Abstract
The related-to-ubiquitin (RUB) protein is post-translationally conjugated to the cullin subunit of the SCF (SKP1, Cullin, F-box) class of ubiquitin protein ligases. Although the precise biochemical function of RUB modification is unclear, studies indicate that the modification is important for SCF function. In Arabidopsis, RUB modification of CUL1 is required for normal function of SCFTIR1, an E3 required for response to the plant hormone auxin. In this report we show that an Arabidopsis protein called RCE1 functions as a RUB-conjugating enzyme in vivo. A mutation in the RCE1 gene results in a phenotype like that of the axr1 mutant. Most strikingly, plants deficient in both RCE1 and AXR1 have an embryonic phenotype similar to mp and bdl mutants, previously shown to be deficient in auxin signaling. Based on these results, we suggest that the RUB-conjugation pathway is required for auxin-dependent pattern formation in the developing embryo. In addition, we show that RCE1 interacts directly with the RING protein RBX1 and is present in a stable complex with SCF. We propose that RBX1 functions as an E3 for RUB modification of CUL1.
Keywords: auxin/CUL1/RCE1/RUB conjugation/RUB modification
Introduction
The ubiquitin–protein conjugation pathway regulates diverse cellular functions by promoting the ubiquitylation of protein substrates (Pickart, 2001). The pathway consists of three proteins, or protein complexes, called ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2) and ubiquitin-protein ligase (E3). The SCFs make up one important class of E3 ligases (Deshaies, 1999). SCFs consists of four subunits: an F-box protein responsible for substrate binding, SKP1 (ASK in Arabidopsis), the RING protein RBX1/ROC1/HRT1 and CUL1 (Gray et al., 1999). Biochemical and structural studies show that the CUL1 subunit functions as a scaffold for the complex (Zheng et al., 2002). SKP1 and the F-box protein form a subcomplex that binds near the N-terminus of CUL1, while the RBX1 subunit binds sequences near the C-terminus (Furukawa et al., 2000; Zheng et al., 2002). RBX1 also binds E2, thus bringing E2 in close proximity to the substrate (Deshaies, 1999; Zheng et al., 2002).
The Arabidopsis genome encodes ∼700 F-box proteins, suggesting a very broad role for SCFs in plants (Gagne et al., 2002). So far, SCF complexes have been directly implicated in response to the plant hormones auxin (SCFTIR1) (Gray et al., 1999) and jasmonate (SCFCOI1) (Xu et al., 2002). In addition, genetic studies have implicated F-box proteins in circadian rhythm (Nelson et al., 2000; Somers et al., 2000), senescence (Woo et al., 2001), apical dominance (Stirnberg et al., 2002), flower and meristem development (Ingram et al., 1997; Samach et al., 1999) and phytochrome signaling (Dieterle et al., 2001).
The related-to-ubiquitin protein (RUB), called Nedd8 in some species, is conjugated to the SCF subunit CUL1 through a series of steps similar to ubiquitin conjugation (Yeh et al., 2000; Hellmann and Estelle, 2002). A single RUB molecule is linked to a specific lysine residue near the C-terminus of the protein (Yeh et al., 2000). Genetic studies in diverse species indicate that cullin modification is required for SCF function. In fission yeast, the modification is essential for viability while in mouse and Caenorhabditis elegans, a defect in the Nedd8 conjugation pathway results in embryonic lethality (Osaka et al., 2000; Tateishi et al., 2001; Kurz et al., 2002). So far, the precise role of RUB modification is unclear. In vitro, the modification increases SCF activity (Morimoto et al., 2000; Podust et al., 2000; Read et al., 2000; Wu et al., 2000). However, genetic studies in Arabidopsis and fungi show that both decreased and increased levels of RUB–CUL1 have a negative effect on SCF function (Lyapina et al., 2001; Schwechheimer et al., 2001; Gray et al., 2002). These results suggest that a cycle of RUB conjugation and removal is required for SCF activity in vivo.
In Arabidopsis, RUB conjugation is accomplished by a heterodimeric RUB-activating enzyme composed of the AXR1 and ECR1 proteins and a RUB-conjugating enzyme called RCE1 (del Pozo and Estelle, 1999; del Pozo et al., 2002; Dharmasiri and Estelle, 2002). Whether or not there is a RUB-specific E3 activity is uncertain. Recent results suggest that the RBX1 protein may provide this function. Overexpression of RBX1 in budding yeast and in Arabidopsis results in an increase in RUB–CUL1 formation (Kamura et al., 1999; Gray et al., 2002).
In this paper, we describe a genetic and biochemical analysis of the RCE1 protein. Our results show that severe disruption of the RUB conjugation pathway in Arabidopsis causes a seedling lethal phenotype characteristic of a defect in auxin signaling (Hardtke and Berleth, 1998; Hamann et al., 2002). In addition, we show that RCE1 interacts directly with RBX1 and is in a stable complex with the SCF. Based on these results, we propose that RBX1 functions as an E3 for RUB modification of CUL1.
Results
Expression of the RCE1 gene
In Arabidopsis, the heterodimeric RUB-activating enzyme is composed of the AXR1 and ECR1 proteins (del Pozo et al., 1998, 2002; del Pozo and Estelle, 1999). Previous studies have shown that AXR1 and ECR1 are expressed throughout the life cycle of the plant with particularly high expression levels in dividing and elongating cells (del Pozo et al., 2002). To investigate the pattern of RCE1 expression, we first examined expression in various plant tissues by RNA blotting. As indicated in Figure 1A, RCE1 RNA accumulates to substantial levels in all plant organs examined. To further characterize RCE1 expression, we constructed an in-frame fusion between RCE1 and the β-glucuronidase (GUS) gene. The construct, including a 1.5 kb DNA fragment from the promoter region of RCE1, was introduced into Arabidopsis plants, and 16 lines were examined for expression of GUS. All of the lines had a similar pattern of GUS staining. In young seedlings, GUS staining was strongest in the elongation zone of the root and at the shoot–hypocotyl junction (Figure 1B). Significant staining was also observed in root hairs. In older seedlings, intense staining was observed at the shoot apex, the root tip and the site of lateral root initiation (Figure 1C–E). In leaves, particularly strong staining was observed in the veins and trichomes (Figure 1F and G). The results of in situ hybridization studies also indicated strong expression in the inflorescence and floral meristems (data not shown). These results indicate that RCE1 is broadly expressed in a manner similar to that of AXR1 and ECR1.
Fig. 1. Pattern of RCE1 gene expression analyzed by RNA blot and GUS staining. (A) RNA blot showing expression of RCE1 in all the tissues examined. The lower panel shows an ethidium bromide stained gel to demonstrate equal loading. (B–G) GUS staining in RCE1::GUS plants. These plants carry a translational fusion between RCE1 and GUS under control of the RCE1 promoter. (B) GUS staining in 4-day-old light grown seedling. (C) Shoot apical region of a 7-day-old light grown seedling. (D) Root apex of a 10-day-old seedling. (E) Root segment from a 10-day-old seedling showing GUS staining in lateral root primordium. (F) Primary leaf from 10-day-old seedling. (G) A magnified view of a trichome from (F) showing intense staining at the basal region.
Loss of RCE1 results in altered morphology
Mutations in the AXR1 subunit of the RUB-activating enzyme result in diverse defects in morphology (del Pozo et al., 1998). Similar defects are observed in plants expressing a dominant-negative form of the ECR1 subunit (del Pozo et al., 2002). To investigate the role of RCE1 in plant growth and development, we searched the available collections of insertion lines for mutations in RCE1. One line from the IMA collection contains a Ds insert 371 nucleotides upstream of the ATG for RCE1. This mutation, named rce1-1, is recessive and confers a characteristic phenotype (described below). RNA blot analysis indicates that RCE1 RNA levels are significantly reduced in plants homozygous for the rce1-1 allele (Figure 2A). However, some transcript is still present, indicating that rce1-1 is not a null mutation. To confirm that the rce1-1 mutation is responsible for the phenotype described below, we constructed a 35S::Myc-RCE1 fusion gene and introduced it into rce1-1 plants. The transgene restored normal morphology to mutant plants indicating that the observed defects are caused by a reduction in RCE1 levels (data not shown).
Fig. 2. A mutation in RCE1 confers morphological defects similar to auxin-resistant mutants. (A) RNA blot showing expression of RCE1 in 6-day-old wild-type and rce1-1 seedlings. (B) Response of 6-day-old seedling roots to gravity. (C) Twenty-one-day-old rosettes. (D) Thirty-six-day-old plants. For both (C) and (D), Ler is on the left and rce1-1 is on the right.
Based on genetic segregation data, the rce1 mutation does not significantly disrupt gametophyte or embryo development. However, rce1 plants exhibit a variety of growth defects throughout development. When grown on the surface of vertically orientated agar medium, the roots of mutant seedlings appeared to wander over the surface of the agar, suggesting a defect in gravitropism. To test this possibility, wild-type and mutant roots were grown in a vertical orientation for 6 days and turned 90° to a horizontal orientation. After 8 h, the angle from the vertical axis was measured. The results in Figure 2B show that rce1-1 seedlings have a reduced response to the change in the gravity vector. The axr1-12 mutant has a similar defect.
Rosette and inflorescence morphology are also altered in the rce1 mutant (Table I; Figure 2C and D). In general, organ length is reduced throughout development. The rosette leaves of rce1 plants are smaller than those of Ler leaves, with shorter petioles and rounder, crinkled leaf blades. The inflorescence is shorter and more highly branched than Ler, a phenotype that is quite similar to axr1 plants. Finally, the length of the floral organs is reduced (data not shown), resulting in the production of a shorter silique (Table I).
Table I. Morphological analysis of rce1-1.
| Ler | rce1-1 | |
|---|---|---|
| Mature plant height (cm) | 22.7 ± 1.8 | 8.3 ± 1.3 |
| Number of primary inflorescences per plant | 2.4 ± 0.9 | 6.1 ± 1.0 |
| Length of leaf blade (cm) | 1.3 ± 0.2 | 0.7 ± 0.1 |
| Length of petiole (cm) | 0.7 ± 0.1 | 0.4 ± 0.07 |
| Length of silique (cm) | 1.3 ± 0.1 | 0.6 ± 0.09 |
RCE1 is required for RUB modification of CUL1
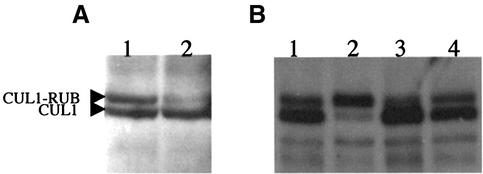
In a previous study, we showed that RCE1 promotes RUB modification of CUL1 in vitro (del Pozo and Estelle, 1999). To show that this is also the case in vivo, we examined the status of CUL1 in wild-type and rce1-1 plants by immunoblotting. Figure 3A shows that the level of RUB–CUL1 in the mutant is significantly reduced compared with Ler plants. To further investigate the effects of loss of RCE1 on the pathway, we crossed rce1-1 into a line that carries the 35S-RBX1 transgene. Overexpression of RBX1 results in a dramatic increase in RUB–CUL1 levels (Gray et al., 2002). The results in Figure 3B show that the loss of RCE1 suppresses the effect of increased RBX1 levels on RUB1–CUL1, confirming that RCE1 acts upstream of RBX1 in the RUB conjugation pathway.
Fig. 3. RCE1 is required for RUB modification of CUL1. Ten micrograms of total proteins were loaded in each lane. Proteins were blotted and treated with α-CUL1 antiserum. (A) Protein blot of Ler (lane 1) and rce1-1 (lane 2). (B) Protein blot of Col-0 (lane 1), 35S::RBX1-2B RCE1 (lane 2), rce1-1 (lane 3) and 35S::RBX1-2B rce1-1 (lane 4).
The rce1 mutants are deficient in auxin and jasmonate response
A number of studies have shown that SCF function is sensitive to the level of RUB–CUL1 modification (Liakopoulos et al., 1999; Morimoto et al., 2000; Osaka et al., 2000; Podust et al., 2000; Read et al., 2000; Gray et al., 2001, 2002; Lyapina et al., 2001; Schwechheimer et al., 2001). In Arabidopsis, the best-characterized SCF complex is SCFTIR1, which is required for auxin response (Gray et al., 1999). To determine whether the rce1-1 plants are deficient in auxin response, we first examined the effects of auxin on root growth. In our standard root growth assay, we found that rce1-1 is significantly resistant to the synthetic auxin 2,4-D compared with the parental Ler line (Figure 4A). The axr1-12 mutant was included in this experiment for comparison. To confirm that the auxin response defect is related to a reduction in RCE1 levels, we also tested rce1-1, 35S::Myc-RCE1 plants for auxin response. Figure 4B shows that the transgene restores normal auxin response to rce1-1 plants. Another well-characterized effect of auxin is the induction of lateral root formation (Himanen et al., 2002). To determine the effect of rce1-1 on this process, we transferred 6-day-old seedlings to medium with 85 nM 2,4-D and counted lateral roots after 3 days. As shown in Figure 4C, rce1-1 seedlings produced fewer lateral roots in response to auxin than the wild type.
Fig. 4. RCE1 is required for auxin and jasmonate response. (A) Inhibition of seedling roots on media containing 2,4-D. Col (diamonds), Ler (triangles), rce1-1 (squares) and axr1-12 (circles). (B) Growth of rce1-1, rce1-1, 35S::Myc-RCE1 and Col-0 roots on medium containing 85 nM 2,4-D. Black line represents the position of the root tip at time of transfer to auxin medium. (C) Lateral root formation in seedling roots in response to 85 nM 2,4-D. (D) RNA blot showing the induction of IAA2 in 6-day-old seedlings treated with or without 20 µM 2,4-D for 60 min. The ethidium bromide stained gel is shown in the bottom panel. (E) GUS staining of 7-day-old seedlings. The seedlings were treated with or without 20 µM 2,4-D for 2 h. (F) Pulse–chase analysis of IAA7/AXR2 in Col-0 and rce1-1 seedlings. The half-lives represent the mean of three separate experiments ± SD. (G) Inhibition of seedling roots by methyl jasmonate. Col-0 (diamonds), Ler (triangles), rce1-1 (squares) and axr1-12 (circles).
To determine whether these growth defects were accompanied by changes in auxin-regulated gene expression, we examined expression of the auxin-response gene IAA2 in wild-type and rce1-1 seedlings. Figure 4D shows that induction of this gene is reduced in rce1-1 plants. Similarly, expression of the auxin-responsive GUS reporter BA3-GUS is nearly absent in the rce1-1 background (Figure 4E). All of these results clearly demonstrate that RCE1 is required for auxin response.
So far, the only known SCF substrates in Arabidopsis are the Aux/IAA proteins (Gray et al., 2001). To determine whether the rce1-1 mutation affects degradation of these proteins, we measured the half-life of the AXR2/IAA7 protein in a pulse–chase experiment. The results of a representative experiment are shown in Figure 4E. Based on three experiments, the half-life of AXR2/IAA7 is 7.99 ± 1.96 min and 12.26 ± 1.97 min for Col and rce1-1, respectively (Figure 4F). These results indicate that degradation of AXR2/IAA7 is impaired in the rce1-1 mutant.
In Arabidopsis, SCFCOI1 is required for response to the plant hormone jasmonic acid (JA) (Xie et al., 1998; Xu et al., 2002). Several recent studies have shown that the axr1 mutants are moderately resistant to JA, suggesting that RUB modification of CUL1 is also required for optimal SCFCOI1 function (Gray et al., 2002; Schwechheimer et al., 2002; Tiryaki and Staswick, 2002; Xu et al., 2002). We examined the response of the rce1-1 mutant and find that it also displays reduced JA response, confirming that RUB modification of CUL1 is required for normal SCFCOI1 function (Figure 4G).
The RUB conjugation pathway is required during embryogenesis
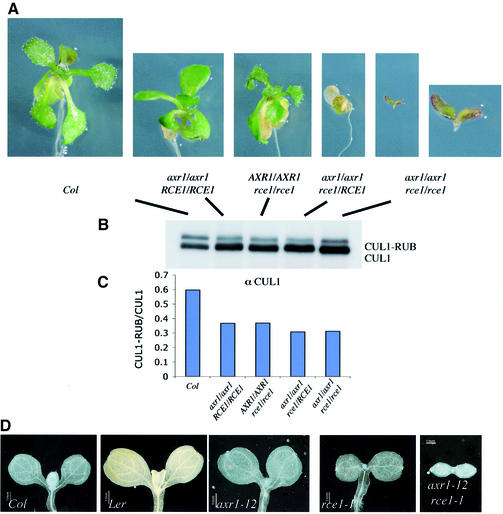
The phenotypes of the axr1 and rce1 mutants indicate that the RUB conjugation pathway has an important role in plant growth and development. However, rce1 is not a null mutation, and a related protein called RCE2 may also provide some RUB E2 activity (S.Dharmasiri and M.Estelle, unpublished data). Similarly, a gene closely related to AXR1 is present in the Arabidopsis genome, suggesting that the axr1-12 mutant may retain some RUB E1 activity (N.Dharmasiri and M.Estelle, unpublished data). Indeed the axr1-12 mutation reduces but does not eliminate RUB modification of CUL1 (Figure 5A). To determine the effects of a more severely impaired RUB conjugation pathway, we crossed the axr1-12 mutant to rce1-1 plants and examined the phenotype of the F2 generation. The results are shown in Figure 5A. Strikingly, the RCE1 gene is dosage sensitive in an axr1 background. Homozygous axr1-12 seedlings with a single RCE1 gene had a seedling lethal phenotype. After germination, these seedlings did not produce any leaves and soon died. Homozygous axr1-12 rce1-1 seedlings had an even more severe phenotype. Double mutant seedlings typically had two cotyledons but lacked all basal structures including the hypocotyl and root. This phenotype is very similar to that of the monopteros (mp), bodenlos (bdl) and auxin resistant 6 (axr6) seedlings (Berleth and Jurgens, 1993; Hamann et al., 1999b; Hobbie et al., 2000). Recent studies implicate MP, BDL and AXR6 in auxin regulation of pattern formation during embryogenesis (Hardtke and Berleth, 1998; Hamann et al., 2002).
Fig. 5. The RUB conjugation pathway is required during embryogenesis. (A) Phenotypes of wild-type and mutant seedlings. The rce1-1 axr1-12 double mutant seedlings were 6 days old when photographed. These seedlings died within 10–12 days of germination. All other seedlings were 12–14 days old when photographed. (B) Protein blot showing the levels of unmodified and modified CUL1 for each genotype. Total proteins were extracted from 6-day-old seedlings and immunoblotted with α-CUL1 antiserum. Ten micrograms of protein was loaded in each lane. (C) Ratio of RUB–CUL1 to CUL1 from (B) determined using NIH image. (D) Vascular patterning in cotyledons of 6-day-old seedlings.
The effects of each genotype on CUL1 modification are shown in Figure 5B. Surprisingly, significant amounts of modified CUL1 are present in each of the lines, including the double mutant. However, the level of unmodified CUL1 increases in the mutant backgrounds so that the relative level of RUB–CUL1 is decreased in each case (Figure 5C). Thus, a defect in RUB conjugation results in an increase in stability of the CUL1 protein.
Auxin is known to play an important role in vascular development (Berleth et al., 2000). To determine whether the axr1-12 rce1-1 double mutants exhibit defects in vascular structure, we examined the cotyledons of these seedlings. Both axr1-12 and rce1-1 cotyledons showed only slightly reduced vascular development (Figure 5D). In contrast, vascular development was severely deficient in axr1-12 rce1-1 plants. Typically, one short vascular strand was visible in each cotyledon of these seedlings. Again, this phenotype is similar to that observed in mp, bdl and axr6 seedlings (Berleth and Jurgens, 1993; Hamann et al., 1999a; Hobbie et al., 2000).
RCE1 interacts directly with RBX1 and is part of a stable complex with the SCF
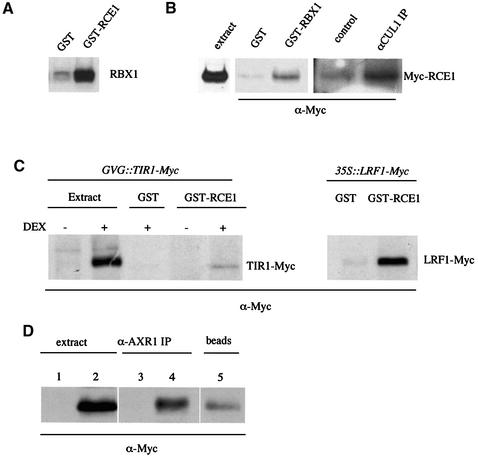
Recent studies have shown that overexpression of RBX1 dramatically increases the level of RUB–CUL1 (Kamura et al., 1999; Gray et al., 2002). Based on these results, we have suggested that RBX1 functions as an E3 for RUB modification of CUL1 (Gray et al., 2002). If this is the case, RBX1 should directly bind RCE1. To test this possibility, we added 32P-labeled RBX to glutathione beads carrying GST–RCE1. The results in Figure 6A show that labeled RBX1 is retained on the beads, indicating a specific interaction between RBX1 and RCE1. To determine whether RCE1 interacts with SCF complexes, we used the 35S::Myc-RCE1 line. As described above, this transgene restored a wild-type phenotype to rce1-1 plants, indicating that the Myc-RCE1 protein is functional. Initially, we used 35S::Myc-RCE1 plants to determine whether GST–RBX1 interacts with Myc-RCE1. The results in Figure 6B show that GST–RBX1 interacts with Myc-RCE1 in plant extracts, consistent with our results using purified proteins. To show that CUL is also present in a complex with RCE1, we immunoprecipitated CUL1 from extracts prepared from 35S::Myc-RCE1 seedlings. Immunoblotting with anti-Myc antibody revealed the presence of Myc-RCE1 in the immunoprecipitate (Figure 6B). Taken together these results indicate that RCE1 interacts with a complex that contains at least the CUL1 and RBX1 proteins.
Fig. 6. RCE1 interacts with the SCF and the RUB activating enzyme. (A) [32P]RBX1 (20 µl) was added to glutathione beads loaded with 3 µg of either GST or GST–RCE1. After washing, proteins retained on the beads were analyzed by SDS–PAGE. (B) Protein extracts were prepared from 35S::Myc-RCE1 seedlings. Proteins were recovered after pull-down with GST–RBX1 or immunoprecipitation with α-CUL1 antiserum and analyzed by immunoblot using α-Myc antiserum. (C) GST–RCE1 pulldowns were performed using protein extracts prepared from GVG::TIR1-Myc or 35S::LRF1-Myc seedlings. Recovered proteins were analyzed by immunoblot using α-Myc antiserum. (D) Protein extracts from 35S::Myc-RCE1 seedlings were immunoprecipitated with α-AXR1 antiserum and immunoblotted with α-Myc antibody. Lanes 1 and 3, Columbia extract; lanes 2, 4 and 5, 35S::Myc-RCE1 extract.
To confirm and extend these results, we performed GST pull-down experiments with GST–RCE1 to demonstrate an interaction with a complex containing an F-box protein. Extracts were prepared from Arabidopsis lines expressing the TIR1-Myc transgene. Figure 6C shows that GST–RCE1 pulled down the TIR1-Myc protein from plant extracts. This experiment was also performed with another F-box protein that is closely related to TIR1 called leucine rich repeat F-box 1 (LRF1). As shown in Figure 6C, GST–RCE1 also pulled down LRF1-Myc from plant extracts. These results indicate that RCE1 interacts with complete SCF complexes.
Since RCE1 is associated with the SCF, we wondered whether AXR1 might also be in this complex. First we asked whether AXR1 interacts with Myc-RCE1 by immunoprecipitating AXR1 from 35S::Myc-RCE1 seedlings. Figure 6D shows that Myc-RCE1 co-immunoprecipitates with AXR1, indicating that RCE1 forms a stable complex with AXR1. We next attempted to demonstrate an interaction between AXR1–ECR1 and the SCF by reciprocal co-immunoprecipitation of AXR1 and CUL1, and GST pull-down experiments. However, we were unable to detect an interaction using either of these approaches.
Discussion
The conjugation of RUB/Nedd8 to cullin proteins is a highly conserved process that occurs in all eukaryotes (Yeh et al., 2000). As in the ubiquitin pathway, RUB conjugation requires a RUB-activating enzyme and a RUB-conjugating enzyme. In Arabidopsis, the activating enzyme is a heterodimer consisting of the AXR1 and ECR1 proteins (del Pozo et al., 1998, 2002). Loss of the AXR1 subunit results in a reduction in RUB-modified CUL1 and a variety of growth defects, many of which appear to be caused by stabilization of SCF substrates. In this study, we show that the RCE1 protein is also required for RUB modification of CUL1, consistent with its proposed role as RUB-E2 (del Pozo and Estelle, 1999). Insertion of a Ds element adjacent to RCE1 results in a phenotype that is very similar to that of the axr1 mutant, including stabilization of Aux/IAA proteins and a reduction in auxin response.
Among animals, RUB is known to be essential for viability in C.elegans, Drosophila melanogaster and mice (Tateishi et al., 2001; Kurz et al., 2002; Ou et al., 2002). Nedd8-deficient Drosophila mutants arrest as first-instar larvae, while in C.elegans and mice, mutations that affect Nedd8 conjugation result in embryo lethality. In Arabidopsis, an assessment of the biological importance of RUB conjugation has been hindered by genetic redundancy. Strong axr1 alleles lack the AXR1 protein, but a closely related gene called AXL1 provides partial RUB-E1 activity (N.Dharmasiri and M.Estelle, unpublished data). Here we show that the combination of axr1-12 and rce1-1 results in an embryonic phenotype that is strikingly similar to that of the mp, bdl and axr6 mutants (Berleth and Jurgens, 1993; Hamann et al., 1999a; Hobbie et al., 2000). In the developing embryo, MP/ARF5 has an essential role in the establishment of the apical–basal axis and in vascular differentiation. Recent studies suggest that MP function depends on degradation of the Aux/IAA protein BDL/IAA12 (Hamann et al., 2002). Thus, either the loss of MP/ARF5 (as in mp) or the stabilization of BDL/IAA12 (as in the gain-of-function bdl alleles) results in similar defects in embryogenesis. Recently, we have shown that the AXR6 gene encodes CUL1 (H.Hellmann, L.Hobbie and M.Estelle, unpublished data), suggesting that the axr6 phenotype is caused by stabilization of BDL/IAA12 and perhaps other Aux/IAA proteins. The fact that axr1-12 rce1-1 seedlings have a similar phenotype is consistent with the proposal that RUB modification of CUL1 is required for degradation of Aux/IAA proteins during embryogenesis.
So far the precise function of RUB modification has remained elusive. In vitro experiments indicate that cullin modification increases activity of the SCF and related cullin-based E3 complexes (Morimoto et al., 2000; Podust et al., 2000; Read et al., 2000; Wu et al., 2000). Other evidence suggests that the modification may be important for recruitment of the E2 enzyme to the complex (Wu et al., 2002). Recent studies have also shown that removal of RUB from CUL1 is required for normal SCF function (Lyapina et al., 2001; Schwechheimer and Deng, 2001; Schwechheimer et al., 2001; Gray et al., 2002). RUB de-conjugation is accomplished by the COP9 signalosome (CSN), a multi-subunit complex related to the lid subcomplex of the proteosome (Schwechheimer and Deng, 2001). A reduction in CSN levels causes accumulation of RUB–CUL1 and a phenotype that is similar to the axr1 mutants (Schwechheimer et al., 2001). These results indicate that in vivo, SCF function requires a cycle of RUB conjugation and removal. Similar results have been obtained in fungal species (Lyapina et al., 2001). In our experiments, we find that the combination of axr1 and rce1 does not dramatically alter the steady-state level of RUB–CUL1. However, because the total amount of CUL protein increases, the relative amount of RUB–CUL1 is decreased. These results suggest that a reduction in activity of the RUB conjugation pathway results in stabilization of CUL1. It is possible that the modification is required for SCF disassembly and CUL1 degradation. Alternatively, a fraction of CUL1 may be degraded during normal SCF function. If the SCF is not functioning properly due to a defect in the RUB pathway, less CUL1 will be consumed. It is also important to note that unknown substrates of RUB conjugation may exist. A reduction in RUB modification of these proteins may account for aspects of the phenotype.
Although the effects of mutations in components of the CSN and the RUB conjugation pathway are similar, they are not identical. Mutants that completely lack the CSN do not have an embryonic defect whereas the axr1 rce1 double mutant has a severe embryonic defect. Apparently RUB conjugation is important very early in the life cycle of the plant whereas RUB de-conjugation and other potential functions of the CSN are not required until after germination.
The basic characteristics of an E3 are the ability to bind the E2 enzyme and the substrate, thus promoting transfer of ubiquitin from one to the other. In the case of RUB modification of CUL1, RBX1 appears to have these characteristics. It binds both RCE1 and CUL1, and when overexpressed, promotes RUB modification of CUL1. However, additional biochemical experiments are required to confirm that RBX1 does function as the RUB E3. Nevertheless, the possibility that RBX1 might be the RUB E3 raises some interesting questions. In the context of SCF E3 function, RBX1 is known to recruit the ubiquitin E2 to the complex. Since RCE1 is closely related to ubiquitin E2s, it seems likely that both proteins bind the same site on RBX1. If this is correct, competition between RCE1 and the ubiquitin E2 may have a role in regulation of the SCF. Our results also indicate that RCE1 is present in a stable complex with the SCF. In the future, it will be interesting to determine whether the ubiquitin E2 and the CSN are in this complex as well, and if they are, how their various activities are coordinated.
Materials and methods
Plant material and growth conditions
The rce1-1 seeds were obtained from the Nottingham Arabidopsis Stock Center (NASC). All other mutants and transgenic lines were in Colombia ecotype. Seeds were surface sterilized and grown on Arabidopsis thaliana medium + 1% sucrose (ATS) plates under 16 h light/8 h dark conditions at 22°C. For root growth assays, 5- to 6-day-old seedlings were transferred onto ATS plates with or without 2,4-D or methyl jasmonate (Bedoukian Research, CT). Root lengths and number of lateral roots were measured after 3–5 days depending on the experiment. All protein extracts were generated using 6- to 8-day-old seedlings grown in liquid ATS medium in flasks under constant shaking.
RCE1 constructs and plant transformation
To prepare the RCE1-GUS reporter gene construct, a 1.5 kb segment from the promoter region immediately upstream of the translation initiation site of the RCE1 gene was ligated to the 0.55 kb RCE1 cDNA carrying the entire open reading frame. This construct was inserted directly into the pBI101.1 plant transformation vector (Jefferson et al., 1987), in frame with the GUS protein at the C-terminus of the fusion. The expression pattern was studied in 16 independent transgenic lines. To construct Myc-tagged RCE1, a Myc cassette was removed from the pGEM vector and ligated to the 5′ end of the RCE1 cDNA lacking the first ATG. This construct was inserted into pROKII binary vector carrying the 35S promoter. Both RCE1-GUS reporter and Myc-RCE1 constructs were transformed into Agrobacterium tumefaciens line GV3101. Plant transformations, and GUS assays were performed as described previously (del Pozo et al., 2002). To confirm function of the Myc-RCE1 fusion protein, the 35S::Myc-RCE1 transgene was introduced into RCE1 (Col-0) by transformation and crossed into rce1-1 (Ler) plants.
The GST-RCE1 construct was prepared by inserting the 0.55 kb RCE1 cDNA into pGEX4T-3 vector. GST–RCE1 was purified using glutathione beads according to standard protocols. Other proteins used in this study were GST–RBX1 (Gray et al., 2002), GST–IAA7 (Gray et al., 2001) and TIR1-myc (Gray et al., 1999).
Pull-down assays, immunoprecipitations and pulse–chase analysis
For pull-down and immunoprecipitation assays, proteins were extracted from 6-day-old seedlings into buffer containing 50 mM Tris (pH 7.5), 150 mM NaCl, 0.25% Tween-20, 1 mM PMSF and 5 mg/ml protease inhibitor cocktail (Roche). All pull-downs and immunoprecipitations were performed using 1 mg total proteins, according to standard protocols. Monoclonal α-Myc was from BabCo (Richmand, CA). The AXR1 and CUL1 antibodies have been described previously (del Pozo et al., 1998; Gray et al., 1999). Protein blot analyses were performed according to standard protocols and detected with ECL (Amersham). Pulse–chase analysis was performed using 6-day-old seedlings as described previously (Gray et al., 2002).
In vitro interaction between RBX1 and RCE1
GST–RBX was labeled with 32P using protein kinase A and treated with thrombin to release labeled RBX. Supernatant containing labeled RBX was incubated with benzamidine beads (to remove thrombin) and glutathione beads (to remove any remaining GST–RBX), then 20 µl of labeled RBX was incubated with 3 µg of GST–RCE1 for 12 h in 200 µl of binding buffer [50 mM Tris–Cl pH 7.5, 150 mM NaCl, 0.5% NP-40, 0.1 mM dithiothreitol (DTT) and protease inhibitor cocktail]. Beads were washed three times with 1 ml of the binding buffer before analysis by SDS–PAGE.
RNA gel blot analysis
To study the expression of RCE1, total RNA was extracted from seedlings growing in liquid culture, or from adult tissues. For the expression of IAA2 gene, 6-day-old rce1-1 and wild-type (Landsberg) seedlings were treated with or without 20 µM 2,4-D for 60 min. Total RNA was extracted using Tri-reagent (Sigma), and 10 µg RNA was loaded on each lane. The entire coding region of RCE1 cDNA or IAA2 cDNA was used as the probe.
Seedling vascular patterns
Seedlings grown on ATS medium for 7 days were fixed in ethanol:acetic acid:water (6:3:1), and cleared in Hoyers solution (Liu and Meinke, 1998). The vascular patterns were photographed using dark field optics.
Acknowledgments
Acknowledgements
The authors would like to thank Dr Seth Davis for drawing our attention to the rce1-1 line and the NASC for providing this line. This work was supported by National Institutes of Health Grant RO1-GM43411 and National Science Foundation Grant 0115870 to M.E.
References
- Berleth T. and Jurgens,G. (1993) The role of the monopteros gene in organising the basal body region of the Arabidopsis embryo. Development, 118, 575–587. [Google Scholar]
- Berleth T., Mattsson,J. and Hardtke,C.S. (2000) Vascular continuity and auxin signals. Trends Plant Sci., 5, 387–393. [DOI] [PubMed] [Google Scholar]
- del Pozo J.C. and Estelle,M. (1999) The Arabidopsis cullin AtCUL1 is modified by the ubiquitin-related protein RUB1. Proc. Natl Acad. Sci. USA, 96, 15342–15347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo J.C., Timpte,C., Tan,S., Callis,J. and Estelle,M. (1998) The ubiquitin-related protein RUB1 and auxin response in Arabidopsis. Science, 280, 1760–1763. [DOI] [PubMed] [Google Scholar]
- del Pozo J.C., Dharmasiri,S., Hellmann,H., Walker,L., Gray,W.M. and Estelle,M. (2002) AXR1-ECR1-dependent conjugation of RUB1 to the Arabidopsis cullin AtCUL1 is required for auxin response. Plant Cell, 14, 421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies R.J. (1999) SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell. Dev. Biol., 15, 435–467. [DOI] [PubMed] [Google Scholar]
- Dharmasiri S. and Estelle,M. (2002) The role of regulated protein degradation in auxin response. Plant Mol. Biol., 49, 401–409. [PubMed] [Google Scholar]
- Dieterle M., Zhou,Y.C., Schafer,E., Funk,M. and Kretsch,T. (2001) EID1, an F-box protein involved in phytochrome A-specific light signaling. Genes Dev., 15, 939–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa M., Zhang,Y., McCarville,J., Ohta,T. and Xiong,Y. (2000) The CUL1 C-terminal sequence and ROC1 are required for efficient nuclear accumulation, NEDD8 modification and ubiquitin ligase activity of CUL1. Mol. Cell. Biol., 20, 8185–8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne J.M., Downes,B.P., Shiu,S.H., Durski,A.M. and Vierstra,R.D. (2002) The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc. Natl Acad. Sci. USA, 99, 11519–11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray W.M. et al. (1999) Identification of an SCF ubiquitin–ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev., 13, 1678–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray W.M., Kepinski,S., Rouse,D., Leyser,O. and Estelle,M. (2001) Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature, 414, 271–276. [DOI] [PubMed] [Google Scholar]
- Gray W.M., Hellmann,H., Dharmasiri,S. and Estelle,M. (2002) Role of the Arabidopsis RING-H2 protein RBX1 in RUB modification and SCF function. Plant Cell, 14, 2137–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann T., Mayer,U. and Jurgens,G. (1999a) The auxin-insensitive bodenlos mutation affects primary root formation and apical–basal patterning in the Arabidopsis embryo. Development, 126, 1387–1395. [DOI] [PubMed] [Google Scholar]
- Hamann T., Mayer,U. and Jurgens,G. (1999b) The auxin-insensitive bodenlos mutation affects primary root formation and apical–basal patterning in the Arabidopsis embryo. Development, 126, 1387–1395. [DOI] [PubMed] [Google Scholar]
- Hamann T., Benkova,E., Baurle,I., Kientz,M. and Jurgens,G. (2002) The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev., 16, 1610–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke C.S. and Berleth,T. (1998) The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J., 17, 1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann H. and Estelle,M. (2002) Plant development: regulation by protein degradation. Science, 297, 793–797. [DOI] [PubMed] [Google Scholar]
- Himanen K., Boucheron,E., Vanneste,S., De Almeida Engler,J., Inze,D. and Beeckman,T. (2002) Auxin-mediated cell cycle activation during early lateral root initiation. Plant Cell, 14, 2339–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbie L., McGovern,M., Hurwitz,L.R., Pierro,A., Liu,N.Y., Bandyopadhyay,A. and Estelle,M. (2000) The axr6 mutants of Arabidopsis thaliana define a gene involved in auxin response and early development. Development, 127, 23–32. [DOI] [PubMed] [Google Scholar]
- Ingram G.C., Doyle,S., Carpenter,R., Schultz,E.A., Simon,R. and Coen,E.S. (1997) Dual role for fimbriata in regulating floral homeotic genes and cell division in Antirrhinum. EMBO J., 16, 6521–6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh,T.A. and Bevan,M.W. (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J., 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamura T., Conrad,M.N., Yan,Q., Conaway,R.C. and Conaway,J.W. (1999) The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes Dev., 13, 2928–2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz T., Pintard,L., Willis,J.H., Hamill,D.R., Gonczy,P., Peter,M. and Bowerman,B. (2002) Cytoskeletal regulation by the Nedd8 ubiquitin-like protein modification pathway. Science, 295, 1294–1298. [DOI] [PubMed] [Google Scholar]
- Liakopoulos D., Busgen,T., Brychzy,A., Jentsch,S. and Pause,A. (1999) Conjugation of the ubiquitin-like protein NEDD8 to cullin-2 is linked to von Hippel-Lindau tumor suppressor function. Proc. Natl Acad. Sci. USA, 96, 5510–5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.M. and Meinke,D.W. (1998) The titan mutants of Arabidopsis are disrupted in mitosis and cell cycle control during seed development. Plant J., 16, 21–31. [DOI] [PubMed] [Google Scholar]
- Lyapina S., Cope,G., Shevchenko,A., Serino,G., Tsuge,T., Zhou,C., Wolf,D.A., Wei,N. and Deshaies,R.J. (2001) Promotion of NEDD8-CUL1 conjugate cleavage by COP9 signalosome. Science, 292, 1382–1385. [DOI] [PubMed] [Google Scholar]
- Morimoto M., Nishida,T., Honda,R. and Yasuda,H. (2000) Modification of cullin-1 by ubiquitin-like protein Nedd8 enhances the activity of SCFskp2 toward p27kip1. Biochem. Biophys. Res. Commun., 270, 1093–1096. [DOI] [PubMed] [Google Scholar]
- Nelson D.C., Lasswell,J., Rogg,L.E., Cohen,M.A. and Bartel,B. (2000) FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell, 101, 331–340. [DOI] [PubMed] [Google Scholar]
- Osaka F. et al. (2000) Covalent modifier NEDD8 is essential for SCF ubiquitin-ligase in fission yeast. EMBO J., 19, 3475–3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou C.Y., Lin,Y.F., Chen,Y.J. and Chien,C.T. (2002) Distinct protein degradation mechanisms mediated by Cul1 and Cul3 controlling Ci stability in Drosophila eye development. Genes Dev., 16, 2403–2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart C.M. (2001) Mechanisms underlying ubiquitination. Annu. Rev. Biochem., 70, 503–533. [DOI] [PubMed] [Google Scholar]
- Podust V.N., Brownell,J.E., Gladysheva,T.B., Luo,R.S., Wang,C., Coggins,M.B., Pierce,J.W., Lightcap,E.S. and Chau,V. (2000) A Nedd8 conjugation pathway is essential for proteolytic targeting of p27Kip1 by ubiquitination. Proc. Natl Acad. Sci. USA, 97, 4579–4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read M.A. et al. (2000) Nedd8 modification of cul-1 activates SCFβTrCP-dependent ubiquitination of IκBα. Mol. Cell. Biol., 20, 2326–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach A., Klenz,J.E., Kohalmi,S.E., Risseeuw,E., Haughn,G.W. and Crosby,W.L. (1999) The UNUSUAL FLORAL ORGANS gene of Arabidopsis thaliana is an F-box protein required for normal patterning and growth in the floral meristem. Plant J., 20, 433–445. [DOI] [PubMed] [Google Scholar]
- Schwechheimer C. and Deng,X.W. (2001) COP9 signalosome revisited: a novel mediator of protein degradation. Trends Cell Biol., 11, 420–426. [DOI] [PubMed] [Google Scholar]
- Schwechheimer C., Serino,G., Callis,J., Crosby,W.L., Lyapina,S., Deshaies,R.J., Gray,W.M., Estelle,M. and Deng,X.W. (2001) Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIR1 in mediating auxin response. Science, 292, 1379–1382. [DOI] [PubMed] [Google Scholar]
- Schwechheimer C., Serino,G. and Deng,X.W. (2002) Multiple ubiquitin ligase-mediated processes require COP9 signalosome and AXR1 function. Plant Cell, 14, 2553–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers D.E., Schultz,T.F., Milnamow,M. and Kay,S.A. (2000) ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell, 101, 319–329. [DOI] [PubMed] [Google Scholar]
- Stirnberg P., van De Sande,K. and Leyser,H.M. (2002) MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development, 129, 1131–1141. [DOI] [PubMed] [Google Scholar]
- Tateishi K., Omata,M., Tanaka,K. and Chiba,T. (2001) The NEDD8 system is essential for cell cycle progression and morphogenetic pathway in mice. J. Cell Biol., 155, 571–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiryaki I. and Staswick,P.E. (2002) An Arabidopsis mutant defective in jasmonate response is allelic to the auxin-signaling mutant axr1. Plant Physiol., 130, 887–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo H.R., Chung,K.M., Park,J.H., Oh,S.A., Ahn,T., Hong,S.H., Jang,S.K. and Nam,H.G. (2001) ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell, 13, 1779–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Chen,A. and Pan,Z.Q. (2000) Conjugation of Nedd8 to CUL1 enhances the ability of the ROC1–CUL1 complex to promote ubiquitin polymerization. J. Biol. Chem., 275, 32317–32324. [DOI] [PubMed] [Google Scholar]
- Wu K., Chen,A., Tan,P. and Pan,Z.Q. (2002) The Nedd8-conjugated ROC1–CUL1 core ubiquitin ligase utilizes Nedd8 charged surface residues for efficient polyubiquitin chain assembly catalyzed by Cdc34. J. Biol. Chem., 277, 516–527. [DOI] [PubMed] [Google Scholar]
- Xie D.X., Feys,B.F., James,S., Nieto-Rostro,M. and Turner,J.G. (1998) COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science, 280, 1091–1094. [DOI] [PubMed] [Google Scholar]
- Xu L., Liu,F., Lechner,E., Genschik,P., Crosby,W.L., Ma,H., Peng,W., Huang,D. and Xie,D. (2002) The SCFCOL1 ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell, 14, 1919–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E.T., Gong,L. and Kamitani,T. (2000) Ubiquitin-like proteins: new wines in new bottles. Gene, 248, 1–14. [DOI] [PubMed] [Google Scholar]
- Zheng N. et al. (2002) Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature, 416, 703–709. [DOI] [PubMed] [Google Scholar]