into the pCMV-globin expression vector and transfected into mouse L929 cells. RNAs from transfected (U87-U5 and U87-U6) or non-transfected (Nt) cells were analysed by RNase A/T1 mapping. For other details, see the legend to Figure 1A. (B) In situ localization of U87-U5 and U87-U6 RNAs. Mouse cells expressing the U87-U5 or U87-U6 RNAs were hybridized with specific fluorescent oligonucleotides. CBs were visualized by co-expression of RFP–coilin. Bar, 10 µm. (C) Mapping of 2′-O-methylated nucleotides and pseudouridines in the U87-U5 RNA. RNA isolated from mouse cells expressing the U87-U5 RNA was analysed by primer extension. (D) Primer extension mapping of 2′-O-methylated nucleotides and pseudouridines in the U87-U6 RNA. For other details, see the legend to Figure 3B.
Sm snRNAs are modified in coilin knockout mice cells
Previous studies indicated that coilin is a crucial structural component of the CB (Bauer and Gall, 1997; Almeida et al., 1998; Tucker et al., 2001). Depletion of coilin by either immunoprecipitation from a Xenopus egg extract (Bauer and Gall, 1997) or genetic depletion in mice (Tucker et al., 2001) results in formation of ‘residual’ CBs that accumulate the nucleolar proteins fibrillarin and Nopp140, but fail to recruit Sm snRNPs and the SMN complex. Recently, a mouse embryonic fibroblast (MEF) cell line carrying a coilin gene that lacks its C-terminal 487 amino acids has been established from homozygous knockout mice (Tucker et al., 2001). The knockout allele, in principle, could express the N-terminal 82 amino acids of coilin, although western blotting or immunofluorescence failed to confirm accumulation of this fragment (Tucker et al., 2001). To assess the effect of coilin depletion and consequently the lack of canonical CBs on snRNA modification, we compared the 2′-O-methylation and pseudouridylation patterns of U5 snRNAs obtained from coilin knockout and control MEF cells. As shown in Figure 6, all 2′-O-methylated nucleotides (Gm37, Um41 and Cm45) and pseudouridines (Ψ43, Ψ46 and Ψ53) reported for the mammalian U5 snRNA (Massenet et al., 1998) were detectable in U5 snRNAs obtained from both the mutant (lanes 4 and 7) and control cells (lanes 2 and 5). Likewise, we found that all 2′-O-methylated nucleotides and pseudouridines known to be present in mammalian U2 snRNAs were also synthesized in the mutant MEF cells (data not shown), demonstrating that depletion of coilin does not impair the internal modification of pol II-specific snRNAs.
Fig. 6. The U5 snRNA is correctly modified in coilin knockout mouse embryonic fibroblast cells. RNAs from coilin knockout (–/–) or control (+/+) cells were subjected to primer extension analysis with a U5-specific primer. For other details, see the legend to Figure 1B and C.
Identification of a novel CB-like structure in coilin knockout cells
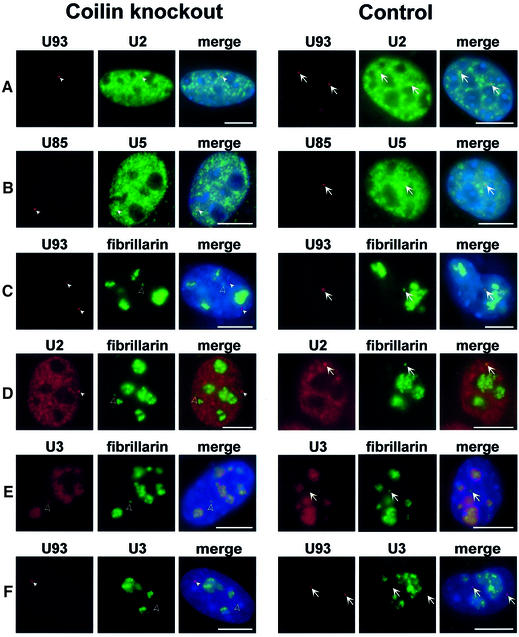
Because recruitment of Sm snRNPs to CB appears to be a coilin-dependent process (Bauer and Gall, 1997; Hebert et al., 2001; Tucker et al., 2001), we investigated where modification of snRNAs might occur in coilin-deficient MEF cells. First, we determined the subnuclear distribution of transiently expressed human U93 and U85 scaRNAs in coilin knockout MEF cells (Figure 7A and B). Interestingly, we found that in the nucleoplasm of transfected mutant MEF cells, the U85 and U93 scaRNAs accumulated in discrete foci (indicated by arrowheads), which were highly reminiscent of the authentic CBs observed in control cells (arrows). More importantly, we found that the U2 and U5 snRNAs, the cognate substrates of the U93 and U85 modification guide scaRNPs, respectively, were also enriched in the scaRNA-containing CB-like structures in both mutant and control cells. We conclude that in coilin knockout MEF cells, the scaRNAs and Sm snRNAs colocalize in CB-sized nucleoplasmic foci.
Fig. 7. Identification of a novel CB-like structure accumulating scaRNAs and spliceosomal snRNAs. MEF cells derived from coilin knockout or control mice were transfected with the pCMV-globin-U93 (A, C and F), pCMV-globin-U85 (B) and/or pfibrillarin-GFP (C, D and E) expression constructs. Distribution of the U93, U85, U2 and U5 RNAs was detected by in situ hybridization with sequence-specific fluorescent oligonucleotide probes. Extranucleolar foci corresponding to canonical CBs (arrows) and ‘residual CBs’ containing fibrillarin and U3 snoRNA (open arrowheads) or scaRNAs and snRNAs (closed arrowheads) are indicated. DAPI staining of nuclear DNA was omitted in (D). Bar, 10 µm.
It has been shown that in the absence of coilin, ‘residual’ CBs are formed that lack Sm snRNPs and SMN, but accumulate the nucleolar proteins fibrillarin and Nopp140 (Bauer and Gall, 1997; Tucker et al., 2001). Indeed, in the coilin knockout MEF cells, transiently expressed fibrillarin–GFP fusion protein accumulated mainly in nucleoli, but was also apparent in extranucleolar foci that correspond to the previously described residual CBs (Tucker et al., 2001; Figure 7C–E, open arrowheads). Co-staining the same cells with fluorescent oligonucleotide probes specific for the U93 scaRNA (C) or the U2 snRNA (D) revealed that none of these RNAs colocalize with the fibrillarin-containing residual CBs. As predicted, U93 scaRNA and U2 snRNA colocalized in CBs in control cells (A, arrows), as marked by the fibrillarin–GFP fusion protein (C and D, arrows). These observations collectively demonstrate that in coilin knockout MEF cells, two different kinds of ‘residual’ CB are formed: the newly discovered scaRNA-containing foci that also accumulate the Sm snRNAs, and the previously reported fibrillarin-rich residual CBs that concentrate the nucleolar epitopes fibrillarin and Nopp140 (Tucker et al., 2001). Interestingly, the SMN complex fails to localize to any of these nuclear foci (Tucker et al., 2001; data not shown), further emphasizing the importance of coilin in SMN recruitment to CBs (Hebert et al., 2001, 2002). It is also noteworthy that, upon overexpression of the U85 box C/D-H/ACA scaRNA in mutant MEF cells, significant amounts of fibrillarin–GFP were detected in the Sm snRNA- and scaRNA-containing residual CBs as well. This indicates that fibrillarin-associated box C/D scaRNAs, as expected, can recruit fibrillarin to the scaRNA-containing residual CBs (data not shown).
Substantial evidence suggests that besides snRNP synthesis, CBs also function in the biogenesis of snoRNPs (Narayanan et al., 1999; Mouaikel et al., 2002; Verheggen et al., 2002). In situ localization of the endogenous U3 snoRNA in coilin knockout MEF cells expressing fibrillarin–GFP demonstrated that the fibrillarin-rich residual CBs accumulated the U3 snoRNA (Figure 7E, open arrowhead) even more efficiently than canonical CBs did in the control cells (arrows). Double labelling of the mutant MEF cells with fluorescent oligonucleotide probes specific for the U93 scaRNA and the U3 snoRNA clearly demonstrated that the scaRNA-containing residual CBs fail to recruit the U3 snoRNA, whereas in control cells, the U3 and U93 RNAs colocalized in canonical CBs (Figure 7F). These results suggest that in coilin knockout MEF cells, the biogenesis of spliceosomal snRNPs and nucleolar snoRNPs is compartmentalized into two different types of residual CBs that specifically accumulate scaRNPs or stockpile the nucleolar proteins fibrillarin and Nopp140, respectively.
Discussion
Post-transcriptional nucleotide modification is an important, but not yet fully understood step in the biogenesis of spliceosomal snRNAs. We have recently reported that 2′-O-methylation and pseudouridylation of the U1, U2, U4 and U5 Sm snRNAs are directed by guide RNAs, which are highly reminiscent of snoRNAs directing the nucleolar modification of rRNAs and the pol III-transcribed U6 snRNA (Jády and Kiss, 2001; Darzacq et al., 2002; Kiss et al., 2002). However, instead of accumulating in the nucleolus, these Sm snRNA guide RNAs specifically localize to CBs, suggesting that these subnuclear organelles may function in snRNA modification (Kiss, 2001; Darzacq et al., 2002). In this study, we have demonstrated that Sm snRNA-specific sequences are efficiently and correctly 2′-O-methylated and pseudouridylated when targeted into the CB. Thus, modification scaRNPs accumulating in CBs are functionally active, making it unlikely that CBs are simply storage sites for scaRNPs. Together with the fact that the CB is the only common nuclear site where snRNAs and scaRNAs are known to accumulate, our findings strongly suggest that CB is the subcellular locale where modification of Sm snRNAs takes place. This conclusion has been further strengthened by recent cell fractionation experiments, which found that most, if not all, of the cellular U85 scaRNA copurifies with the CB fraction of HeLa nuclei (P.Richard, X.Darzacq, C.Verheggen, B.E.Jády, E.Bertrand and T.Kiss, manuscript in preparation).
Previous microinjection experiments in Xenopus oocytes favoured the nucleolus as the site of snRNA modification, although they did not exclude the possibility that Sm snRNA modification might also occur in CBs (Lange and Gerbi, 2000; Yu et al., 2001; Gerbi and Lange, 2002). In this work, we have demonstrated that, in cultured mouse cells, the nucleolus lacks modification activities responsible for 2′-O-methylation and pseudouridylation of the U5 snRNA. In contrast, activities mediating modification of the RNA pol III-specific U6 snRNA and 5.8S rRNA were readily detectable within the nucleolus. Together with our earlier observation that U2-specific sequences remain unmodified when expressed in the nucleolus (Ganot et al., 1999), it seems very unlikely that the mammalian nucleolus has a function in the post-transcriptional modification of Sm-class snRNAs.
Furthermore, the observation that cytoplasmic pre-U2 snRNA lacks internal modifications led us to conclude that cytoplasmic RNP assembly, 3′-end maturation, cap hypermethylation and nuclear re-import precede the nuclear modification events carried out by the scaRNPs. The Lamond laboratory has shown that after re-entering the nucleus, nascent Sm snRNPs (marked by GFP-tagged Sm proteins) transiently accumulate in CBs before concentrating in speckles (Sleeman and Lamond, 1999a). Consistent with this view, previous localization studies concluded that spliceosomal snRNAs associated with CBs possess a TMG cap and are complexed with Sm proteins, indicating that they have completed the cytoplasmic maturation steps (reviewed in Matera, 1999). Thus, the available data are most consistent with a model in which newly assembled snRNPs re-enter the nucleus and pass through CBs, where they undergo scaRNP-mediated 2′-O-methylation and pseudouridylation.
The importance of the Sm binding site for U2 modification has been confirmed both in transfected COS-7 cells (this work) and in microinjected Xenopus eggs (Yu et al., 2001). However, our discovery that short fragments of U5 and U2 snRNAs embedded in the U87 scaRNA are correctly modified when targeted into the CB demonstrates that neither the Sm binding site nor the associated Sm proteins are required for the scaRNP-mediated 2′-O-methylation and pseudouridylation reactions. Hence, we propose that the major, if not the only, function of the Sm motif in snRNA modification is to facilitate the nuclear import of Sm snRNPs. Our results further support the notion that, apart from the short target sequence, the snoRNA- and scaRNA-directed modification reactions are independent of both the primary sequence and secondary structure of the substrate RNA. Targeting substrate RNAs into the appropriate subnuclear compartments where their cognate modification guide RNPs accumulate may thus be one of the most important controlling elements of 2′-O-methylation and pseudouridylation of cellular RNAs. We envisage that sequestering of snRNA and rRNA modification machineries into the nucleolus and CB can prevent undesired modification events in snRNAs, rRNAs and also in mRNAs. These considerations further emphasize the importance of the structural and functional compartmentalization of the nucleus (Lamond and Earnshaw, 1998; Matera, 1999; Sleeman and Lamond, 1999b; Lewis and Tollervey, 2000; Dundr and Misteli, 2001).
Since its identification by autoimmune patient sera (Andrade et al., 1991; Raska et al., 1991), p80 coilin has been considered as an unambiguous protein marker of the CB. As such, coilin was assumed to have a crucial role in defining CB structure via recruitment of its molecular components (Bauer and Gall, 1997; Almeida et al., 1998; Bellini, 2000). In fact, coilin has been demonstrated to specifically interact with SMN (Hebert et al., 2001) and to associate, either directly or indirectly, with the U1 snRNP (Müller et al., 2000). Consistent with these findings, previous characterization of coilin knockout MEF cells revealed that in the absence of the full-length coilin protein, residual CBs are formed that fail to accumulate Sm snRNPs and the SMN protein, but concentrate the nucleolar epitopes fibrillarin and Nopp140 (Tucker et al., 2001). These fibrillarin-rich residual CBs, as demonstrated by this work, also accumulate the U3 snoRNA. Furthermore, in situ localization of the U85 and U93 scaRNAs in the coilin knockout MEF cells revealed a novel type of residual CB that specifically accumulates scaRNAs and contains Sm snRNAs, but fails to recruit detectable amounts of fibrillarin, U3 snoRNA or SMN. These results strengthen the idea that coilin tethers SMN to CBs (Hebert et al., 2001, 2002) and demonstrate that in the absence of full-length coilin, Sm snRNPs can accumulate within scaRNA-containing residual CBs, where internal modification of Sm snRNAs most likely occurs. Finally, the demonstration that two different kinds of residual CB are formed in coilin knockout MEF cells, which accumulate either Sm snRNAs or the U3 snoRNA, indicates that coilin functions to couple aspects of the biogenesis of spliceosomal snRNPs and nucleolar snoRNPs into a single nuclear subdomain. This idea is further supported by the fact that coilin can interact with both pre-mRNA processing factors such as the SMN and SmB proteins (Hebert et al., 2001) as well as with the nucleolar protein Nopp140 (Isaac et al., 1998).
A remarkable feature of the scaRNA- and fibrillarin-containing residual CBs formed in coilin knockout MEF cells is that, based on their size and shape in the light microscope, they are indistinguishable from the canonical CBs detected in control mouse cells. It is tempting to speculate that although the two kinds of residual CB contain fairly different components, they may be defined by a common structural organizer that is apparently different from coilin. The finding that at least two different kinds of residual CB are formed in the absence of coilin, coupled with the fact that these structures contain components of two distinct RNA processing pathways further supports the emerging view that the CB is an unexpectedly dynamic organelle with multiple cellular functions (Gall, 2000; Carmo-Fonseca, 2002; Ogg and Lamond, 2002).
Materials and methods
General procedures
Manipulation of DNA, RNA and oligodeoxynucleotides was carried out according to standard laboratory protocols (Sambrook et al., 1989). The identity of all constructions was verified by sequence analysis. The following oligodeoxynucleotides were used in this study: 1, TTT CTCGAGTACGAACAAGGAAGTGG; 2, TTTGGTACCTCAGGGA AGCAGTTAAGTC; 3, GGACAATATATTAAATGACCGCGGAGA GCAGGGAGATGGAAT; 4, CTAGCTTTTACTAAAGATTT; 5, TCG AAAATCTTTAGTAAAAG; 6, CTAGAGATGACACGCAAATTCGT GAAGCGC; 7, TCGAGCGCTTCACGAATTTGCGTGTCATCT; 8, CTAGATTAATGTGAATTGCAGGACACATGACTAGTC; 9, TGCA GACTAGTCATGTGTCCTGCAATTCACATTAAT; 10, ATAATCGA TTCAAATGATGTAATAAC; 11, ATACTCGAGTTCATCAGCTTAC TAAAGATTTATTGGTATGTCTCATTC; 12, ATACTCGAGGGTG ACCAAACCTTTTACCC; 13, CGAGTATCCAACAAAACGAGCT TTTACTAAAGATTTCTGGAGTGGCAGTCATGGGT; 14, CGAGT ATCCAACAAAACGATGACACGCAAATTCGTGAAGCGAGTGG CAGTCATGGGT; 15, ATAATCGATAGTCCCACTCCACTCCTGTG; 16, CCCGGAGGGGGTGCACC; 17, TCCCTGCTCTCCGCGGT; 18, TACCAGGTCGATGCGTG; 19, CCAGTGATTTTTTTCTCCATTT TAGC; 20, CACCATCACACCCATGACT; 21, TGGTTTAAGACTCA GAG; 22, AT*CCATTTTAGCTTCCT*TAGCTCCTGAAAAT*CTC GCCAAGCTGT*A; 23 AT*CATCAGCTTACTAAAGAT*TTATTG GTATGTCTCAT*A; 24, AT*CAGAAATCTTTAGTAAAAGCT*C GTTTTGTTGGATACTCGT*A; 25, AT*TCACGAATTTGCGTGT CAT*CGTTTTGTTGGATACTCGT*A; 26, AT*CTCGAGGGTACCA CCAAT*CCTGTGCGGCTCCAT*A; 27, CT*GGGCTTAGCTAAAC CAACT*GAATCACAACAGCCTTGAT*A; 28, AT*ACTGATAAG AACAGATACT*ACACTTGATCTTAGCCAT*A; 29, AT*CCACGG AAATCTTTAGT*AAAAGGCGAAAGATTTATGCGAT*A; 30, TT* GCACAGAAGCAGCACCT*AGAGCCGGCTTCACGCTT*T. Amino-allyl-modified T nucleotides are marked by asterisks.
Plasmid construction and transfection of mammalian cells
To generate the pU2sm– expression construct, the human U2 snRNA gene (DDBJ/EMBL/GenBank accession No. K03023) carrying 326 bp upstream and 120 bp downstream sequences was PCR-amplified using human HeLa genomic DNA as a template and oligonucleotides 1 and 2 as upstream and downstream primers, respectively. The obtained PCR fragment was digested with the XhoI and KpnI restriction endonucleases and introduced into the same sites of the pBluescript (Stratagene) cloning vector, resulting in pU2. Construction of pU2sm– was performed by a two-step PCR. First, the 3′ half of the U2 gene was amplified with oligonucleotides 2 and 3. Utilization of oligonucleotide 3 resulted in introduction of an altered Sm binding site (98-ACCGCGGA-105). The resulting DNA fragment was used in the second PCR as a mutagenic megaprimer in combination with oligonucleotide 1. The amplified DNA carrying the Sm mutant U2 gene was digested with XhoI and KpnI and was cloned into the same sites of pBluescript. To obtain pW-U5, oligonucleotides 4 and 5 were annealed and ligated into the XbaI and XhoI sites of the mouse ribosomal minigene construct, pW (Hadjiolova et al., 1994). The same approach was used for generation of pW-U6 and pW-5.8S, except that oligonucleotides 6/7 and 8/9, respectively, were inserted into the XbaI/XhoI-cut pW expression plasmid. To generate pCMV-globin-yU24m, the coding region of the yeast U24 snoRNA (DDBJ/EMBL/GenBank accession No. Z48760) was PCR-amplified using yeast genomic DNA as a template and oligonucleotides 10 and 11 as 5′- and 3′-specific primers, respectively. Utilization of oligo nucleotide 11 introduced a novel substrate recognition sequence (69-AAATCTTTAGTAAG-82) into the yeast U24 snoRNA. The amplified DNA fragment carrying the mutant U24 gene was digested with endonucleases ClaI and XhoI and inserted into the same sites of the pCMV-globin expression vector (Darzacq et al., 2002). To construct pCMV-globin-U87-U5 and pCMV-globin-U87-U6, the 3′ half of the human U87 scaRNA gene was PCR-amplified using the pCMV-globin-U87 plasmid as a template (Darzacq et al., 2002) and oligonucleotide 12 as a common 3′-terminal primer in combination with either oligonucleotide 13 or 14 as internal primers specific for the U87-U5 and U87-U6 RNAs, respectively. The resulting U87-U5- and U87-U6-specific fragments were used as megaprimers in the second PCRs in which oligonucleotide 15 was used as a 5′-end-specific primer and the pCMV-globin-U87 construct as a template. The amplified DNA fragments containing the U87-U5 and U87-U6 RNA genes were digested with ClaI and XhoI and inserted into the same sites of the pCMV-globin vector. Construction of pRFP-coilin (Boulon et al., 2002) and pfibrillarin-GFP (Dundr et al., 2000) expression vectors has been described. Transfection of simian COS-7 and mouse L929 and MEF cells was performed with Fugene 6 (Roche) transfection reagent according to the manufacturer’s instructions.
RNA analysis
RNA from cultured cells was isolated by the guanidine thiocyanate– phenol–chloroform extraction procedure (Goodall et al., 1990). Cell fractionation and RNase A/T1 protection analysis have been reported (Goodall et al., 1990; Darzacq et al., 2002). To obtain a sequence-specific antisense RNA probe for the human U2sm– RNA, the pU2sm– plasmid was digested with the XhoI endonuclease and used as a template for in vitro RNA synthesis by the T3 RNA polymerase. To obtain antisense probes for the human U87-U5 and U87-U6 RNAs and the yeast U24m RNA, the pCMV-globin-U87-U5, pCMV-globin-U87-U6 and pCMV-globin-U24m expression vectors were linearized with HindIII and transcribed with SP6 RNA polymerase. Mapping of 2′-O-ribose-methylated nucleotides was achieved by the dNTP concentration-dependent primer-extension analysis (Maden et al., 1995). Pseudouridines were visualized by primer extension after reacting the test RNA with CMC (Bakin and Ofengand, 1993). Terminally labelled oligonucleotides were used as primers for mapping of human pre-U2 (oligonucleotide 16), U2sm– (oligonucleotide 17) and U2 (oligonucleotide 18) snRNAs, the W-U5, W-U6 and W-5.8S minigene transcripts (oligonucleotide 19), the U87-U5 and U87-U6 scaRNAs (oligonucleotide 20) and the mouse U5 snRNA (oligonucleotide 21). The labelled primers and antisense RNA probes were purified on 20 and 6% sequencing gels, respectively.
Fluorescent in situ hybridization
In situ hybridization of mouse and COS-7 cells with fluorescent oligonucleotide probes was performed as has been described (http://singerlab.aecom.yu.edu; Darzacq et al., 2002). Sequence-specific oligonucleotides containing aminoallyl-T nucleotides were used for preparation of fluorescent probes for in situ localization of the W-U5 minigene transcript (oligonucleotide 22), the yeast U24m snoRNA (oligonucleotide 23), the human U87-U5 (oligonucleotide 24), U87-U6 (oligonucleotide 25), U93 (oligonucleotide 26) and U85 (oligo nucleotide 27) scaRNAs, the U2 (oligonucleotide 28) and U5 snRNAs (oligonucleotide 29) as well as the U3 snoRNA (oligonucleotide 30). Oligonucleotides 22, 23, 26 and 27 were labelled with FluoroLink Cy3 monofunctional reactive dye (Amersham), whereas the oligonucleo tides 24, 25, 28, 29 and 30 were labelled with FluoroLink Cy5 monofunctional reactive dye (Amersham). Nuclear DNA was stained by 0.1 µg/ml DAPI.
Acknowledgments
Acknowledgements
We are grateful to Y.de Preval for synthesis of oligonucleotides. B.E.J. is supported by a long term EMBO fellowship. X.D. was funded by la Fondation pour la Recherche Médicale. Our work was supported by grants from la Ligue Nationale contre le Cancer, Association pour la Recherche contre le Cancer and the French MNRT (ACI), the National Institutes of Health (NIH) and the Muscular Dystrophy Association (MDA).
References
- Almeida F., Saffrich,R., Ansorge,W. and Carmo-Fonseca,M. (1998) Microinjection of anti-coilin antibodies affects the structure of coiled bodies. J. Cell Biol., 142, 899–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade L.E., Chan,E.K., Raska,I., Peebles,C.L., Roos,G. and Tan,E.M. (1991) Human autoantibody to a novel protein of the nuclear coiled body: immunological characterization and cDNA cloning of p80-coilin. J. Exp. Med., 173, 1407–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ares M. Jr, Mangin,M. and Weiner,A.M. (1985) Orientation-dependent transcriptional activator upstream of a human U2 snRNA gene. Mol. Cell. Biol., 5, 1560–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakin A. and Ofengand,J. (1993) Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: analysis by the application of a new sequencing technique. Biochemistry, 32, 9754–9762. [DOI] [PubMed] [Google Scholar]
- Bauer D.W. and Gall,J.G. (1997) Coiled bodies without coilin. Mol. Biol. Cell, 8, 73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellini M. (2000) Coilin, more than a molecular marker of the cajal (coiled) body. BioEssays, 22, 861–867. [DOI] [PubMed] [Google Scholar]
- Boulon S., Basyuk,E., Blanchard,J.M., Bertrand,E. and Verheggen,C. (2002) Intra-nuclear trafficking: insight from live cell imaging. Biochimie, 84, 805–813. [DOI] [PubMed] [Google Scholar]
- Carmo-Fonseca M. (2002) New clues to the function of the Cajal body. EMBO rep., 3, 726–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X., Jády,B.E., Verheggen,C., Kiss,A.M., Bertrand,E. and Kiss,T. (2002) Cajal body-specific small nuclear RNAs: a novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J., 21, 2746–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M. and Misteli,T. (2001) Functional architecture in the cell nucleus. Biochem. J., 356, 297–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M., Misteli,T. and Olson,M.O. (2000) The dynamics of postmitotic reassembly of the nucleolus. J. Cell Biol., 150, 433–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W. and Pogacic,V. (2002) Biogenesis of small nucleolar ribonucleoproteins. Curr. Opin. Cell Biol., 14, 319–327. [DOI] [PubMed] [Google Scholar]
- Gall J.G. (2000) Cajal bodies: the first 100 years. Annu. Rev. Cell Dev. Biol., 16, 273–300. [DOI] [PubMed] [Google Scholar]
- Ganot P., Bortolin,M.L. and Kiss,T. (1997a) Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell, 89, 799–809. [DOI] [PubMed] [Google Scholar]
- Ganot P., Caizergues-Ferrer,M. and Kiss,T. (1997b) The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev., 11, 941–956. [DOI] [PubMed] [Google Scholar]
- Ganot P., Jády,B.E., Bortolin,M.L., Darzacq,X. and Kiss,T. (1999) Nucleolar factors direct the 2′-O-ribose methylation and pseudouridylation of U6 spliceosomal RNA. Mol. Cell. Biol., 19, 6906–6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbi S.A. and Lange,T.S. (2002) All small nuclear RNAs (snRNAs) of the [U4/U6.U5] Tri-snRNP localize to nucleoli; identification of the nucleolar localization element of U6 snRNA. Mol. Biol. Cell., 13, 3123–3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodall G.J., Wiebauer,K. and Filipowicz,W. (1990) Analysis of pre-mRNA processing in transfected plant protoplasts. Methods Enzymol., 181, 148–161. [DOI] [PubMed] [Google Scholar]
- Hadjiolova K.V., Normann,A., Cavaille,J., Soupene,E., Mazan,S., Hadjiolov,A.A. and Bachellerie,J.P. (1994) Processing of truncated mouse or human rRNA transcribed from ribosomal minigenes transfected into mouse cells. Mol. Cell. Biol., 14, 4044–4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert M.D., Szymczyk,P.W., Shpargel,K.B. and Matera,A.G. (2001) Coilin forms the bridge between Cajal bodies and SMN, the spinal muscular atrophy protein. Genes Dev., 15, 2720–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert M.D., Shpargel,K.B., Ospina,J.K., Tucker,K.E. and Matera,A.G. (2002) Coilin methylation regulates nuclear body formation. Dev. Cell, 3, 329–337. [DOI] [PubMed] [Google Scholar]
- Isaac C., Yang,Y. and Meier,U.T. (1998) Nopp140 functions as a molecular link between the nucleolus and the coiled bodies. J. Cell Biol., 142, 319–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jády B.E. and Kiss,T. (2001) A small nucleolar guide RNA functions both in 2′-O-ribose methylation and pseudouridylation of the U5 spliceosomal RNA. EMBO J., 20, 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T. (2001) Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J., 20, 3617–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T. (2002) Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell, 109, 145–148. [DOI] [PubMed] [Google Scholar]
- Kiss A.M., Jády,B.E., Darzacq,X., Verheggen,C., Bertrand,E. and Kiss,T. (2002) A Cajal body-specific pseudouridylation guide RNA is composed of two box H/ACA snoRNA-like domains. Nucleic Acids Res., 30, 4643–4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond A.I. and Earnshaw,W.C. (1998) Structure and function in the nucleus. Science, 280, 547–553. [DOI] [PubMed] [Google Scholar]
- Lange T.S. and Gerbi,S.A. (2000) Transient nucleolar localization of U6 small nuclear RNA in Xenopus laevis oocytes. Mol. Biol. Cell, 11, 2419–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapeyre B., Mariottini,P., Mathieu,C., Ferrer,P., Amaldi,F., Amalric,F. and Caizergues-Ferrer,M. (1990) Molecular cloning of Xenopus fibrillarin, a conserved U3 small nuclear ribonucleoprotein recognized by antisera from humans with autoimmune disease. Mol. Cell. Biol., 10, 430–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J.D. and Tollervey,D. (2000) Like attracts like: getting RNA processing together in the nucleus. Science, 288, 1385–1389. [DOI] [PubMed] [Google Scholar]
- Lyon C.E., Bohmann,K., Sleeman,J. and Lamond,A.I. (1997) Inhibition of protein dephosphorylation results in the accumulation of splicing snRNPs and coiled bodies within the nucleolus. Exp. Cell Res., 230, 84–93. [DOI] [PubMed] [Google Scholar]
- Maden B.E., Corbett,M.E., Heeney,P.A., Pugh,K. and Ajuh,P.M. (1995) Classical and novel approaches to the detection and localization of the numerous modified nucleotides in eukaryotic ribosomal RNA. Biochimie, 77, 22–29. [DOI] [PubMed] [Google Scholar]
- Massenet S., Mougin,A. and Branlant,C. (1998) Posttrancriptional modification in the U small nuclear RNAs. In Grosjean,H. and Benne,R. (eds), Modification and Editing of RNA. ASM Press, Washington, DC, pp. 201–227.
- Matera A.G. (1999) Nuclear bodies: multifaceted subdomains of the interchromatin space. Trends Cell Biol., 9, 302–309. [DOI] [PubMed] [Google Scholar]
- Mattaj I.W. (1986) Cap trimethylation of U snRNA is cytoplasmic and dependent on U snRNP protein binding. Cell, 46, 905–911. [DOI] [PubMed] [Google Scholar]
- Moss T. and Stefanovsky,V.Y. (1995) Promotion and regulation of ribosomal transcription in eukaryotes by RNA polymerase I. Prog. Nucleic Acid Res. Mol. Biol., 50, 25–66. [DOI] [PubMed] [Google Scholar]
- Mouaikel J., Verheggen,C., Bertrand,E., Tazi,J. and Bordonne,R. (2002) Hypermethylation of the cap structure of both yeast snRNAs and snoRNAs requires a conserved methyltransferase that is localized to the nucleolus. Mol. Cell, 9, 891–901. [DOI] [PubMed] [Google Scholar]
- Muller B., Link,J. and Smythe,C. (2000) Assembly of U7 small nuclear ribonucleoprotein particle and histone RNA 3′ processing in Xenopus egg extracts. J. Biol. Chem., 275, 24284–24293. [DOI] [PubMed] [Google Scholar]
- Narayanan A., Speckmann,W., Terns,R. and Terns,M.P. (1999) Role of the box C/D motif in localization of small nucleolar RNAs to coiled bodies and nucleoli. Mol. Biol. Cell, 10, 2131–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogg S.C. and Lamond,A.I. (2002) Cajal bodies and coilin—moving towards function. J. Cell Biol., 159, 17–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raska I., Andrade,L.E., Ochs,R.L., Chan,E.K., Chang,C.M., Roos,G. and Tan,E.M. (1991) Immunological and ultrastructural studies of the nuclear coiled body with autoimmune antibodies. Exp. Cell Res., 195, 27–37. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sleeman J.E. and Lamond,A.I. (1999a) Newly assembled snRNPs associate with coiled bodies before speckles, suggesting a nuclear snRNP maturation pathway. Curr. Biol., 9, 1065–1074. [DOI] [PubMed] [Google Scholar]
- Sleeman J.E. and Lamond,A.I. (1999b) Nuclear organization of pre-mRNA splicing factors. Curr. Opin. Cell Biol., 11, 372–377. [DOI] [PubMed] [Google Scholar]
- Terns M.P. and Terns,R.M. (2002) Small nucleolar RNAs: versatile trans-acting molecules of ancient evolutionary origin. Gene Expr., 10, 17–39. [PMC free article] [PubMed] [Google Scholar]
- Tucker K.E. et al. (2001) Residual Cajal bodies in coilin knockout mice fail to recruit Sm snRNPs and SMN, the spinal muscular atrophy gene product. J. Cell Biol., 154, 293–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski K.T., You,Z.H., Graham,P.J. and Steitz,J.A. (1998) Modification of U6 spliceosomal RNA is guided by other small RNAs. Mol. Cell, 2, 629–638. [DOI] [PubMed] [Google Scholar]
- Verheggen C., Lafontaine,D.L., Samarsky,D., Mouaikel,J., Blanchard,J.M., Bordonne,R. and Bertrand,E. (2002) Mammalian and yeast U3 snoRNPs are matured in specific and related nuclear compartments. EMBO J., 21, 2736–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa T., Pleiss,J.A. and Guthrie,C. (2002) Spliceosomal snRNAs: Mg2+-dependent chemistry at the catalytic core? Cell, 109, 149–152. [DOI] [PubMed] [Google Scholar]
- Will C.L. and Lührmann,R. (2001) Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol., 13, 290–301. [DOI] [PubMed] [Google Scholar]
- Yu Y.T., Shu,M.D. and Steitz,J.A. (1998) Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. EMBO J., 17, 5783–5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y.T., Shu,M.D., Narayanan,A., Terns,R.M., Terns,M.P. and Steitz,J.A. (2001) Internal modification of U2 small nuclear (sn)RNA occurs in nucleoli of Xenopus oocytes. J. Cell Biol., 152, 1279–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]